Presenting the Secrets: Exploring Endogenous Defense Mechanisms in Chrysanthemums against Aphids
Abstract
1. Introduction
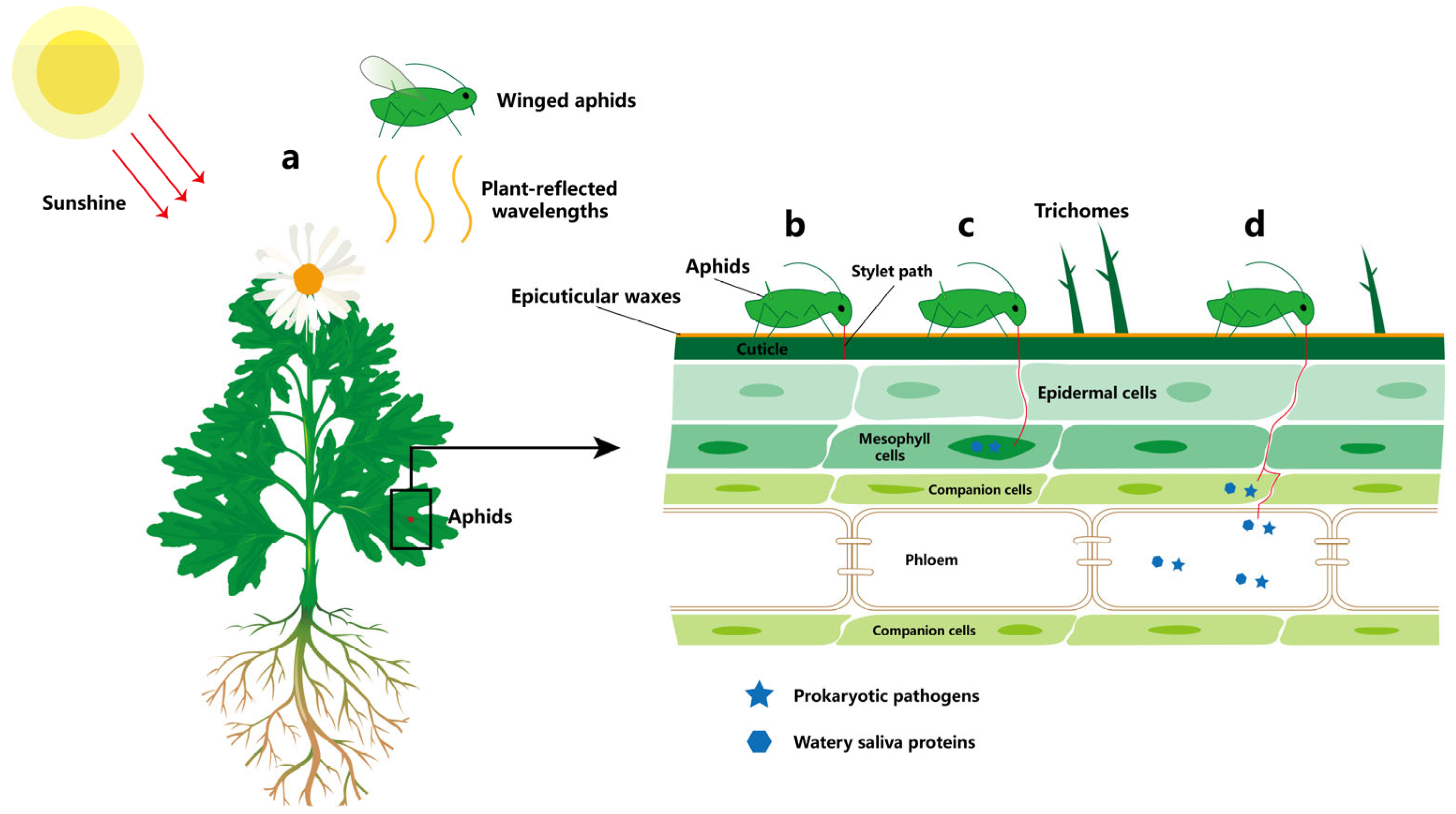
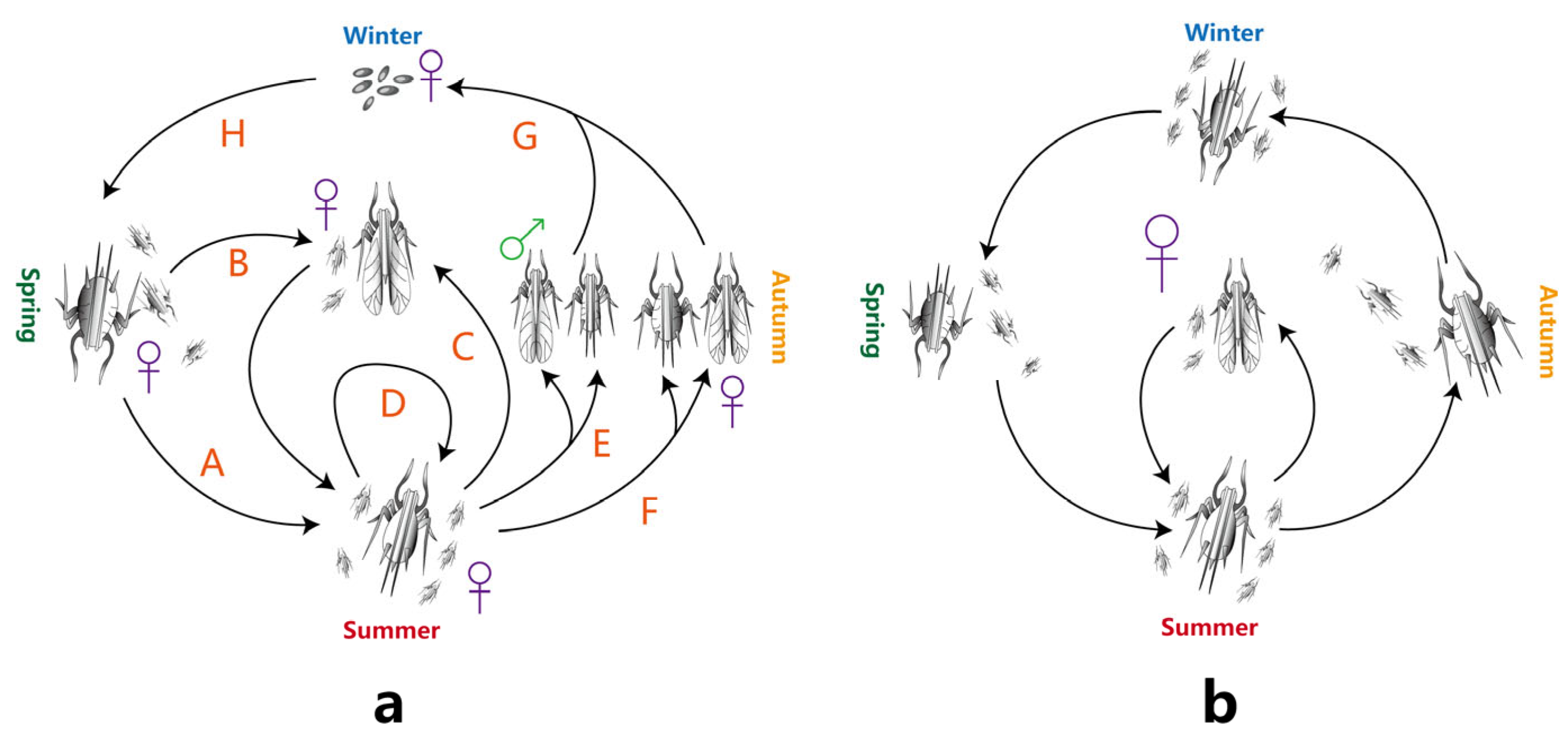
2. Damage Caused by Aphids to Chrysanthemums
2.1. Aphid Feeding
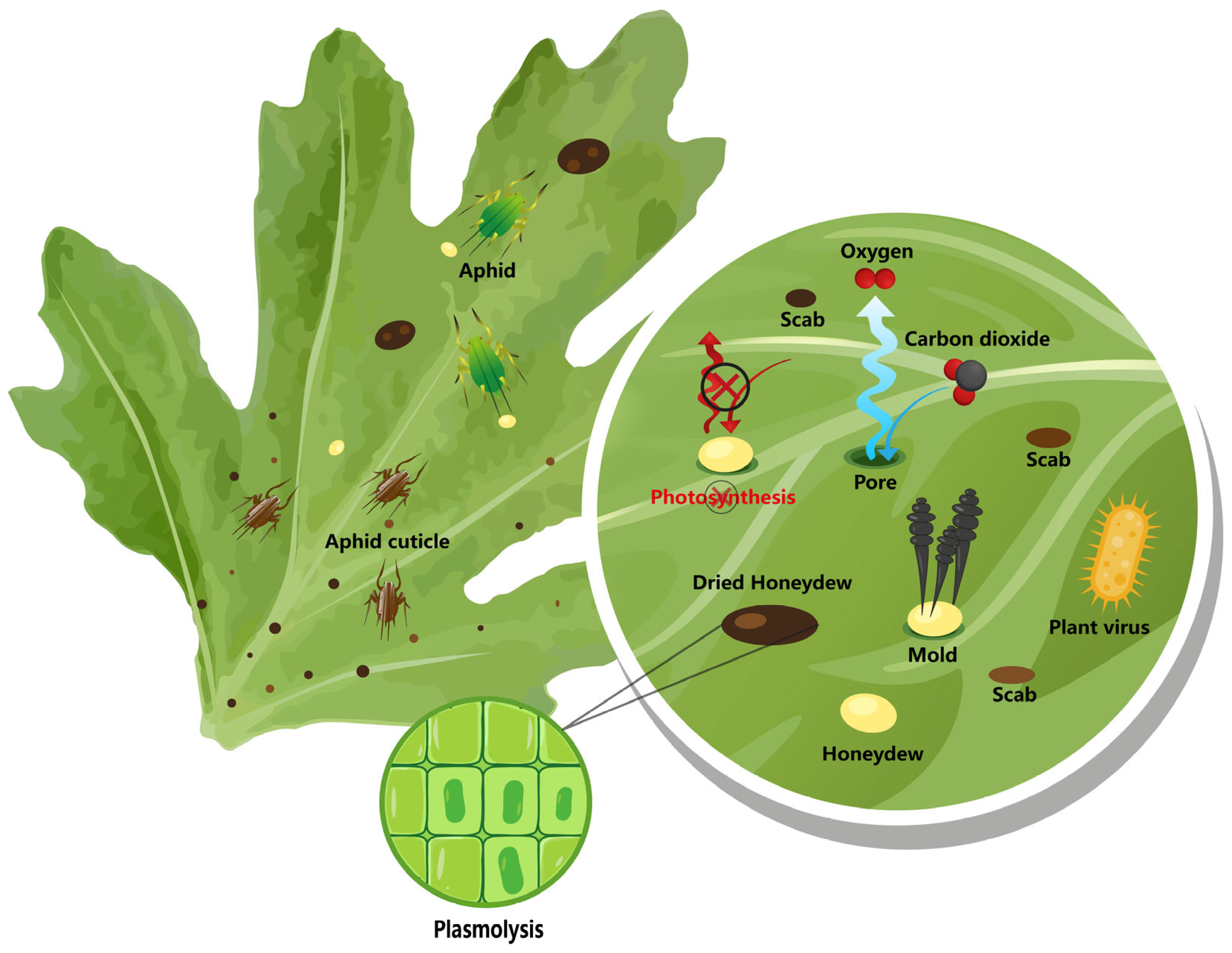
2.2. Aphid Secretions
2.3. Aphid-Borne Viruses
3. Constitutive Defense
3.1. Physical Defense
3.1.1. Leaf Structure
3.1.2. Leaf Color
3.1.3. Leaf Surface Barriers
3.2. Chemical Defense
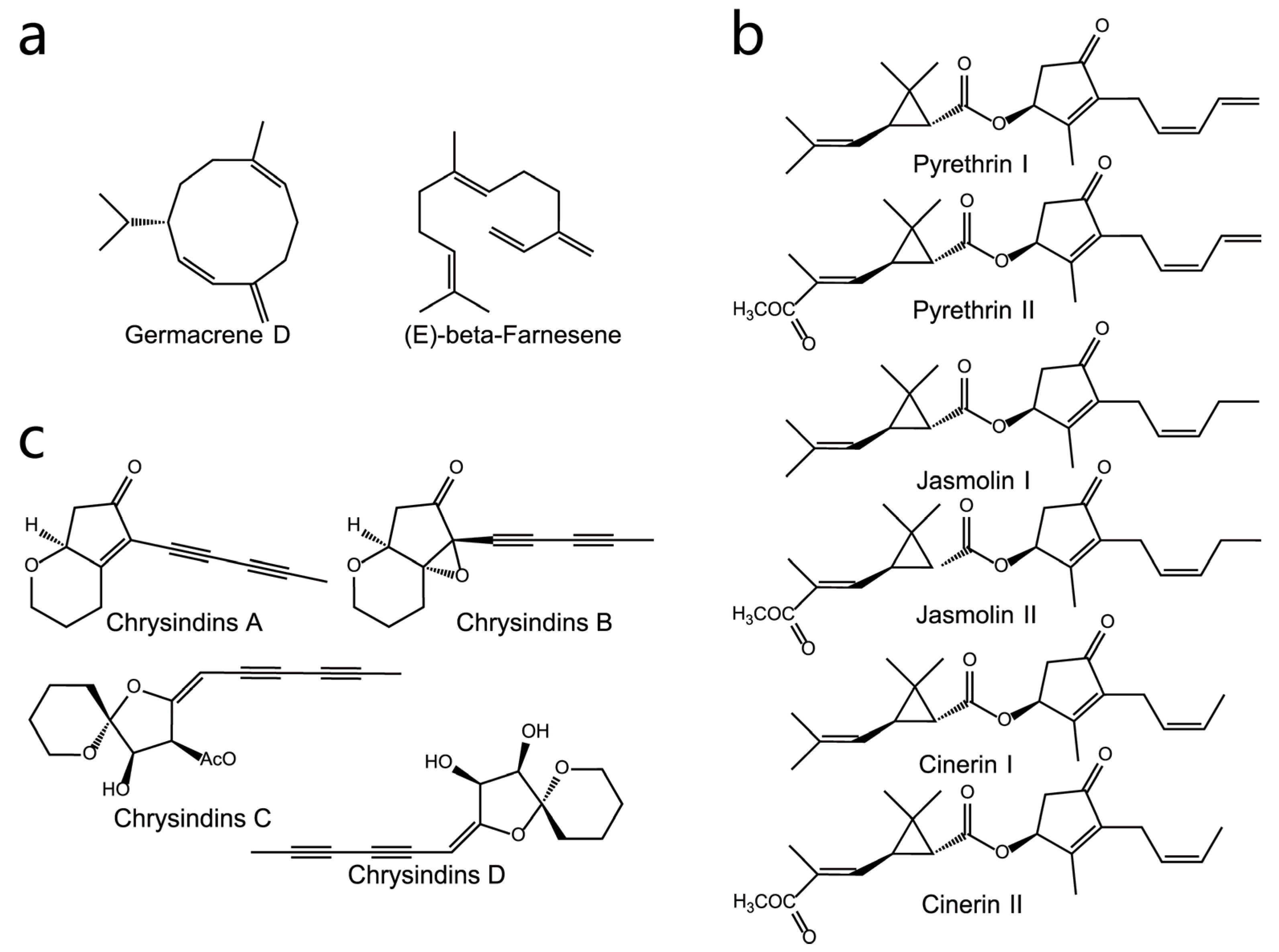
3.2.1. Volatile Compounds
3.2.2. Non-Volatile Compounds
4. Induced Defense
4.1. Nutrients and Reactive Oxygen Species
4.2. Defense Proteins
4.3. Cell Wall Modifications
4.4. Plant Hormones
5. Aphid-Resistance Genes
6. Concluding Remarks
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Mekapogu, M.; Kwon, O.-K.; Song, H.-Y.; Jung, J.-A. Towards the Improvement of Ornamental Attributes in Chrysanthemum: Recent Progress in Biotechnological Advances. Int. J. Mol. Sci. 2022, 23, 12284. [Google Scholar] [CrossRef] [PubMed]
- Teixeira da Silva, J.A.; Shinoyama, H.; Aida, R.; Matsushita, Y.; Raj, S.K.; Chen, F. Chrysanthemum Biotechnology: Quo Vadis? Crit. Rev. Plant Sci. 2013, 32, 21–52. [Google Scholar] [CrossRef]
- Nalam, V.; Louis, J.; Shah, J. Plant defense against aphids, the pest extraordinaire. Plant Sci. 2019, 279, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Blackman, R.; Eastop, V. Taxonomic Issues; CABI International: Wallingford, UK, 2017; pp. 1–36. [Google Scholar]
