Plastome Data of Red Currant and Gooseberry Reveal Potential Taxonomical Issues within the Ribes Genus (Grossulariaceae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, DNA Extraction, and Sequencing
2.2. Chloroplast Genome Assembly and Annotation
2.3. Genome Comparison and Adaptive Evolution Analysis
2.4. Simple Sequence Repeats Analysis
2.5. Phylogenetic Analysis
3. Results
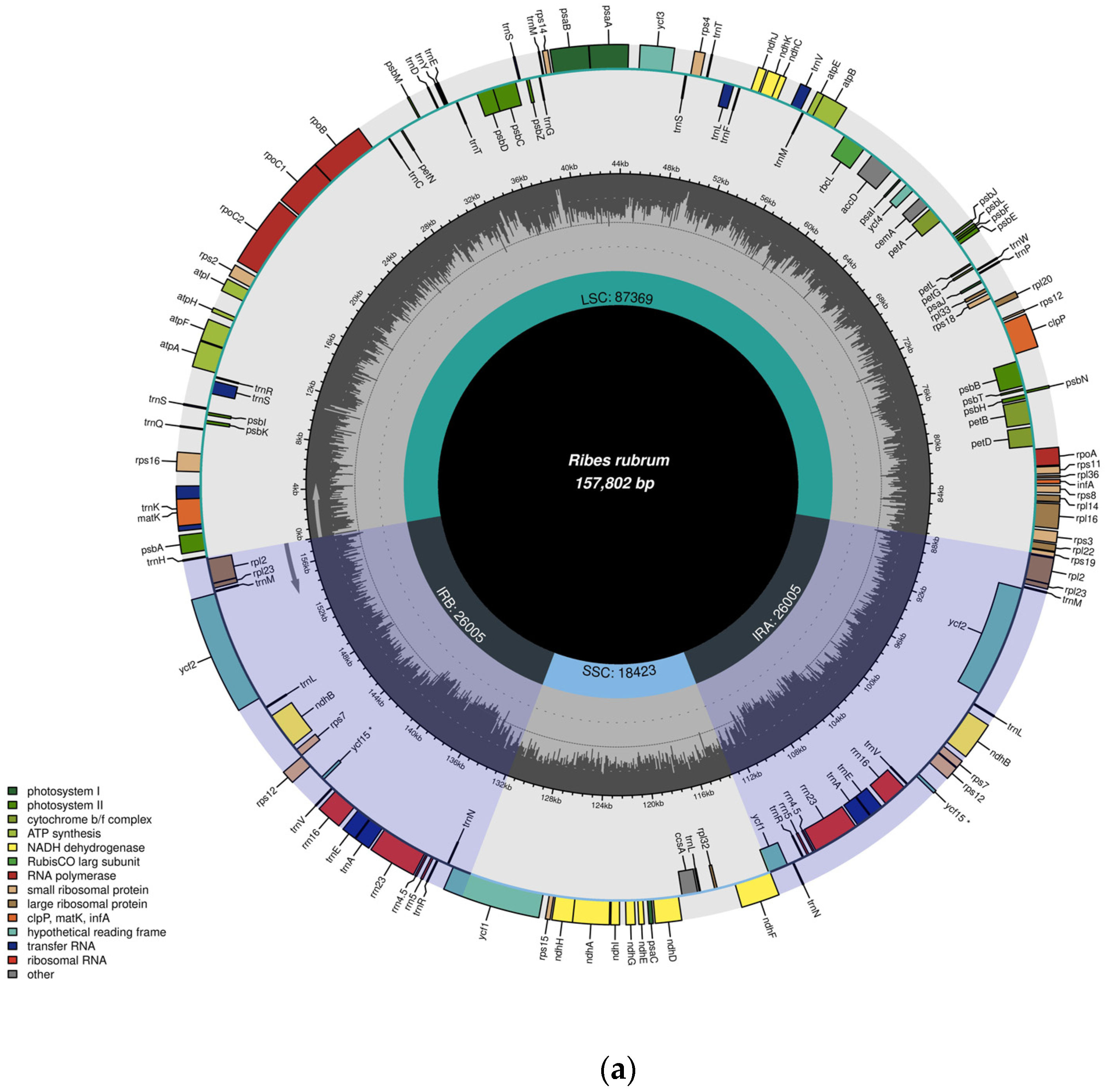
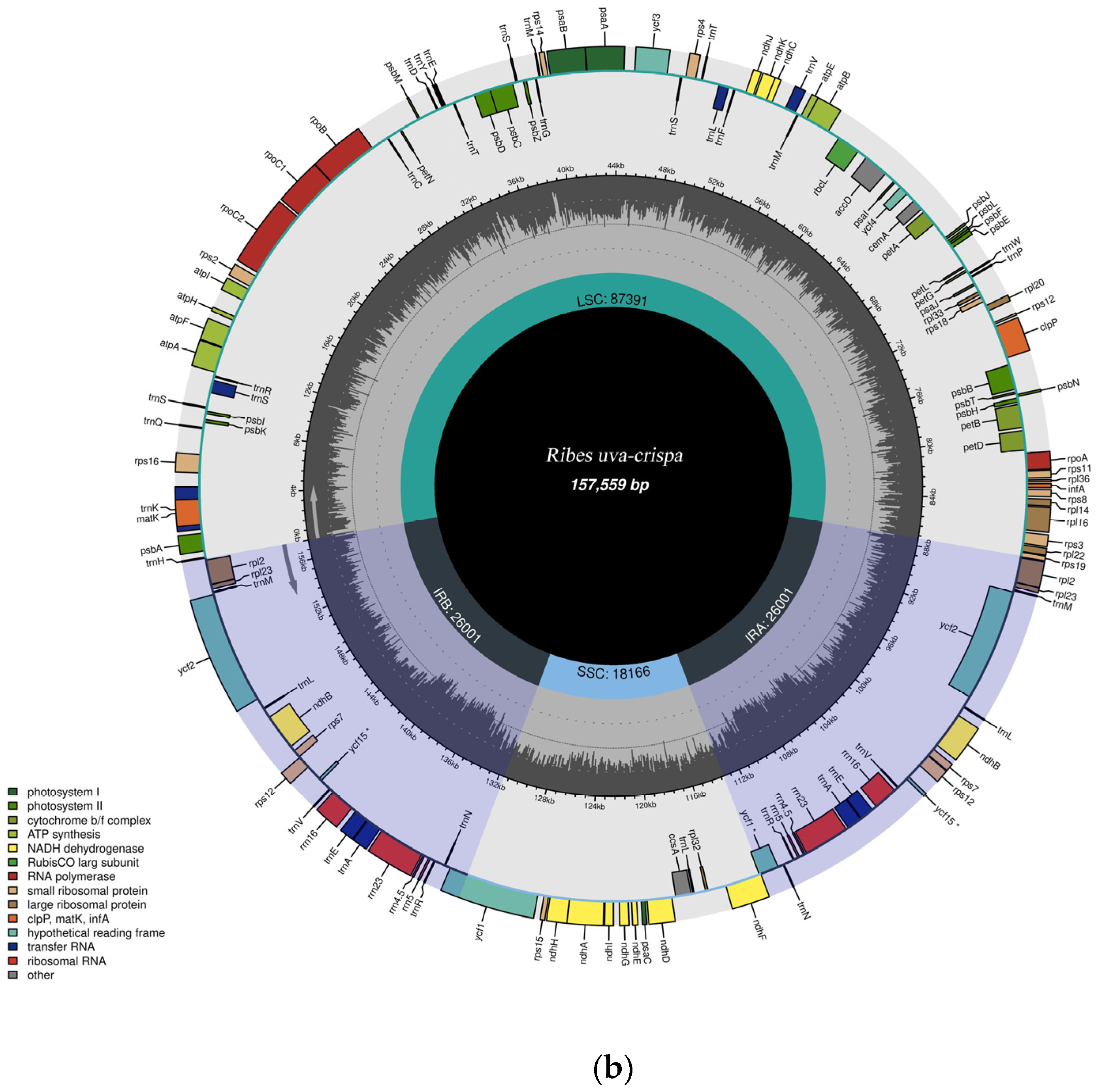
3.1. General Features of the R. rubrum and R. uva-crispa Chloroplast Genomes
3.2. SSR Analysis
3.3. Genome Comparison and Adaptive Evolution Analysis
3.4. Phylogenetic Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sun, Q.; Wang, N.; Xu, W.; Zhou, H. Genus Ribes Linn. (Grossulariaceae): A comprehensive review of traditional uses, phytochemistry, pharmacology and clinical applications. J. Ethnopharmacol. 2021, 276, 114166. [Google Scholar] [CrossRef] [PubMed]
- Knyazev, S.D.; Ogol’tsova, T.P. Selektsiya Smorodiny Chernoi na Sovremennom Etape; Orel State Agrarian University: Oryol, Russia, 2004; 238p. [Google Scholar]
- Blackcurrant-iba. Available online: https://www.blackcurrant-iba.com/wp-content/uploads/2021/12/Global-BC-Production-2020-21.pdf (accessed on 5 July 2023).
