Cytogenetic Study of Five Varieties of Callisia repens (Jacq.) L. (Commelinaceae) from Laos
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Mitotic and Karyotype Analyses
2.3. Statistical Analysis
3. Results
- PCA score plot for karyotype formula
- PCA score plot for CI
- PCA score plot for leaf color
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- POWO. Plant of the World Online, Facilitated by the Royal Botanic Gardens, Kew. Available online: http://www.plantsoftheworldonline.org/ (accessed on 12 June 2023).
- Cronquist, A. An Integrated System of Classification of the Flowering Plants; Columbia University Press: New York, UY, USA, 1981. [Google Scholar]
- Faden, R.B. The morphology and taxonomy of Aneilema R. Brown (Commelinaceae). In Smithsonian Contributions to Botany; Smithsonian Institution Press: Washington, DC, USA, 1991. [Google Scholar]
- AONA, L.Y.S. Revisão Taxonomica e Analise Cladistica do Genero Dichorisandra J.C. Mikan (Commelinaceae). Ph.D. Thesis, Universidade Estadual de Campinas, Campinas, Brazil, 2008. [Google Scholar]
- Mabberley, D.J. Mabberley’s Plant Book, 3rd ed.; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Angiosperm Phylogeny Group; Chase, M.W.; Christenhusz, M.J.; Fay, M.F.; Byng, J.W.; Judd, W.S.; Stevens, P.F. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar] [CrossRef]
- Rahman, A.H.M.M.; Sultana, Z.; Rani, R.; Islam, A.K.M.R. Taxonomic Studies of the Family Commelinaceae at Rajshahi, Bangladesh. Int. J. Adv. Res. 2015, 3, 978–989. [Google Scholar]
- Łuczaj, Ł.; Lamxay, V.; Tongchan, K.; Xayphakatsa, K.; Phimmakong, K.; Radavanh, S.; Kanyasone, V.; Pietras, M.; Karbarz, M. Wild food plants and fungi sold in the markets of Luang Prabang, Lao PDR. J. Ethnobiol. Ethnomed. 2021, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Santos Moraes, Q.D.; de Oliveira Pellegrini, M.O.; Alves-Araújo, A. Flora of Espírito Santo: Commelinaceae. Rodriguésia 2022, 73, e00522021. [Google Scholar] [CrossRef]
- Long, C.L.; Li, R. Ethnobotanical studies on medicinal plants used by the red-headed yao people in Jinping, Yunnan Province, China. J. Ethnopharmacol. 2004, 90, 389–395. [Google Scholar] [CrossRef]
- Delang, C.O. Ecological Succession of Usable Plants in an Eleven-Year Fallow Cycle in Northern Lao P.D.R. Ethnobot. Res. Appl. 2007, 5, 331–350. [Google Scholar] [CrossRef]
