Whole Genome Characterization of Prunus Necrotic Ringspot Virus and Prune Dwarf Virus Infecting Stone Fruits in Russia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. High-Throughput Sequencing (HTS)
2.3. Sequence Analyses
2.4. Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
3. Results
3.1. Plant Material and Virus Detection
3.2. HTS Results
3.3. Characterization of the PDV Genome
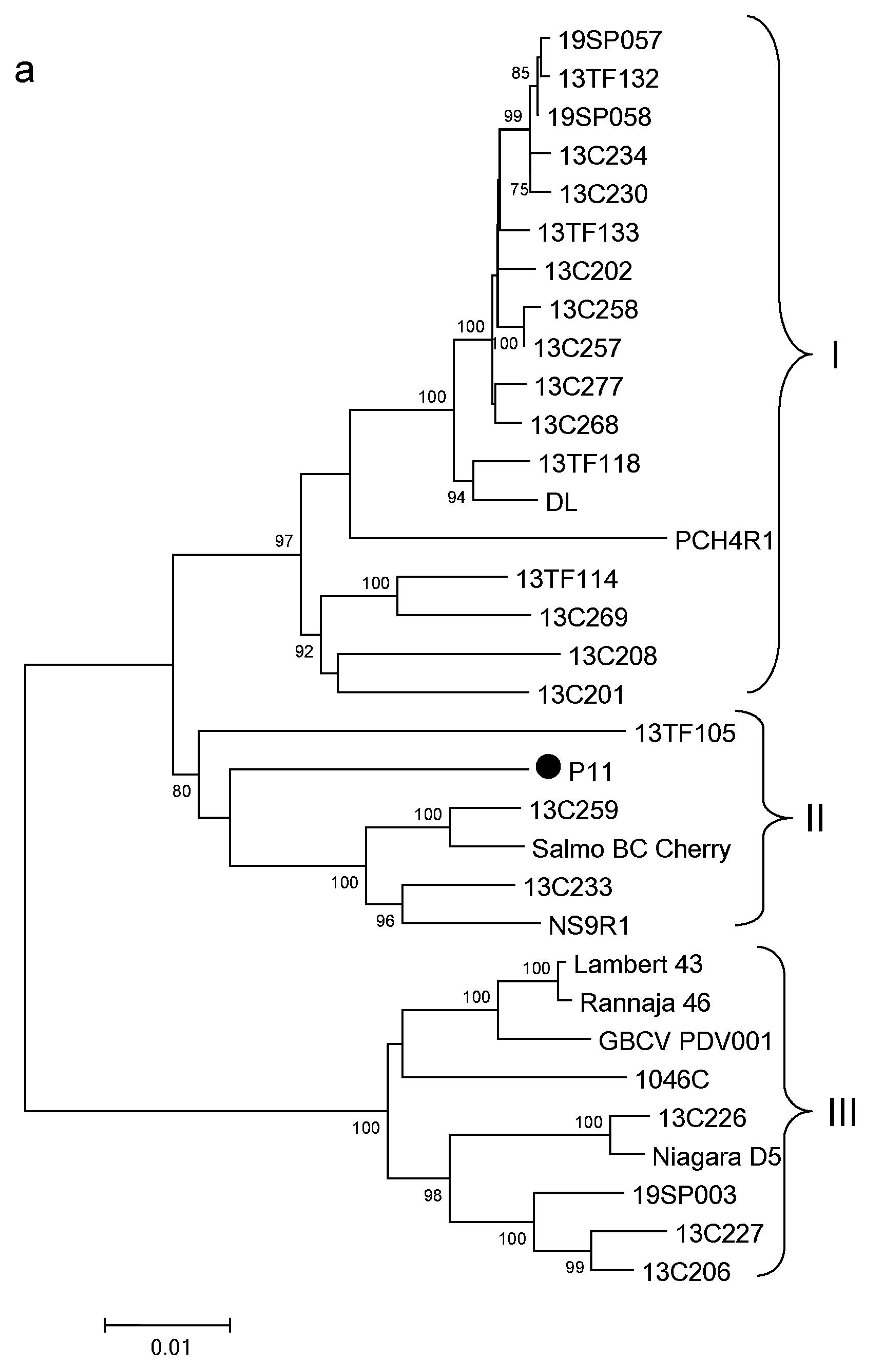
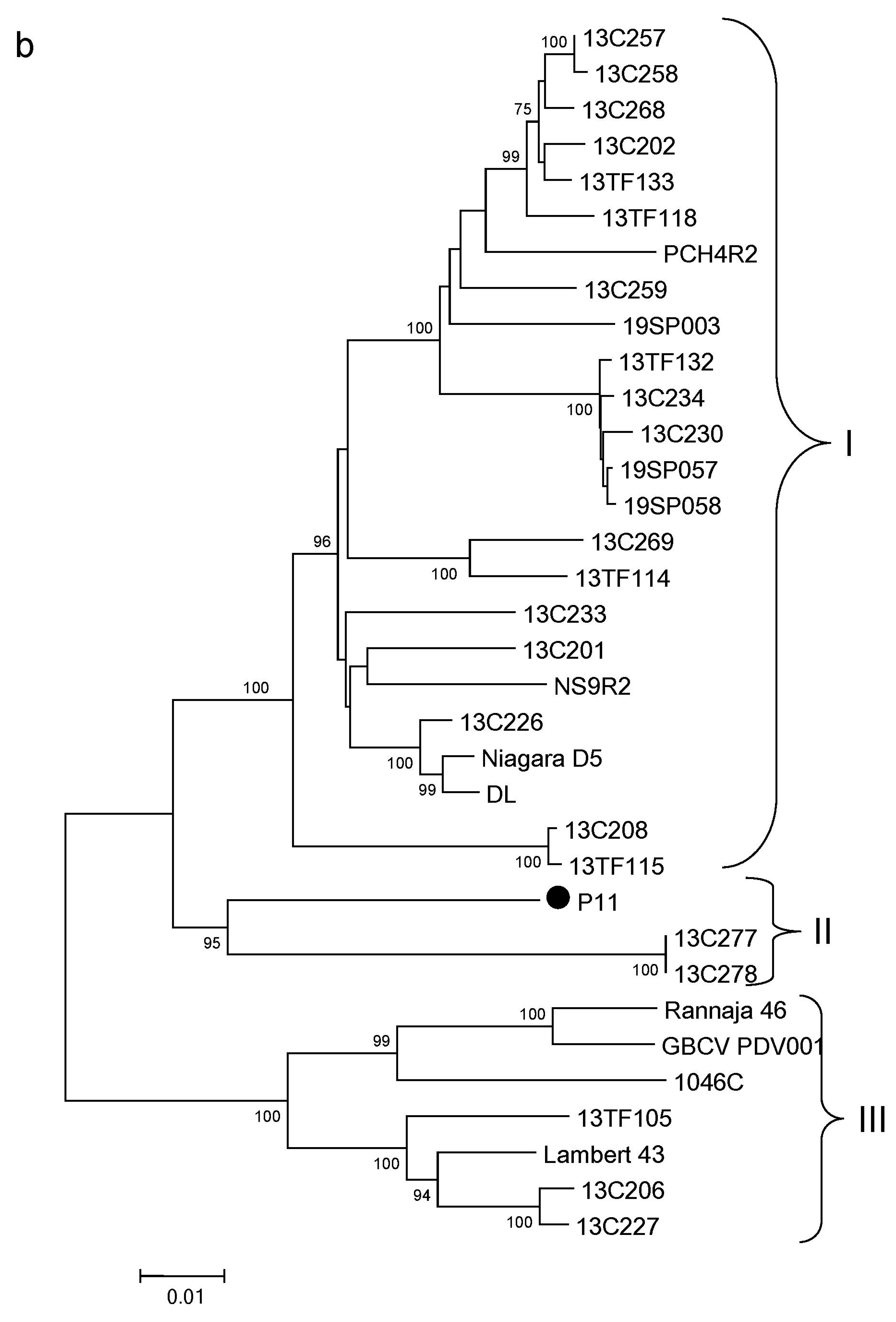
3.4. Phylogenetic Analysis of PDV
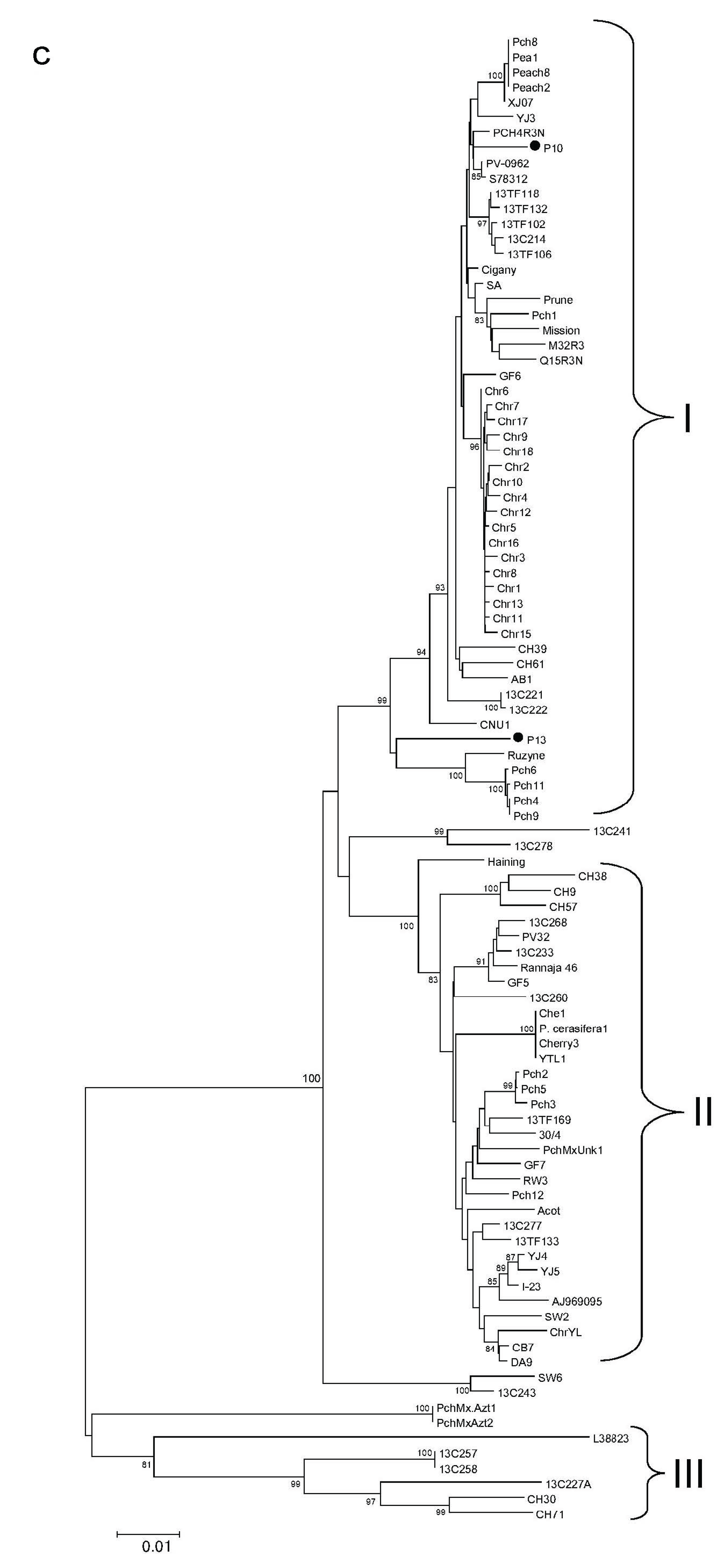
3.5. Characterization of the PNRSV Genomes
3.6. Phylogenetic Analysis of PNRSV
3.7. Recombination in the PDV and PNRSV Complete Genomes
3.8. Partial Characterization of Ourmia-like Virus Genome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hadidi, A.; Barba, M.; Candresse, T.; Jelkmann, W. Virus and Virus-Like Diseases of Pome and Stone Fruits; APS Press: St. Paul, MN, USA, 2011. [Google Scholar]
- Maliogka, V.; Minafra, A.; Saldarelli, P.; Ruiz-García, A.; Glasa, M.; Katis, N.; Olmos, A. Recent advances on detection and characterization of fruit tree viruses using high-throughput sequencing technologies. Viruses 2018, 10, 436. [Google Scholar] [CrossRef] [PubMed]
- Rubio, M.; Martinez-Gomez, P.; Marais, A.; Sanches-Navarro, J.A.; Pallas, V.; Candresse, T. Recent advances and prospect in Prunus virology. Ann. Appl. Biol. 2017, 171, 125–138. [Google Scholar] [CrossRef]