- Hummer, K.E.; Barney, D.L. Crop reports. Hort-Technology 2002, 12, 377–387. [Google Scholar] [CrossRef]
- Tridge. Available online: https://www.tridge.com/intelligences/gooseberry/production (accessed on 5 July 2023).
- Wiethold, J. Des fruits d’ici et d’ailleurs—Regards sur l’histoire de quelques fruits consommés en Europe-Introduction. In Historic and Archaeobotanical Evidence; Ruas, M.-P., Ed.; Edition Omniscience: Geneva, Switzerland, 2016; pp. 11–38. Available online: https://www.researchgate.net/publication/307631956_Red_currant_and_black_current_new_cultivated_fruits_in_late_medieval_and_early_modern_Europe_Historic_and_archaeobotanical_evidence (accessed on 5 July 2023).
- Latałowa, M.; Badura, M.; Jarosińska, J.; Święta-Musznicka, J. Useful plants in medieval and postmedieval archaeobotanical material from the Hanseatic towns of Northern Poland (Kołobrzeg, Gdańsk and Elbląg). In Medieval Food traditions in Northern Europe; National Museum: Copenhagen, Denmark, 2007; Volume 12, pp. 39–72. [Google Scholar]
- Hedrick, U.P. The Small Fruits of New York. Report for the Year; Forgotten Books: London, UK, 1925. [Google Scholar]
- Ogoltsova, T.P. Centers of Origin, Evolution and Taxonomy of the Genus Ribes L.; Sedov, E.N., Ed.; Russian Federation: Oryol, Russia, 2009; Volume IV, pp. 7–15. [Google Scholar]
- Bradford, A. Field Guide to Edible Wild Plants; Stackpole Books: Harrisburg, PA, USA, 1974; p. 68. [Google Scholar]
- Rozanova, M.A. Jagodovedenie i Jagodovodstvo (Berry Science and Berry Growing); Sel’hozgiz: Moscow, Russia, 1935; p. 295. [Google Scholar]
- Harmat, L.; Porpaczy, A.; Himelrick, D.G.; Galletta, G.J. Currant and gooseberry management. In Small Fruit Crop Management; Prentice Hall: Englewood Cliffs, NJ, USA, 1990; pp. 245–272. [Google Scholar]
- Arena, M.E.; Bernini, M.; Vater, G. Growth and fruiting of Ribes magellanicum in Tierra del Fuego, Argentina. N. Z. J. Crop Hortic. Sci. 2007, 35, 61–66. [Google Scholar] [CrossRef]
- Pikunova, A.; Goryunova, S.; Goryunov, D.; Golyaeva, O.; Dolzhikova, M.; Pavlenko, A. Genetic Diversity and Pedigree Analysis of Red Currant Germplasm. Plants 2022, 11, 1623. [Google Scholar] [CrossRef] [PubMed]
- Coville, F.V.; Britton, N.L. North American Flora. Bot. Gaz. 1908, 46, 64. Available online: http://biodiversitylibrary.org/item/15436 (accessed on 28 August 2923).
