Identification of Loci for Four Important Agronomic Traits in Loose-Curd Cauliflower Based on Genome-Wide Association Studies
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Phenotyping and Resequencing
2.2. Sequence Mapping and SNP Calling
2.3. Population Structure and Linkage Disequilibrium Analysis
2.4. GWAS
2.5. Phylogenetic and Transcriptome Analyses
2.6. Statistical Analysis and Availability of Data
3. Results
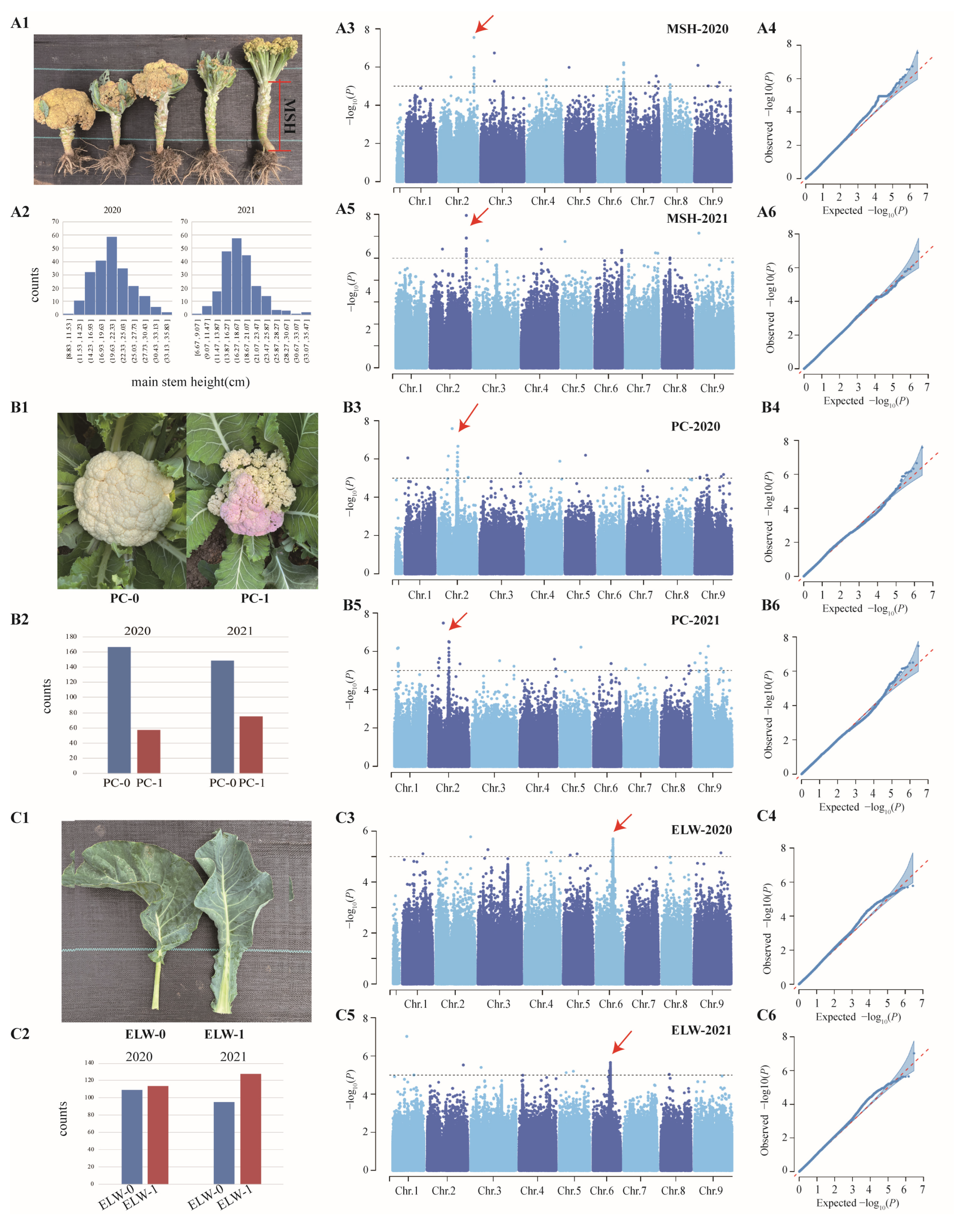
3.1. Plant Material Collection, Phenotype Survey, and DNA Sequencing
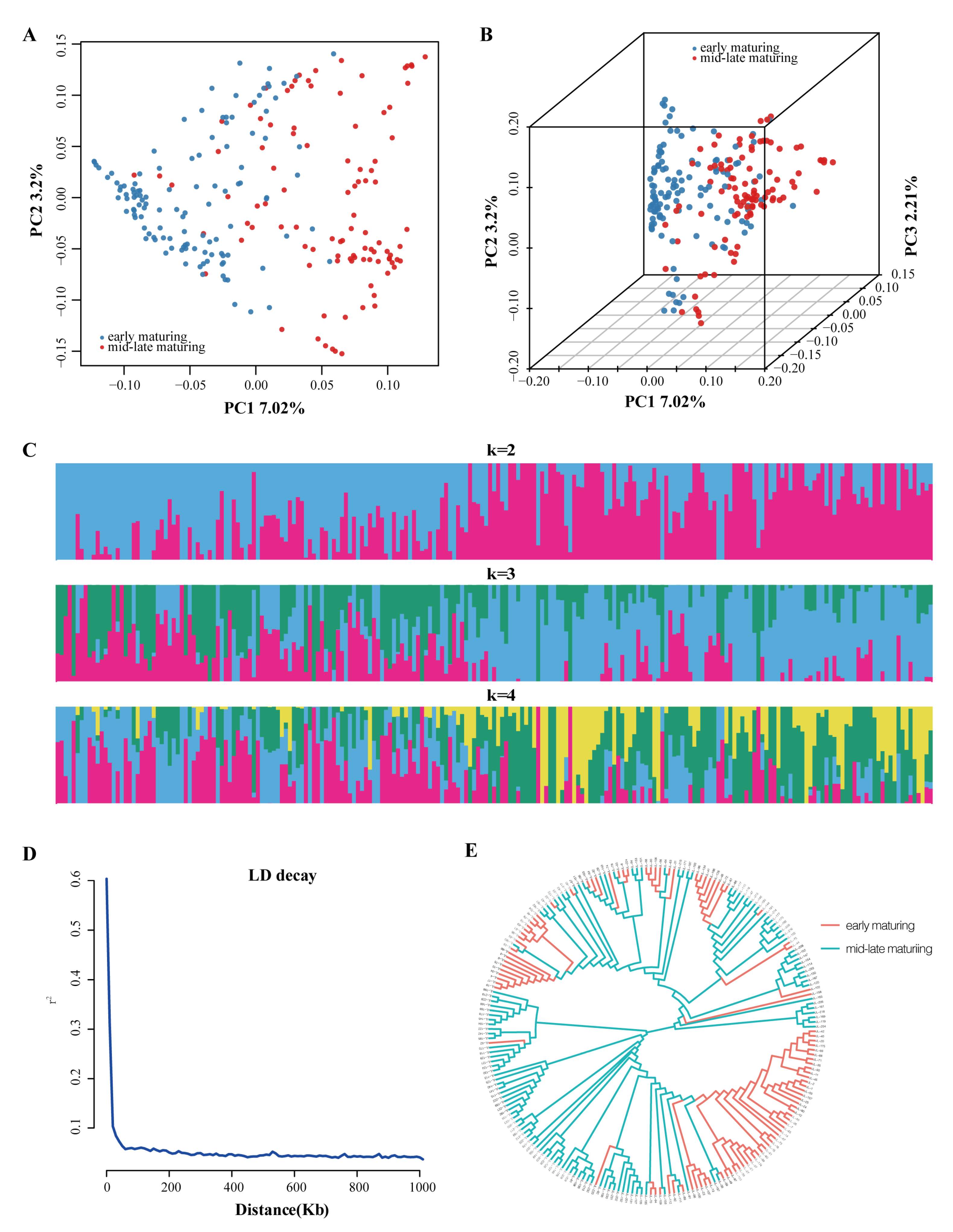
3.2. SNP Identification, Genetic Diversity and Population Structure
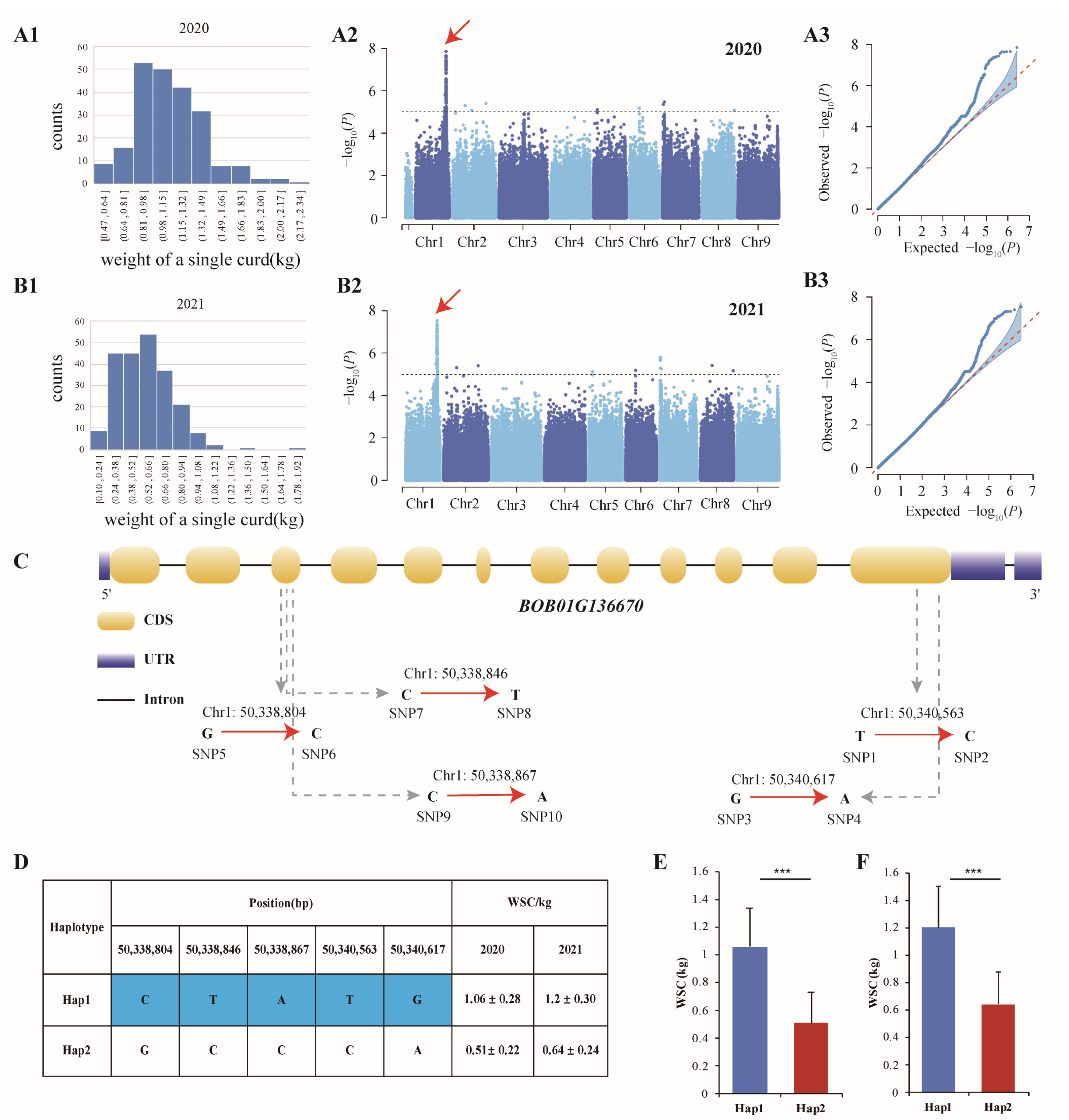
3.3. Genome-Wide Association Analysis of Three Important Agronomic Traits
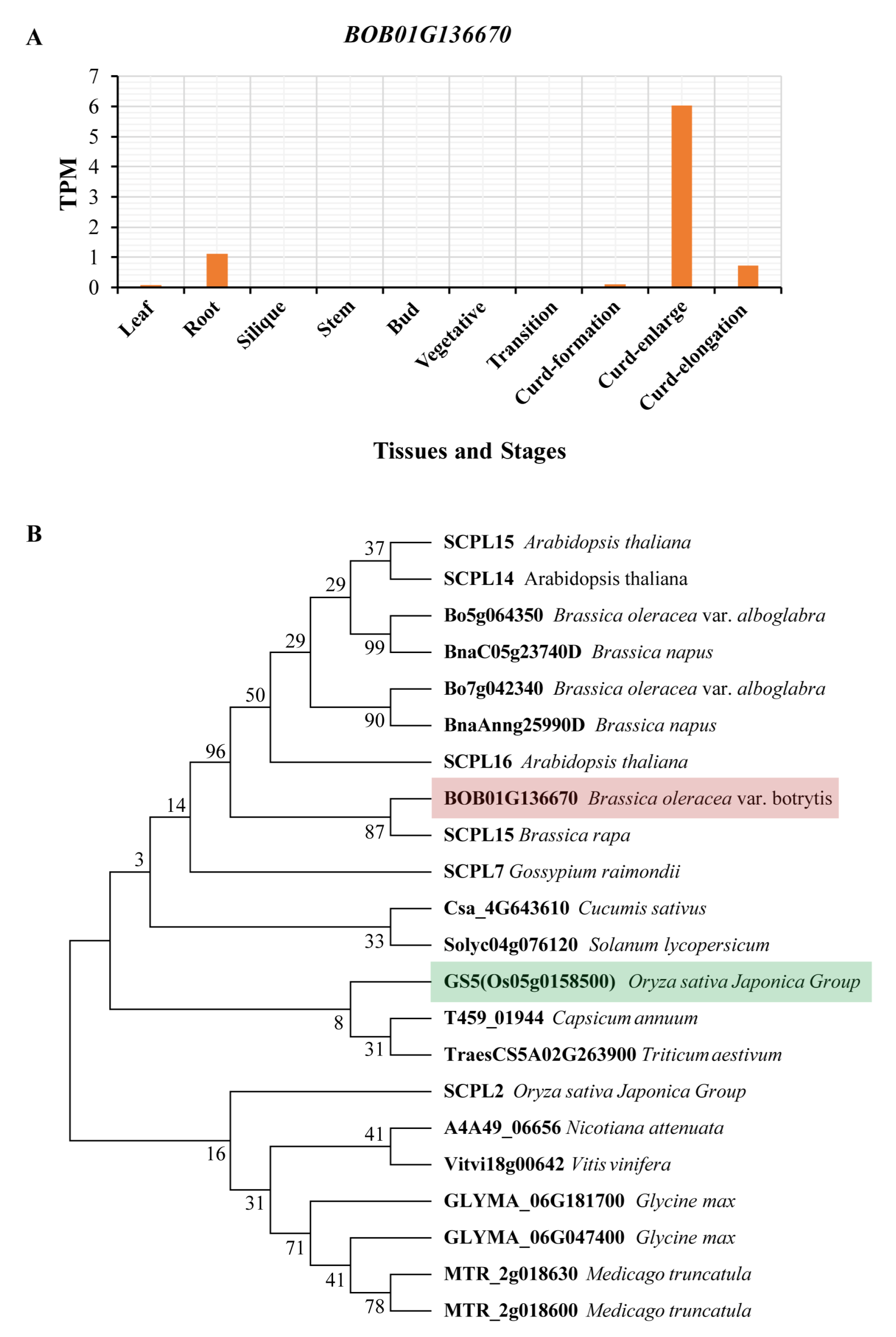
3.4. BOB01G136670 Regulates the Weight of a Single Curd
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| WSC | weight of a single curd |
| MSH | main stem height |
| ELW | external leaf wing |
| PC | purplish curd |
| TPM | transcripts per kilobase per million mapped reads |
References
- Dixon, G. Origins and diversity of Brassica and its relatives. In Vegetable Brassicas and Related Crucifers; CABI: Wallingford, UK, 2006; pp. 1–33. [Google Scholar] [CrossRef]
- Cheng, F.; Wu, J.; Wang, X. Genome triplication drove the diversification of Brassica plants. Hortic. Res. 2014, 1, 14024. [Google Scholar] [CrossRef]
- Liu, S.; Liu, Y.; Yang, X.; Tong, C.; Edwards, D.; Parkin, I.A.; Zhao, M.; Ma, J.; Yu, J.; Huang, S.; et al. The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 2014, 5, 3930. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, Q.; Zhang, Q.; Qin, E.; Jin, C.; Wang, Y.; Wu, M.; Shen, G.; Chen, C.; Song, W. Curd development associated gene (CDAG1) in cauliflower (Brassica oleracea L. var. botrytis) could result in enlarged organ size and increased biomass . Plant Sci. 2017, 254, 82–94. [Google Scholar] [CrossRef] [PubMed]