- Vehrs, S.L.C.; Walker, G.P.; Parrella, M.P. Comparison of Population Growth Rate and Within-Plant Distribution between Aphis gossypii and Myzus persicae (Homoptera: Aphididae) Reared on Potted Chrysanthemums. J. Econ. Entomol. 1992, 85, 799–807. [Google Scholar] [CrossRef]
- Zhong, J.; Wang, Y.; Lu, Y.; Ma, X.; Zhang, Q.; Wang, X.; Zhang, Q.; Sun, M. Identification and Expression Analysis of Chemosensory Genes in the Antennal Transcriptome of Chrysanthemum Aphid Macrosiphoniella sanborni. Insects 2022, 13, 597. [Google Scholar] [CrossRef] [PubMed]
- Storer, J.R.; Emden, H.F. Antibiosis and antixenosis of chrysanthemum cultivars to the aphid Aphis gossypii. Entomol. Exp. Appl. 1995, 77, 307–314. [Google Scholar] [CrossRef]
- Guldemond, J.A.; Tigges, W.T.; De Vrijer, P.W.F. Host Races of Aphis gossypii (Homoptera: Aphididae) on Cucumber and Chrysanthemum. Environ. Entomol. 1994, 23, 1235–1240. [Google Scholar] [CrossRef]
- Simon, J.C.; Stoeckel, S.; Tagu, D. Evolutionary and functional insights into reproductive strategies of aphids. C. R. Biol. 2010, 333, 488–496. [Google Scholar] [CrossRef]
- Thompson, G.A.; Goggin, F.L. Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J. Exp. Bot. 2006, 57, 755–766. [Google Scholar] [CrossRef]
- Goggin, F.L. Plant-aphid interactions: Molecular and ecological perspectives. Curr. Opin. Plant Biol. 2007, 10, 399–408. [Google Scholar] [CrossRef]
- Walling, L.L. Avoiding Effective Defenses: Strategies Employed by Phloem-Feeding Insects. Plant Physiol. 2008, 146, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Powell, G.; Tosh, C.R.; Hardie, J. Host plant selection by aphids: Behavioral, evolutionary, and applied perspectives. Annu. Rev. Entomol. 2006, 51, 309–330. [Google Scholar] [CrossRef] [PubMed]