- Panyaphu, K.; On, T.V.; Sirisa-ard, P.; Srisa-nga, P.; ChansaKaow, S.; Nathakarnkitkul, S. Medicinal plants of the Mien (Yao) in Northern Thailand and their potential value in the primary healthcare of postpartum women. J. Ethnopharmacol. 2011, 135, 226–237. [Google Scholar] [CrossRef]
- Khuankaew, S.; Srithi, K.; Tiansawat, P.; Jampeetong, A.; Inta, A.; Wangpakapattanawong, P. Ethnobotanical study of medicinal plants used by Tai Yai in Northern Thailand. J. Ethnopharmacol. 2014, 151, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Sui, X.; Ahmed, S.; Wang, Z.; Long, C. Ethnobotany and diversity of medicinal plants used by the Buyi in eastern Yunnan, China. Plant Divers. 2020, 42, 401–414. [Google Scholar] [CrossRef] [PubMed]
- Saensouk, P.; Saensouk, S. Karyological study of four species in Callisia and Cyanotis (Commelinaceae) from Thailand. Cytologia 2021, 86, 339–343. [Google Scholar] [CrossRef]
- Saensouk, S.; Saensouk, P. A karyological study of six Commelinaceae species from Thailand. Cytologia 2020, 85, 57–62. [Google Scholar] [CrossRef]
- Gledhill, D. The Names of Plants, 4th ed.; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Cao, J.; Hu, S.; Dong, Q.; Liu, L.; Wang, Z. Green roof cooling contributed by plant species with different photosynthetic strategies. Energy Build. 2019, 195, 45–50. [Google Scholar] [CrossRef]
- Beltrão, G.T.d.A.; Guerra, M. Citogenética de angiospermas coletadas em Pernambuco—III. Ciênc. Cult. 1990, 42, 839–845. [Google Scholar]
- Pitrez, S.R.; Felix, L.P.; Barreto, R.; Guerra, M. Númeroscromossômicos de espécies de Commelinaceae R. Br. ocorrentes no nordeste do Brasil. Bol. Bot. Univ. São Paulo 2001, 19, 7–14. [Google Scholar]
- Grabiele, M.; Daviña, J.R.; Honfi, A.I. Cytogenetic analyses as clarifying tools for taxonomy of the genus Callisia Loefl. (Commelinaceae). Gayana Bot. 2015, 72, 34–41. [Google Scholar] [CrossRef]
- Saensouk, P.; Saensouk, S. Karyological Study of Four Genera and Seven Species (Commelinaceae) in Thailand. Cytologia 2022, 87, 41–47. [Google Scholar] [CrossRef]
- Saensouk, P.; Saensouk, S. Cytogenetics of five species of genus Murdannia (Commelinaceae) from North-eastern Thailand. Biologia 2022, 78, 23–30. [Google Scholar] [CrossRef]
- Senavongse, R.; Saensouk, S.; Saensouk, P. Comparative karyotype analysis in five morphological forms of Bon or Colocasia esculenta (L.) Schott (Araceae) in Thailand. Cytologia 2018, 83, 169–173. [Google Scholar] [CrossRef]