- Anthony, R.G.; James, P.E.; Jordan, B.R. The cDNA sequence of a cauliflower apetala-1/squamosa homolog. Plant Physiol. 1995, 108, 441–442. [Google Scholar] [CrossRef][Green Version]
- Anthony, R.G.; James, P.E.; Jordan, B.R. Cauliflower (Brassica oleracea var. botrytis L.) curd development: The expression of meristem identity genes. J. Exp. Bot. 1996, 47, 181–188. [Google Scholar] [CrossRef][Green Version]
- Cheung, K.L.; Kong, A.N. Molecular targets of dietary phenethyl isothiocyanate and sulforaphane for cancer chemoprevention. AAPS J. 2010, 12, 87–97. [Google Scholar] [CrossRef]
- Gu, H.H.; Jin, C.L.; Zhao, Z.Q.; Sheng, X.G.; Yu, H.F.; Wang, J.S. Analysis of the present situation and prospect of Chinese cauliflower industry. China Veg. 2012, 23, 1–5. [Google Scholar]
- Shan, X.Z.; Zhang, X.L.; Wen, Z.H.; Liu, L.L.; Yao, X.W.; Jiang, H.M.; Niu, G.B.; Sun, D.L. Status, development trend and countermeasure analysis of cauliflower industry in Beijing-Tianjin-Hebei Region. Vegetables 2019, 3, 43–46. [Google Scholar]
- Zhao, Z.Q.; Sheng, X.G.; Yu, H.F.; Wang, J.S.; Shen, Y.S.; Gu, H.H. Identification of QTLs associated with curd architecture in cauliflower. BMC Plant Biol. 2020, 20, 177. [Google Scholar] [CrossRef]
- Hasan, Y.; Briggs, W.; Matschegewski, C.; Ordon, F.; Stützel, H.; Zetzsche, H.; Groen, S.; Uptmoor, R. Quantitative trait loci controlling leaf appearance and curd initiation of cauliflower in relation to temperature. Theor. Appl. Genet. 2016, 129, 1273–1288. [Google Scholar] [CrossRef] [PubMed]