- Hewer, A.; Becker, A.; van Bel, A.J. An aphid’s Odyssey–The cortical quest for the vascular bundle. J. Exp. Biol. 2011, 214, 3868–3879. [Google Scholar] [CrossRef] [PubMed]
- Döring, T.F.; Kirchner, S. A model for colour preference behaviour of spring migrant aphids. Philos. Trans. R. Soc. B Biol. Sci. 2022, 377, 20210283. [Google Scholar] [CrossRef] [PubMed]
- Hardie, J. Spectral specificity for targeted flight in the black bean aphid, Aphis fabae. J. Insect Physiol. 1989, 35, 619–626. [Google Scholar] [CrossRef]
- Zhou, J.J.; He, X.L.; Liu, R.; Field, L.M.; Davies, T.G.E.; Williamson, M.S. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010, 8, e1000313. [Google Scholar]
- Shu-xian, W. The Summary of Aphids’ Life Cycle. Heilongjiang Agric. Sci. 2009, 2, 74–75. [Google Scholar]
- Kempel, A.; Schädler, M.; Chrobock, T.; Fischer, M.; van Kleunen, M. Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc. Natl. Acad. Sci. USA 2011, 108, 5685–5689. [Google Scholar] [CrossRef]
- Agrawal, A.A. Induced Responses to Herbivory in Wild Radish: Effects on Several Herbivores and Plant Fitness. Ecology 1999, 80, 1713–1723. [Google Scholar] [CrossRef]
- War, A.R.; Paulraj, M.G.; Ahmad, T.; Buhroo, A.A.; Hussain, B.; Ignacimuthu, S.; Sharma, H.C. Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 2012, 7, 1306–1320. [Google Scholar] [CrossRef]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Bak, S. Plant defense against insect herbivores. Int. J. Mol. Sci. 2013, 14, 10242–10297. [Google Scholar] [CrossRef]
- Vincent, C.; Weintraub, P.; Hallman, G. Chapter 200—Physical Control of Insect Pests. In Encyclopedia of Insects (Second Edition); Resh, V.H., Cardé, R.T., Eds.; Academic Press: San Diego, CA, USA, 2009; pp. 794–798. [Google Scholar]