- Berger, A. A taxonomic review of currants and gooseberries. Bull. N. Y. State Agric. Exp. Stn. 1924, 31, 167–169. [Google Scholar]
- Komarov, V.L. (Ed.) Flora of the Former USSR Volume IX; Ribesioideae Engl. Keter, London; Koeltz Botanical Books: Glashütten, Germany, 1971; pp. 175–208. [Google Scholar]
- Janczewski, E. Mongraphie des Grosseilliers Ribes L.; Mémoires de la Société de Physique et d’hist. Naturelle de Genève; Impr. W. Kündig: Geneve, Switherland, 1907; Volume 35, pp. 199–517. [Google Scholar]
- Sinnott, Q.P. A revision of Ribes L. subg. Grossularia (Mill.) Pers. sect. Grossularia (Mill.) Nutt. (Grossulariaceae) in North America. Rhodora 1985, 87, 187–286. [Google Scholar]
- Keep, E. Satellite and nucleolar number in hybrids between Ribes nigrum and R. grossularia and in their backcrosses. Can. J. Genet. Cytol. 1962, 4, 206–218. [Google Scholar] [CrossRef]
- Sharma, G.; Lata, S.; Yadav, A. Currants. In Temperate Fruit Crop Breeding Domestication to Cultivar Development; JAYA Publishing House: Delhi, India, 2020; pp. 255–288. [Google Scholar]
- USDA-ARS Germplasm Resources Information Network (GRIN). Available online: https://www.ars-grin.gov/ (accessed on 5 July 2023).
- Daniel, H.; Jin, S.; Zhu, X.G.; Gitzendanner, M.A.; Soltis, D.E.; Soltis, P.S. Green giant—A tiny chloroplast genome with mighty power to produce high-value proteins: History and phylogeny. Plant Biotechnol. J. 2021, 19, 430–447. [Google Scholar] [CrossRef]
- Singh, B.P.; Kumar, A.; Kaur, H.; Singh, H.; Nagpal, A.K. CpGDB: A comprehensive database of chloroplast genomes. Bioinformation 2020, 16, 171. [Google Scholar] [CrossRef]
- Reith, M.; Munholland, J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 1995, 13, 333–335. [Google Scholar] [CrossRef]
- Banerjee, A.; Stefanovic, S. Caught in action: Fine-scale plastome evolution in the parasitic plants of Cuscuta section Ceratophorae (Convolvulaceae). Plant Mol. Biol. 2019, 100, 621–634. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.-Y.; Yu, Y.; Deng, Y.Q.; Li, J.; Huang, Z.X.; Zhou, S.D. The Chloroplast Genome of Lilium henrici: Genome Structure and Comparative Analysis. Molecules 2018, 23, 1276. [Google Scholar] [CrossRef] [PubMed]
- VNIISPK Bioresource Collection (VNIISPK BRC). Available online: https://vniispk.ru/pages/unu (accessed on 3 July 2023).