- Rosen, A.; Hasan, Y.; Briggs, W.; Uptmoor, R. Genome-based prediction of time to curd induction in cauliflower. Front. Plant Sci. 2018, 9, 78. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Lu, W.; Quan, M.; Xiao, L.; Song, F.; Li, P.; Zhou, D.; Xie, J.; Wang, L.; Zhang, D. Genome-wide association studies to improve wood properties: Challenges and prospects. Front. Plant Sci. 2018, 9, 1912. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, J.; Han, B.; Huang, X. Advances in genome-wide association studies of complex traits in rice. Theor. Appl. Genet. 2020, 133, 1415–1425. [Google Scholar] [CrossRef] [PubMed]
- Cortes, L.T.; Zhang, Z.; Yu, J. Status and prospects of genome-wide association studies in plants. Plant Genome 2021, 14, e20077. [Google Scholar] [CrossRef]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Li, H.; Peng, Z.; Yang, X.; Wang, W.; Fu, J.; Wang, J.; Han, Y.; Chai, Y.; Guo, T.; Yang, N.; et al. Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat. Genet. 2013, 45, 43–50. [Google Scholar] [CrossRef]
- Wang, M.; Yan, J.; Zhao, J.; Song, W.; Zhang, X.; Xiao, Y.; Zheng, Y. Genome-wide association study (GWAS) of resistance to head smut in maize. Plant Sci. 2012, 196, 125–131. [Google Scholar] [CrossRef]
- Liu, Y.; Shen, K.; Yin, C.; Xu, X.; Yu, X.; Ye, B.; Sun, Z.; Dong, J.; Bi, A.; Zhao, X.; et al. Genetic basis of geographical differentiation and breeding selection for wheat plant architecture traits. Genome Biol. 2023, 24, 114. [Google Scholar] [CrossRef]