- Tang, F.H.M.; Lenzen, M.; McBratney, A.; Maggi, F. Risk of pesticide pollution at the global scale. Nat. Geosci. 2021, 14, 206–210. [Google Scholar] [CrossRef]
- Cherqui, A.; Tjallingii, W.F. Salivary proteins of aphids, a pilot study on identification, separation and immunolocalisation. J. Insect Physiol. 2000, 46, 1177–1186. [Google Scholar] [CrossRef] [PubMed]
- Harmel, N.; Létocart, E.; Cherqui, A.; Giordanengo, P.; Mazzucchelli, G.; Guillonneau, F.; De Pauw, E.; Haubruge, E.; Francis, F. Identification of aphid salivary proteins: A proteomic investigation of Myzus persicae. Insect Mol. Biol. 2008, 17, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Urbanska, A.; Tjallingii, W.F.; Dixon, A.F.G.; Leszczynski, B. Phenol oxidising enzymes in the grain aphid’s saliva. Entomol. Exp. Appl. 1998, 86, 197–203. [Google Scholar] [CrossRef]
- Furch, A.C.; van Bel, A.J.; Will, T. Aphid salivary proteases are capable of degrading sieve-tube proteins. J. Exp. Bot. 2015, 66, 533–539. [Google Scholar] [CrossRef]
- Will, T.; Tjallingii, W.F.; Thönnessen, A.; van Bel, A.J. Molecular sabotage of plant defense by aphid saliva. Proc. Natl. Acad. Sci. USA 2007, 104, 10536–10541. [Google Scholar] [CrossRef] [PubMed]
- Giordanengo, P.; Brunissen, L.; Rusterucci, C.; Vincent, C.; van Bel, A.; Dinant, S.; Girousse, C.; Faucher, M.; Bonnemain, J.L. Compatible plant-aphid interactions: How aphids manipulate plant responses. C. R. Biol. 2010, 333, 516–523. [Google Scholar] [CrossRef]