- Rott, M.; Xiang, Y.; Boyes, I.; Belton, M.; Saeed, H.; Kesanakurti, P.; Hayes, S.; Lawrence, T.; Birch, C.; Bhagwat, B.; et al. Application of next generation sequencing for diagnostic testing of tree fruit viruses and viroids. Plant Dis. 2017, 101, 1489–1499. [Google Scholar] [CrossRef]
- Martelli, G.; Flores, R.; Schneider, B. Classification of pome and stone fruit viruses, viroids and phytoplasmas. In Virus and Virus-Like Disease of Pome and Stone Fruits; Hadidi, A., Barba, M., Candresse, T., Jelkman, W., Eds.; APS Press: St. Paul, MN, USA, 2011; pp. 13–16. [Google Scholar]
- Hammond, R.W. Prunus necrotic ringspot virus. In Virus and Virus-Like Disease of Pome and Stone Fruits; Hadidi, A., Barba, M., Candresse, T., Jelkman, W., Eds.; APS Press: St. Paul, MN, USA, 2011; pp. 207–213. [Google Scholar]
- Caglayan, K.; Ulubas-Serce, C.; Gazel, M.; Varvery, C. Prune dwarf virus. In Virus and Virus-like Disease of Pome and Stone Fruits; Hadidi, A., Barba, M., Candresse, T., Jelkman, W., Eds.; APS Press: St. Paul, MN, USA, 2011; pp. 199–205. [Google Scholar]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Sanchez-Navarro, J.A.; Scott, S.W. The molecular biology of ilarviruses. Adv. Vir. Res. 2013, 87, 139–181. [Google Scholar]
- Tayal, M.; Wilson, C.; Cieniewicz, E. Bees and thrips carry virus-positive pollen in peach orchards in South Carolina, United States. J. Econ. Entomol. 2023, 116, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Koziel, E.; Bujarski, J.J.; Otulak, K. Molecular biology of Prune dwarf virus—A lesser known member of the Bromoviridae but a vital component in the dynamic virus-host cell interaction network. Int. J. Mol. Sci. 2017, 18, 2733. [Google Scholar] [CrossRef]