- Zhou, Z.; Jiang, Y.; Wang, Z.; Gou, Z.; Lyu, J.; Li, W.; Yu, Y.; Shu, L.; Zhao, Y.; Ma, Y.; et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat. Biotechnol. 2015, 33, 408–414. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Ge, X.; Yuan, Y.; Jin, Y.; Wang, Y.; Zhao, L.; Han, X.; Hu, W.; Yang, L.; et al. Genome-wide association analysis reveals a novel pathway mediated by a dual-TIR domain protein for pathogen resistance in cotton. Genome Biol. 2023, 24, 111. [Google Scholar] [CrossRef]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1266. [Google Scholar] [CrossRef]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.J.; Wang, P.; Julca, I.; et al. A comprehensive genome variation map of melon identifies multiple domestication events and loci influencing agronomic traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhao, S.; Sun, H.; Wang, X.; Wu, S.; Lin, T.; Ren, Y.; Gao, L.; Deng, Y.; Zhang, J.; et al. Resequencing of 414 cultivated and wild watermelon accessions identifies selection for fruit quality traits. Nat. Genet. 2019, 51, 1616–1623. [Google Scholar] [CrossRef] [PubMed]
- Gajardo, H.A.; Wittkop, B.; Soto-Cerda, B.; Higgins, E.E.; Parkin, I.A.P.; Snowdon, R.J.; Federico, M.L.; Iniguez-Luy, F.L. Association mapping of seed quality traits in Brassica napus L. using GWAS and candidate QTL approaches. Mol. Breed. 2015, 35, 143. [Google Scholar] [CrossRef]
- Liu, S.; Huang, H.; Yi, X.; Zhang, Y.; Yang, Q.; Zhang, C.; Fan, C.; Zhou, Y. Dissection of genetic architecture for glucosinolate accumulations in leaves and seeds of Brassica napus by genome-wide association study. Plant Biotechnol. J. 2019, 18, 1472–1484. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Guan, M.; Zhang, Z.; Zhang, Q.; Cui, Y.; Chen, H.; Liu, W.U.; Jan, H.; Voss-Fels, K.P.; Werner, C.R.; et al. GWAS and co-expression network combination uncovers multigenes with close linkage effects on the oleic acid content accumulation in Brassica napus. BMC Genom. 2020, 21, 320. [Google Scholar] [CrossRef]