- Chirkov, S.N.; Sheveleva, A.; Snezhkina, A.; Kudryavtseva, A.; Krasnov, G.; Zakubanskiy, A.; Mitrofanova, I. Highly divergent isolates of chrysanthemum virus B and chrysanthemum virus R infecting chrysanthemum in Russia. PeerJ 2022, 10, e12607. [Google Scholar] [CrossRef]
- Colvin, J.; Omongo, C.A.; Govindappa, M.R.; Stevenson, P.C.; Maruthi, M.N.; Gibson, G.; Seal, S.E.; Muniyappa, V. Host-Plant Viral Infection Effects on Arthropod-Vector Population Growth, Development and Behaviour: Management and Epidemiological Implications. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2006; Volume 67, pp. 419–452. [Google Scholar]
- Zhang, X.Y.; Sun, X.Z.; Zhang, S.; Yang, J.H.; Liu, F.F.; Fan, J. Comprehensive transcriptome analysis of grafting onto Artemisia scoparia W. to affect the aphid resistance of chrysanthemum (Chrysanthemum morifolium T.). BMC Genomics 2019, 20, 776. [Google Scholar] [CrossRef]
- Gao, L.L.; Kamphuis, L.G.; Kakar, K.; Edwards, O.R.; Udvardi, M.K.; Singh, K.B. Identification of potential early regulators of aphid resistance in Medicago truncatula via transcription factor expression profiling. New Phytol. 2010, 186, 980–994. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, N.; Chamila Darshanee, H.L.; Fu, W.Y.; Hu, X.S.; Fan, Y.; Liu, T.X. Resistance of Seven Cabbage Cultivars to Green Peach Aphid (Hemiptera: Aphididae). J. Econ. Entomol. 2018, 111, 909–916. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.Y.; Kong, X.Y.; Pan, H. A study on the structure of micromarphology and its resistant to aphis of chrysanthemum. J. South China Agric. Univ. 1992, S1, 66–68. [Google Scholar]