- Pallas, V.; Aparicio, F.; Herranz, M.C.; Amari, K.; Sanches-Pina, M.A.; Myrta, A.; Sanchez-Navarro, J.A. Ilarviruses in Prunus spp.: A continued concern for fruit trees. Phytopathology 2012, 102, 1108–1120. [Google Scholar] [CrossRef]
- Mitrofanova, I.V.; Smykov, A.V.; Mitrofanova, O.V.; Lesnikova-Sedoshenko, N.P.; Chirkov, S.N.; Zhdanova, I.V. Using in vitro embryo culture for obtaining new breeding forms of peach. Acta Hortic. 2020, 1289, 159–165. [Google Scholar] [CrossRef]
- Prikhod’ko, Y.N.; Chirkov, S.N.; Metlitskaya, K.V.; Tsubera, L.V. Distribution of viral diseases of stone fruits in European part of Russia. Agric. Biol. 2008, 1, 26–32. (In Russian) [Google Scholar]
- Trifonova, A.A.; Boris, K.V.; Mesyats, N.V.; Tsiupka, V.A.; Smykov, A.V.; Mitrofanova, I.V. Genetic diversity of peach cultivars from the collection of the Nikita Botanical Garden based on SSR markers. Plants 2021, 10, 2609. [Google Scholar] [CrossRef] [PubMed]
- Gambino, G.; Perrone, I.; Gribaudo, I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 2008, 19, 520–525. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrel, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombinant pattern in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Untiveros, M.; Perez-Egusquiza, Z.; Clover, G. PCR assays for the detection