- Senavongse, R.; Saensouk, S.; Saensouk, P. Karyological study in three native species of genus Alocasia (Araceae) in the northeast of Thailand. Nucleus 2020, 63, 81–85. [Google Scholar] [CrossRef]
- Bro, R.; Smilde, A.K. Principal component analysis. Anal. Methods 2014, 6, 2812–2831. [Google Scholar] [CrossRef]

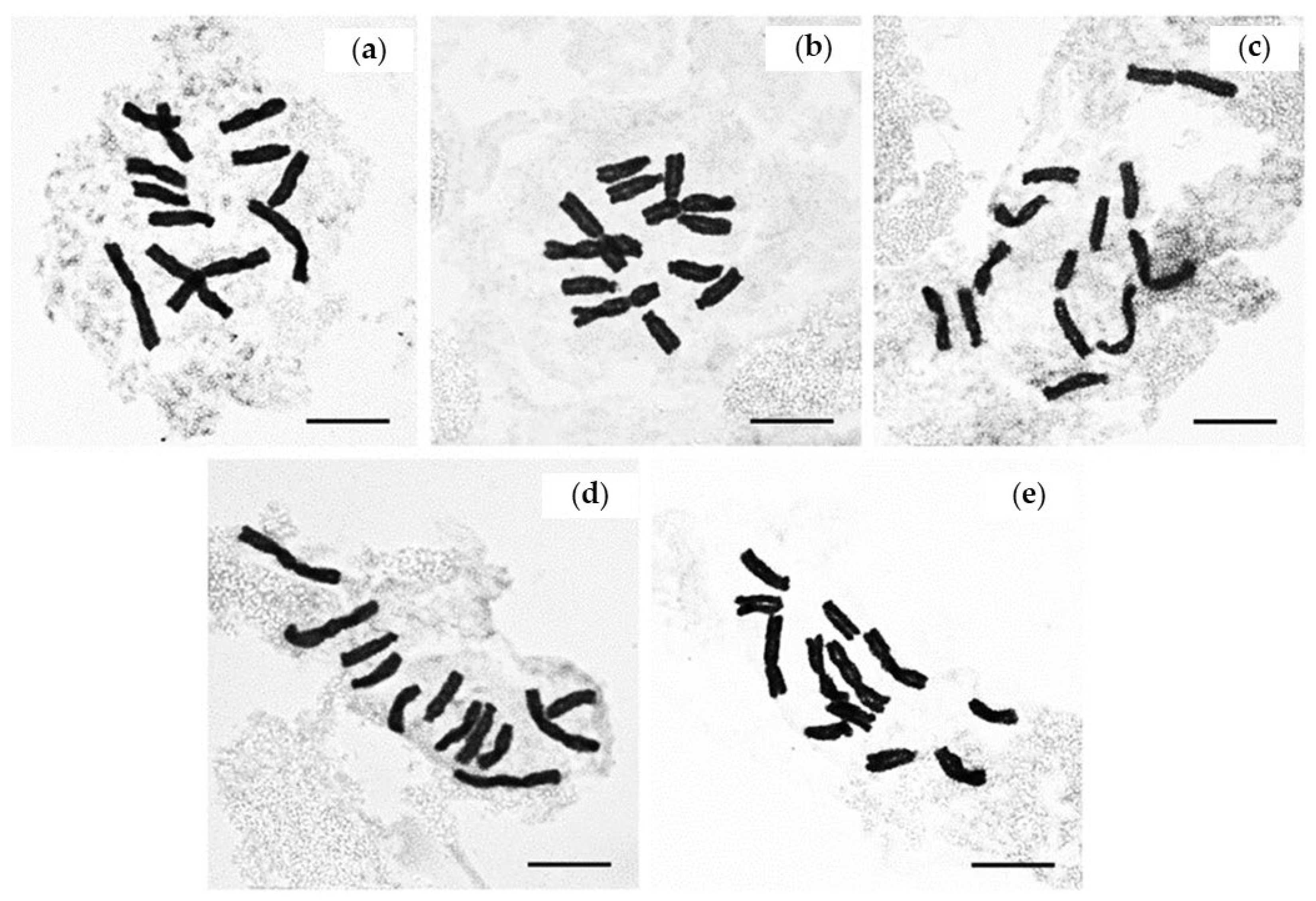


| Species | 2n | n | Karyotype Formula | Origin | Previous Research |
|---|---|---|---|---|---|
| C. repens | 12 | - | - | Brazil | [19] |
| 12 | - | - | Brazil | [20] | |
| 12 | 6 | 2m + 2sm + 6st + 2t | Argentina | [21] | |
| 12 | - | 4sm + 8st | Thailand (Maha Sarakham province) | [15] | |
| C. repens ‘turtle vine’ | 12 | - | 2m + 2sm + 8st | Laos (province of Vientiane) | Present research |
| C. repens ‘green’ | 12 | - | 2m + 10st | Laos (province of Vientiane) | Present research |
| C. repens ‘pink lady’ | 12 | - | 8m + 4st | Laos (province of Vientiane) | Present research |
| C. repens ‘gold’ | 12 | - | 2m + 4sm + 6st | Laos (province of Vientiane) | Present research |
| C. repens ‘Bianca’ | 12 | - | 2m + 2sm + 8st | Laos (province of Vientiane) | Present research |
| Variety | Mean Length of Chromosome Pairs 1–6, RL, and CI | Chromosome Type | ||||
|---|---|---|---|---|---|---|
| Ls ± SD (µm) | Ll ± SD (µm) | LT ± SD (µm) | RL (%) | CI | ||
| Turtle vine | 5.79 ± 0.02 | 8.43 ± 0.03 | 14.21 ± 0.05 | 24.38 ± 0.01 | 0.59 ± 0.02 | Metacentric |
| 4.59 ± 0.01 | 8.34 ± 0.03 | 12.94 ± 0.04 | 22.19 ± 0.01 | 0.64 ± 0.02 | Submetacentric | |
| 1.02 ± 0.01 | 7.31 ± 0.03 | 8.33 ± 0.03 | 14.28 ± 0.01 | 0.88 ± 0.01 | Subtelocentric | |
| 1.09 ± 0.02 | 6.90 ± 0.02 | 7.99 ± 0.03 | 13.70 ± 0.02 | 0.86 ± 0.01 | Subtelocentric | |
| 0.92 ± 0.02 | 6.64 ± 0.03 | 7.56 ± 0.04 | 12.96 ± 0.01 | 0.88 ± 0.02 | Subtelocentric | |
| 0.89 ± 0.01 | 6.39 ± 0.02 | 7.28 ± 0.02 | 12.49 ± 0.01 | 0.88 ± 0.01 | Subtelocentric | |
| Green | 3.42 ± 0.02 | 8.09 ± 0.03 | 11.51 ± 0.05 | 23.45 ± 0.01 | 0.70 ± 0.01 | Subtelocentric |
| 4.65 ± 0.01 | 6.75 ± 0.03 | 11.40 ± 0.04 | 23.22 ± 0.01 | 0.59 ± 0.01 | Metacentric | |
| 0.92 ± 0.02 | 6.15 ± 0.02 | 7.07 ± 0.03 | 14.39 ± 0.02 | 0.87 ± 0.01 | Subtelocentric | |
| 0.92 ± 0.01 | 5.81 ± 0.02 | 6.73 ± 0.02 | 13.70 ± 0.02 | 0.86 ± 0.01 | Subtelocentric | |
| 0.72 ± 0.01 | 5.96 ± 0.02 | 6.69 ± 0.02 | 13.62 ± 0.01 | 0.89 ± 0.01 | Subtelocentric | |
| 0.83 ± 0.02 | 4.86 ± 0.02 | 5.70 ± 0.02 | 11.60 ± 0.01 | 0.85 ± 0.02 | Subtelocentric | |