- He, J.; Chen, F.; Chen, S.; Lv, G.; Deng, Y.; Fang, W.; Liu, Z.; Guan, Z.; He, C. Chrysanthemum leaf epidermal surface morphology and antioxidant and defense enzyme activity in response to aphid infestation. J. Plant Physiol. 2011, 168, 687–693. [Google Scholar] [CrossRef]
- Sumitomo, K.; Nishijima, T.; Onozaki, T.; Shibata, M. Density, Length and Development of Non-Glandular Trichome on the Leaves of Wild Chrysanthemums and Chrysanthemum Cultivars. Hort. Res. 2006, 5, 351–356. [Google Scholar] [CrossRef][Green Version]
- Ramirez, A.M.; Stoopen, G.; Menzel, T.R.; Gols, R.; Bouwmeester, H.J.; Dicke, M.; Jongsma, M.A. Bidirectional Secretions from Glandular Trichomes of Pyrethrum Enable Immunization of Seedlings. Plant Cell 2012, 24, 4252–4265. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, S.; Lu, A.; Chen, F.; Tang, F.; Guan, Z.; Teng, N. Production and characterisation of the intergeneric hybrids between Dendranthema morifolium and Artemisia vulgaris exhibiting enhanced resistance to chrysanthemum aphid (Macrosiphoniellasanbourni). Planta 2010, 231, 693–703. [Google Scholar] [CrossRef]
- Bruce, T.J.; Birkett, M.A.; Blande, J.; Hooper, A.M.; Martin, J.L.; Khambay, B.; Prosser, I.; Smart, L.E.; Wadhams, L.J. Response of economically important aphids to components of Hemizygia petiolata essential oil. Pest Manag. Sci. 2005, 61, 1115–1121. [Google Scholar] [CrossRef]
- Li, J.; Hu, H.; Chen, Y.; Xie, J.; Li, J.; Zeng, T.; Wang, M.; Luo, J.; Zheng, R.; Jongsma, M.A.; et al. Tissue specificity of (E)-β-farnesene and germacrene D accumulation in pyrethrum flowers. Phytochemistry 2021, 187, 112768. [Google Scholar] [CrossRef]