of members of the genus Ilarvirus and family Bromoviridae. J. Virol. Methods 2010, 165, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Predajna, L.; Shihelska, N.; Benedictova, D.; Soltys, K.; Candresse, T.; Glasa, M. Molecular characterization of Prune dwarf virus cherry isolates from Slovakia shows their substantial variability and reveals recombination events in PDV RNA3. Eur. J. Plant Pathol. 2017, 147, 877–885. [Google Scholar] [CrossRef]
- Bonilla, F.R.; Cieniewicz, E. Distribution and diversity of prunus necrotic ringspot virus, prune dwarf virus, and peach latent mosaic viroid in wild Prunus spp. in South Carolina and Georgia. PhytoFrontiers 2022, 2, 363. [Google Scholar] [CrossRef]
- Kamenova, I.L.; Borisova, A.Z. Biological and molecular properties of Prune dwarf virus cherry isolates from Bulgaria. J. Plant Dis. Prot. 2022, 129, 301–311. [Google Scholar] [CrossRef]
- Ulubas Serce, C.; Ertunc, F.; Ozturk, A. Identification and genomic variability of Prune dwarf virus variants infecting stone fruit trees in Turkey. J. Phytopathol. 2009, 157, 298–305. [Google Scholar] [CrossRef]
- Vaskova, D.; Petrzik, K.; Spak, J. Molecular variability of the capsid protein of the prune dwarf virus. Eur. J. Plant Pathol. 2000, 106, 573–580. [Google Scholar] [CrossRef]
- Noorani, M.S.; Baid, S.B.; Khan, J.A.; Pravej, A. Whole genome characterization and diagnostics of prunus necrotic ringspot virus (PNRSV) infecting apricot in India. Sci. Rep. 2023, 13, 4393. [Google Scholar] [CrossRef] [PubMed]