- Wei, L.; Jian, H.; Lu, K.; Filardo, F.; Yin, N.; Liu, L.; Qu, C.; Li, W.; Du, H.; Li, J. Genome-wide association analysis and differential expression analysis of resistance to sclerotinia stem rot in Brassica napus. Plant Biotechnol. J. 2016, 14, 1368–1380. [Google Scholar] [CrossRef]
- Thorwarth, P.; Yousef, E.A.A.; Schmid, K.J. Genomic prediction and association mapping of curd-related traits in gene bank accessions of cauliflower. G3 Genes Genomes Genet. 2018, 8, 707–718. [Google Scholar] [CrossRef]
- Matschegewski, C.; Zetzsche, H.; Hasan, Y.; Leibeguth, L.; Briggs, W.; Ordon, F.; Uptmoor, R. Genetic variation of temperature-regulated curd induction in cauliflower: Elucidation of floral transition by genome-wide association mapping and gene expression analysis. Front. Plant Sci. 2015, 6, 720. [Google Scholar] [CrossRef]
- Allen, G.C.; Flores-Vergara, M.; Krasynanski, S.; Kumar, S.; Thompson, W. A modified protocol. for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat. Protoc. 2006, 1, 2320–2325. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Huang, G.; He, S.; Yang, Z.; Sun, G.; Ma, X.; Li, N.; Zhang, X.; Sun, J.; Liu, M.; et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat. Genet. 2018, 50, 796–802. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic. Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.M. Admixture, population structure, and F-statistics. Genetics 2016, 202, 1485–1501. [Google Scholar] [CrossRef]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.; Xu, J.; He, W.; Yang, T. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef]