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 3 July 2023).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Dierckxsens, N.; Mardulyn, P.; Smits, G. NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 2016, 45, 18. [Google Scholar] [CrossRef]
- Kearse, M.; Moir, R.; Wilson, A.; Stones-Havas, S.; Cheung, M.; Sturrock, S.; Buxton, S.; Cooper, A.; Markowitz, S.; Duran, C.; et al. Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012, 28, 1647–1649. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Chan, P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016, 44, W54–W57. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef]
- Jühling, F.; Mörl, M.; Hartmann, R.K.; Sprinzl, M.; Stadler, P.F.; Pütz, J. tRNAdb 2009: Compilation of tRNA sequences and tRNA genes. Nucleic Acids Res. 2009, 37, D159–D162. [Google Scholar] [CrossRef]
- Shi, L.; Chen, H.; Jiang, M.; Wang, L.; Wu, X.; Huang, L.; Liu, C. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 2019, 47, W65–W73. [Google Scholar] [CrossRef]
- Zheng, S.; Poczai, P.; Hyvönen, J.; Tang, J.; Amiryousefi, A. Chloroplot: An online program for the versatile plotting of organelle genomes. Front. Genet. 2020, 11, 1123. [Google Scholar] [CrossRef]
- Brudno, M.; Do, C.B.; Cooper, G.M.; Kim, M.F.; Davydov, E.; Green, E.D.; Sidow, A.; Batzoglou, S. NISC Comparative Sequencing Program. LAGAN and Multi-LAGAN: Efficient tools for large-scale multiple alignment of genomic DNA. Genome Res. 2003, 13, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L. GMATA: An Integrated Software Package for Genome-Scale SSR Mining, Marker Development and Viewing. Front. Plant Sci. 2016, 7, 1350. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Huelsenbeck, J.P.; Ronquist, F. MRBAYES: Bayesian inference of phylogeny. Bioinformatics 2001, 17, 754–755. [Google Scholar] [CrossRef] [PubMed]
- Bellot, S.; Cusimano, N.; Luo, S.; Sun, G.; Zarre, S.; Gröger, A.; Temsch, E.; Renner, S.S. Assembled Plastid and Mitochondrial Genomes, as well as Nuclear Genes, Place the Parasite Family Cynomoriaceae in the Saxifragales. Genome Biol. Evol. 2016, 8, 2214–2230. [Google Scholar] [CrossRef] [PubMed]
- VNIISPK. Available online: https://vniispk.ru/varieties/belaya-potapenko (accessed on 3 July 2023).
- Plants of the World. Available online: https://powo.science.kew.org/ (accessed on 5 July 2023).
- World Flora Online. Available online: http://www.worldfloraonline.org/ (accessed on 5 July 2023).
- VNIISPK. Available online: https://vniispk.ru/varieties/nekrasovskii (accessed on 3 July 2023).
- Fan, W.-B.; Wu, Y.; Yang, J.; Shahzad, K.; Li, Z.-H. Comparative Chloroplast Genomics of Dipsacales Species: Insights into Sequence Variation, Adaptive Evolution, and Phylogenetic Relationships. Front. Plant Sci. 2018, 9, 689. [Google Scholar] [CrossRef]
- Soltis, D.E.; Mort, M.E.; Latvis, M.; Mavrodiev, E.V.; O’meara, B.C.; Soltis, P.S.; Burleigh, J.G.; de Casas, R.R. Phylogenetic relationships and character evolution analysis of Saxifragales using a supermatrix approach. Am. J. Bot. 2013, 100, 916–929. [Google Scholar] [CrossRef]
- Li, H.-T.; Yi, T.-S.; Gao, L.-M.; Ma, P.-F.; Zhang, T.; Yang, J.-B.; Gitzendanner, M.A.; Fritsch, P.W.; Cai, J.; Luo, Y.; et al. Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 2019, 5, 461–470. [Google Scholar] [CrossRef]
- Suzuki, C.K.; Rep, M.; van Dijl, J.M.; Suda, K.; Grivell, L.A.; Schatz, G. ATP-dependent proteases that also chaperone protein biogenesis. Trends Biochem. Sci. 1997, 22, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.S.; Riaz, R.; Majid, M.; Mehmood, K.; Mustafa, G.; Joyia, F.A. The tobacco chloroplast YCF4 gene is essential for transcriptional gene regulation and plants photoautotrophic growth. Front. Plant Sci. 2022, 13, 1014236. [Google Scholar] [CrossRef] [PubMed]
- Gui, L.; Jiang, S.; Xie, D.; Yu, L.; Huang, Y.; Zhang, Z.; Liu, Y. Analysis of complete chloroplast genomes of Curcuma and the contribution to phylogeny and adaptive evolution. Gene 2020, 732, 144355. [Google Scholar] [CrossRef]
- Wang, L.; Liang, J.; Sa, W.; Wang, L. Sequencing and comparative analysis of the chloroplast genome of Ribes odoratum provide insights for marker development and phylogenetics in Ribes. Physiol. Mol. Biol. Plants 2021, 27, 81–92. [Google Scholar] [CrossRef]
- Li, H.-T.; Luo, Y.; Gan, L.; Ma, P.-F.; Gao, L.-M.; Yang, J.-B.; Cai, J.; Gitzendanner, M.A.; Fritsch, P.W.; Zhang, T.; et al. Plastid phylogenomic insights into relationships of all flowering plant families. BMC Biol. 2021, 19, 232. [Google Scholar] [CrossRef]
- Angiosperm Phylogeny Website. Available online: http://www.mobot.org/MOBOT/research/APweb (accessed on 3 July 2023).