- Kamenova, I.L.; Borisova, A.Z. Molecular variability of the coat protein gene of prunus necrotic ringspot virus on sweet and sour cherry in Bulgaria. J. Plant Pathol. 2021, 103, 97–104. [Google Scholar] [CrossRef]
- Ulubas Serce, C.; Ertunc, F. RT-PCR detection and molecular characterization of Prunus necrotic ringspot virus isolates occurring in Turkey. J. Phytopathol. 2004, 152, 498–502. [Google Scholar] [CrossRef]
- Fiore, N.; Fajardo, T.V.; Prodan, S.; Herranz, M.C.; Aparicio, F.; Montealegre, J.; Elena, S.F.; Pallas, V.; Sanchez-Navarro, J. Genetic diversity of the movement and coat protein genes of South American isolates of Prunus necrotic ringspot virus. Arch. Virol. 2008, 153, 909–919. [Google Scholar] [CrossRef] [PubMed]
- Chare, E.R.; Holmes, E.C. A phylogenetic survey of recombination in plant viruses. Arch. Virol. 2006, 151, 933–946. [Google Scholar] [CrossRef]
- Ravelonandro, M. Reliable metodologies and impactful tools to control fruit tree viruses. Crops 2021, 1, 32–41. [Google Scholar] [CrossRef]
- Pita, J.S.; Roossink, M.J. Fixation of emerging interviral recombination in Cucumber mosaic virus populations. J. Virol. 2013, 87, 1264–1269. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, J.; Zhang, H.; Tang, X.; Du, Z. Molecular evidence and sequence analysis of a natural reassortant between Cucumber mosaic virus subgroup IA and II strains. Virus Genes 2007, 35, 405–413. [Google Scholar] [CrossRef]