| Pink lady | 6.02 ± 0.02 | 8.18 ± 0.03 | 14.20 ± 0.05 | 24.58 ± 0.02 | 0.58 ± 0.02 | Metacentric |
| 5.81 ± 0.01 | 6.85 ± 0.03 | 12.66 ± 0.04 | 22.13 ± 0.02 | 0.54 ± 0.02 | Metacentric | |
| 4.08 ± 0.02 | 4.16 ± 0.02 | 8.23 ± 0.03 | 14.38 ± 0.01 | 0.51 ± 0.03 | Metacentric | |
| 0.97 ± 0.01 | 6.82 ± 0.02 | 7.80 ± 0.03 | 13.63 ± 0.01 | 0.88 ± 0.01 | Subtelocentric | |
| 0.89 ± 0.02 | 6.35 ± 0.02 | 7.24 ± 0.02 | 12.66 ± 0.02 | 0.88 ± 0.01 | Subtelocentric | |
| 3.19 ± 0.01 | 3.90 ± 0.02 | 7.08 ± 0.02 | 12.38 ± 0.01 | 0.55 ± 0.01 | Metacentric | |
| Gold | 5.34 ± 0.02 | 8.22 ± 0.03 | 13.56 ± 0.05 | 24.40 ± 0.01 | 0.61 ± 0.01 | Submetacentric |
| 5.06 ± 0.01 | 7.56 ± 0.03 | 12.62 ± 0.04 | 22.71 ± 0.02 | 0.60 ± 0.02 | Submetacentric | |
| 1.08 ± 0.01 | 7.10 ± 0.02 | 8.17 ± 0.03 | 14.71 ± 0.02 | 0.87 ± 0.01 | Subtelocentric | |
| 0.90 ± 0.02 | 6.61 ± 0.02 | 7.52 ± 0.03 | 13.53 ± 0.02 | 0.88 ± 0.01 | Subtelocentric | |
| 1.45 ± 0.01 | 5.63 ± 0.02 | 7.08 ± 0.02 | 12.74 ± 0.01 | 0.81 ± 0.02 | Subtelocentric | |
| 2.81 ± 0.01 | 3.82 ± 0.02 | 6.63 ± 0.02 | 11.92 ± 0.01 | 0.58 ± 0.02 | Metacentric | |
| Bianca | 4.15 ± 0.02 | 6.29 ± 0.03 | 10.44 ± 0.04 | 23.29 ± 0.01 | 0.60 ± 0.01 | Submetacentric |
| 4.13 ± 0.01 | 5.54 ± 0.03 | 9.67 ± 0.04 | 21.56 ± 0.01 | 0.57 ± 0.01 | Metacentric | |
| 0.92 ± 0.01 | 6.03 ± 0.02 | 6.95 ± 0.03 | 15.49 ± 0.02 | 0.87 ± 0.01 | Subtelocentric | |
| 1.68 ± 0.01 | 4.41 ± 0.02 | 6.09 ± 0.02 | 13.59 ± 0.02 | 0.72 ± 0.02 | Subtelocentric | |
| 0.97 ± 0.02 | 5.05 ± 0.02 | 6.02 ± 0.02 | 13.43 ± 0.01 | 0.84 ± 0.02 | Subtelocentric | |
| 0.47 ± 0.02 | 5.21 ± 0.02 | 5.68 ± 0.02 | 12.65 ± 0.01 | 0.92 ± 0.02 | Subtelocentric | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sengthong, A.; Saensouk, S.; Saensouk, P.; Souladeth, P. Cytogenetic Study of Five Varieties of Callisia repens (Jacq.) L. (Commelinaceae) from Laos. Horticulturae 2023, 9, 1050. https://doi.org/10.3390/horticulturae9091050
Sengthong A, Saensouk S, Saensouk P, Souladeth P. Cytogenetic Study of Five Varieties of Callisia repens (Jacq.) L. (Commelinaceae) from Laos. Horticulturae. 2023; 9(9):1050. https://doi.org/10.3390/horticulturae9091050
Chicago/Turabian StyleSengthong, Anousone, Surapon Saensouk, Piyaporn Saensouk, and Phetlasy Souladeth. 2023. "Cytogenetic Study of Five Varieties of Callisia repens (Jacq.) L. (Commelinaceae) from Laos" Horticulturae 9, no. 9: 1050. https://doi.org/10.3390/horticulturae9091050