- Zoubarev, A.; Hamer, K.M.; Keshav, K.D.; McCarthy, E.L.; Santos, J.R.C.; Van Rossum, T.; McDonald, C.; Hall, A.; Wan, X.; Lim, R. Gemma: A resource for the reuse, sharing and meta-analysis of expression profiling data. Bioinformatics 2012, 28, 2272–2273. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290. [Google Scholar] [CrossRef] [PubMed]
- Guo, N.; Wang, S.; Gao, L.; Liu, Y.; Wang, X.; Lai, E.; Duan, M.; Wang, G.; Li, J.; Yang, M.; et al. Genome sequencing sheds light. on the contribution of structural variants to Brassica oleracea diversification. BMC Biol. 2021, 19, 93. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Deng, M.; Guo, H.; Raihan, S.; Luo, J.; Xu, Y.; Dong, X.; Yan, J. Maize orthologs of rice GS5 and their transregulator are associated with kernel development. J. Integr. Plant Biol. 2015, 57, 943–953. [Google Scholar] [CrossRef]
- Ma, L.; Li, T.; Hao, C.; Wang, Y.; Chen, X.; Zhang, X. TaGS5-3A, a grain size gene selected during wheat improvement for larger kernel and yield. Plant Biotechnol. J. 2016, 14, 1269–1280. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.I.; Brown, M.A.; Yang, J. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef]
- Sebastian, R.L.; Kearsey, M.J.; King, G.J. Identification of quantitative trait loci controlling developmental characteristics of Brassica oleracea L. Theor. Appl. Genet. 2002, 104, 601–609. [Google Scholar] [CrossRef]
- Lang, L.; Niu, G.; Dan, X.; Zhang, X.; Wen, Z.; Jiang, H. Determination of metabolites and principal component analysis of purplish curd in white cauliflower. China Cucurbits Veg. 2021, 34, 57–61. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Takano-Kai, N.; Jiang, H.; Kubo, T.; Sweeney, M.; Matsumoto, T.; Kanamori, H.; Padhukasahasram, B.; Bustamante, C.; Yoshimura, A.; Doi, K.; et al. Evolutionary history of GS3, a gene conferring grain size in rice. Genetics 2009, 182, 1323–1334. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Huang, W.; Shi, M.; Zhu, M.; Lin, H.A. QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Gu, S.; Wan, X.; Gao, H.; Guo, T.; Su, N.; Lei, C.; Zhang, X.; Cheng, Z.; Guo, X.; et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008, 18, 1199–1209. [Google Scholar] [CrossRef]
- Chen, W.; Chen, L.; Zhang, X.; Yang, N.; Guo, J.; Wang, M.; Ji, S.; Zhao, X.; Yin, P.; Cai, L.; et al. Convergent selection of a WD40 protein that enhances grain yield in maize and rice. Science 2022, 375, 1372. [Google Scholar] [CrossRef]
- Frary, A.; Nesbitt, T.C.; Grandillo, S.; Knaap, E.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B.; Tanksley, S.D. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef]
- Cong, B.; Liu, J.; Tanksley, S. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 13606–13611. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Munos, S.; Blanca, J.; Canizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef]
- Li, M.; Wang, X.; Li, C.; Li, H.; Zhang, J.; Ye, Z. Silencing GRAS2 reduces fruit weight in tomato. J. Integr. Plant Biol. 2018, 60, 498–513. [Google Scholar] [CrossRef]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.; Huang, S.; Yang, T.; Tang, Y.; Yang, S.; Aisimutuola, P.; et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef]
- Smith, L.B.; King, G.J. The distribution of BoCAL-a alleles in Brassica oleracea is consistent with a genetic model for curd development and domestication of the cauliflower. Mol. Breed. 2000, 6, 603–613. [Google Scholar] [CrossRef]