- Codoner, F.; Elena, S.F. The promiscuous evolutionary history of the family Bromoviridae. J. Gen. Virol. 2008, 89, 1739–1747. [Google Scholar] [CrossRef]
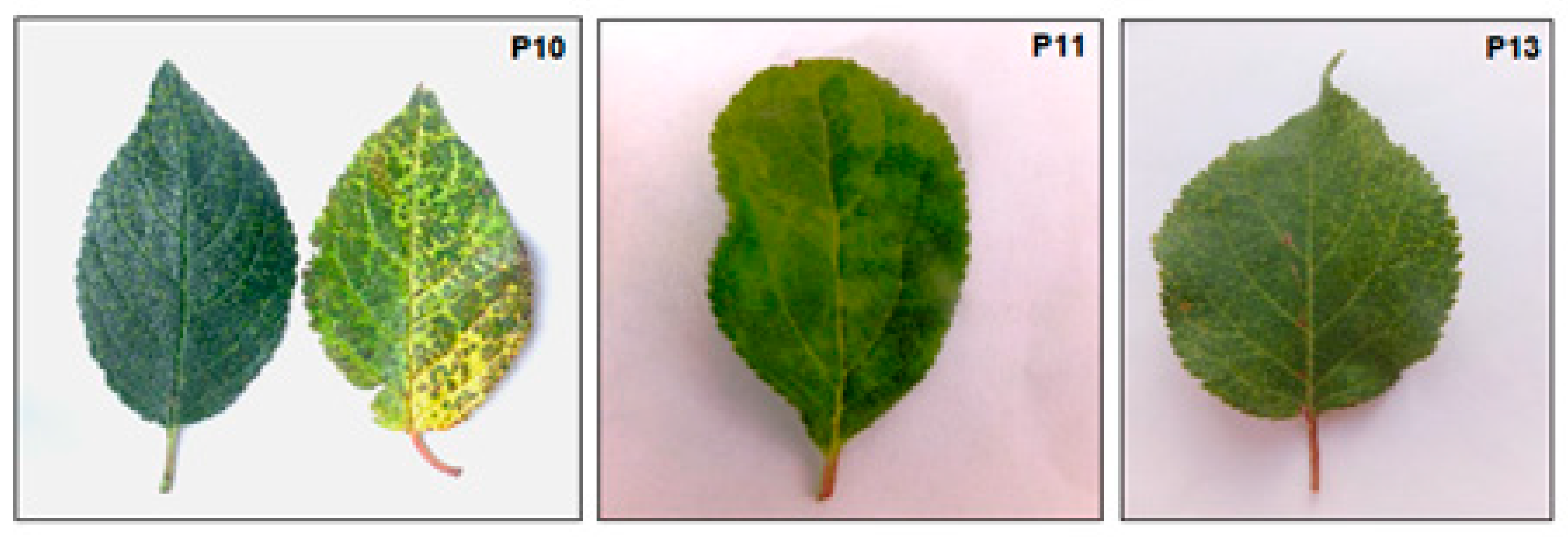



| Sample | Number of Clean Reads a per Sample | Virus Detected | Number of Virus- Specific Reads b |
|---|---|---|---|
| P10 | 23657640 | Prunus necrotic ringspot virus | 2710 (RNA1) 1956 (RNA2) 4368 (RNA3) |
| Ourmia-like virus | 58 | ||
| P11 | 17497020 | Prune dwarf virus | 3645 (RNA1) 3146 (RNA2) 7272 (RNA3) |
| P13 | 2903382 | Prunus necrotic ringspot virus | 7444 (RNA1) 3424 (RNA2) 1129 (RNA3) |
| Virus | Genome Region | Recombinant | Beginning Breakpoint | Ending Breakpoint | Major Parent | Minor Parent | p Values * |
|---|---|---|---|---|---|---|---|
| PDV | ORF1 | PCH4R1 | 600 | 1101 | 13TF118 | Salmo BC Cherry | 10−5 |
| 3118 | 3317 | 13TF118 | Salmo BC Cherry | 10−7 | |||
| ORF3a-IR-ORF3b | PA78 | 963 | 1614 | SWM1 | 13TF114 | 10−8 | |
| PE247 | 963 | 1613 | Rannaja 46 | SOF17P17 | 10−18 | ||
| SOF15P11 | 1 | 933 | SWM1 | SOF17P17 | 10−7 | ||
| 13C257 | 47 | 561 | 13C226 | 13C278 | 10−9 | ||
| 13C258 | 47 | 561 | 13C226 | 13C278 | 10−9 | ||
| PNRSV | ORF1 | Che1 | 1268 | 2090 | P. cerasifera 1 | Rannaja 46 | 10−28 |
| Che2 | 2090 | 3138 | CH57 | P. cerasifera 1 | 10−35 | ||
| Q15R1N | 1 | 790 | 13TF132 | CH57 | 10−33 | ||
| TNpeach5 | 844 | 1542 | 13C243 | Q15R1N | 10−15 | ||
| TNpeach5 | 32 | 624 | 13C243 | SCh-Taian | 10−7 | ||
| ORF2 | Che1 | 1243 | ORF2 end | PV-0962 | P. cerasifera 1 | 10−14 | |