- Senters, A.E.; Soltis, D.E. Phylogenetic relationships in Ribes (Grossulariaceae) inferred from ITS sequence data. Taxon 2003, 52, 51–66. [Google Scholar] [CrossRef]
- Weigend, M.; Mohr, O.; Motley, T.J. Phylogeny and classification of the genus Ribes (Grossulariaceae) based on 5S-NTS sequences and morphological and anatomical data. Jahrbücher Syst. Pflanzengesch. Pflanzengeogr. 2002, 124, 163–182. [Google Scholar] [CrossRef]
- Messinger, W.; Hummer, K.; Liston, A. Ribes (Grossulariaceae) phylogeny as indicated by restriction-site polymorphisms of PCR-amplified chloroplast DNA. Plant Syst. Evol. 1999, 217, 185–195. [Google Scholar] [CrossRef]
- Schultheis, L.; Donoghue, M.J. Molecular phylogeny and biogeography of ribes (Grossulariaceae), with an emphasis on gooseberries (subg. Grossularia). Syst. Bot. 2004, 29, 77–96. [Google Scholar] [CrossRef]
- Pikunova, A.V.; Martirosian, E.V.; Kniazev, S.D.; Ryzhova, N.N. Application of the RAPD-analysis for the study of genetic polymorphism and phylogenetic relationships in the Ribes L. genus. Russ. J. Genet. Appl. Res. 2012, 2, 141–151. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pikunova, A.; Goryunova, S.; Golyaeva, O.; Dolzhikova, M.; Pavlenko, A.; Kurashev, O.; Sotnikova, E.; Polivanova, O.; Sivolapova, A.; Kazakov, O.; et al. Plastome Data of Red Currant and Gooseberry Reveal Potential Taxonomical Issues within the Ribes Genus (Grossulariaceae). Horticulturae 2023, 9, 972. https://doi.org/10.3390/horticulturae9090972
Pikunova A, Goryunova S, Golyaeva O, Dolzhikova M, Pavlenko A, Kurashev O, Sotnikova E, Polivanova O, Sivolapova A, Kazakov O, et al. Plastome Data of Red Currant and Gooseberry Reveal Potential Taxonomical Issues within the Ribes Genus (Grossulariaceae). Horticulturae. 2023; 9(9):972. https://doi.org/10.3390/horticulturae9090972
Chicago/Turabian StylePikunova, Anna, Svetlana Goryunova, Olga Golyaeva, Maria Dolzhikova, Anna Pavlenko, Oleg Kurashev, Evgeniia Sotnikova, Oksana Polivanova, Anastasia Sivolapova, Oleg Kazakov, and et al. 2023. "Plastome Data of Red Currant and Gooseberry Reveal Potential Taxonomical Issues within the Ribes Genus (Grossulariaceae)" Horticulturae 9, no. 9: 972. https://doi.org/10.3390/horticulturae9090972
APA StylePikunova, A., Goryunova, S., Golyaeva, O., Dolzhikova, M., Pavlenko, A., Kurashev, O., Sotnikova, E., Polivanova, O., Sivolapova, A., Kazakov, O., & Goryunov, D. (2023). Plastome Data of Red Currant and Gooseberry Reveal Potential Taxonomical Issues within the Ribes Genus (Grossulariaceae). Horticulturae, 9(9), 972. https://doi.org/10.3390/horticulturae9090972