- Liu, Y.; Ce, F.; Tang, H.; Tian, G.; Yang, L.; Qian, W.; Dong, H. Genome-wide analysis of the serine carboxypeptidase-like (SCPL) proteins in Brassica napus L. Plant Physiol. Bioch. 2022, 186, 310–321. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, L.; Zhao, W.; Fu, L.; Han, Y.; Wang, K.; Yan, L.; Li, Y.; Zhang, X.; Min, D. Genome-wide analysis of the serine carboxypeptidase-like protein family in Triticum aestivum reveals TaSCPL184-6D is involved in abiotic stress response. BMC Genom. 2021, 22, 350. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Wen, Z.; Jiang, H.; Niu, G.; Liu, L.; Yao, X.; Sun, D.; Shan, X. Identification of Loci for Four Important Agronomic Traits in Loose-Curd Cauliflower Based on Genome-Wide Association Studies. Horticulturae 2023, 9, 970. https://doi.org/10.3390/horticulturae9090970
Zhang X, Wen Z, Jiang H, Niu G, Liu L, Yao X, Sun D, Shan X. Identification of Loci for Four Important Agronomic Traits in Loose-Curd Cauliflower Based on Genome-Wide Association Studies. Horticulturae. 2023; 9(9):970. https://doi.org/10.3390/horticulturae9090970
Chicago/Turabian StyleZhang, Xiaoli, Zhenghua Wen, Hanmin Jiang, Guobao Niu, Lili Liu, Xingwei Yao, Deling Sun, and Xiaozheng Shan. 2023. "Identification of Loci for Four Important Agronomic Traits in Loose-Curd Cauliflower Based on Genome-Wide Association Studies" Horticulturae 9, no. 9: 970. https://doi.org/10.3390/horticulturae9090970
APA StyleZhang, X., Wen, Z., Jiang, H., Niu, G., Liu, L., Yao, X., Sun, D., & Shan, X. (2023). Identification of Loci for Four Important Agronomic Traits in Loose-Curd Cauliflower Based on Genome-Wide Association Studies. Horticulturae, 9(9), 970. https://doi.org/10.3390/horticulturae9090970