| Che2 | 1324 | ORF2 end | PV-0962 | P. cerasifera 1 | 10−26 | ||
| ORF3a-IR-ORF3b | 13C257 | 48 | 264 | 13C227A | 13C260 | 10−15 | |
| 13C258 | 48 | 264 | 13C227A | 13C260 | 10−15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chirkov, S.; Sheveleva, A.; Tsygankova, S.; Slobodova, N.; Sharko, F.; Petrova, K.; Mitrofanova, I. Whole Genome Characterization of Prunus Necrotic Ringspot Virus and Prune Dwarf Virus Infecting Stone Fruits in Russia. Horticulturae 2023, 9, 941. https://doi.org/10.3390/horticulturae9080941
Chirkov S, Sheveleva A, Tsygankova S, Slobodova N, Sharko F, Petrova K, Mitrofanova I. Whole Genome Characterization of Prunus Necrotic Ringspot Virus and Prune Dwarf Virus Infecting Stone Fruits in Russia. Horticulturae. 2023; 9(8):941. https://doi.org/10.3390/horticulturae9080941
Chicago/Turabian StyleChirkov, Sergei, Anna Sheveleva, Svetlana Tsygankova, Natalia Slobodova, Fedor Sharko, Kristina Petrova, and Irina Mitrofanova. 2023. "Whole Genome Characterization of Prunus Necrotic Ringspot Virus and Prune Dwarf Virus Infecting Stone Fruits in Russia" Horticulturae 9, no. 8: 941. https://doi.org/10.3390/horticulturae9080941
APA StyleChirkov, S., Sheveleva, A., Tsygankova, S., Slobodova, N., Sharko, F., Petrova, K., & Mitrofanova, I. (2023). Whole Genome Characterization of Prunus Necrotic Ringspot Virus and Prune Dwarf Virus Infecting Stone Fruits in Russia. Horticulturae, 9(8), 941. https://doi.org/10.3390/horticulturae9080941