- Aquilino, K.M.; Cardinale, B.J.; Ives, A.R. Reciprocal effects of host plant and natural enemy diversity on herbivore suppression: An empirical study of a model tritrophic system. Oikos 2005, 108, 275–282. [Google Scholar] [CrossRef]
- Zhang, Y.M.; Huang, Y.F.; Tao, L.; Huang, J.W. Chemical components of essential oils from Flos chrysanthemi Indici in different areas. Chin. J. Chin. Mater. Medica 2002, 27, 265–267. [Google Scholar]
- Li, J.; Hu, H.; Mao, J.; Yu, L.; Stoopen, G.; Wang, M.; Mumm, R.; de Ruijter, N.C.A.; Dicke, M.; Jongsma, M.A.; et al. Defense of pyrethrum flowers: Repelling herbivores and recruiting carnivores by producing aphid alarm pheromone. N. Phytol. 2019, 223, 1607–1620. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, K. Pyrethrin biosynthesis and its regulation in Chrysanthemum cinerariaefolium. Top. Curr. Chem. 2012, 314, 73–81. [Google Scholar] [PubMed]
- Meng, F.; Wang, Z.; Zang, Z.; Song, X.P.; Sun, J.; Li, W.B. Relationship between Isoflavanones Content and Soybean Resistance to Soybean Aphid. Crops 2011, 3, 11–15. [Google Scholar]
- Fu, P.P.; Xia, Q.; Lin, G.; Chou, M.W. Pyrrolizidine Alkaloids—Genotoxicity, Metabolism Enzymes, Metabolic Activation, and Mechanisms. Drug Metab. Rev. 2004, 36, 1–55. [Google Scholar] [CrossRef] [PubMed]
- Lindigkeit, R.; Biller, A.; Buch, M.; Schiebel, H.M.; Boppré, M.; Hartmann, T. The two facies of pyrrolizidine alkaloids: The role of the tertiary amine and its N-oxide in chemical defense of insects with acquired plant alkaloids. Eur. J. Biochem. 1997, 245, 626–636. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Wang, R.; Shi, Y.P. Chrysindins A–D, polyacetylenes from the flowers of Chrysanthemum indicum. Planta Med. 2011, 77, 1806–1810. [Google Scholar] [CrossRef]
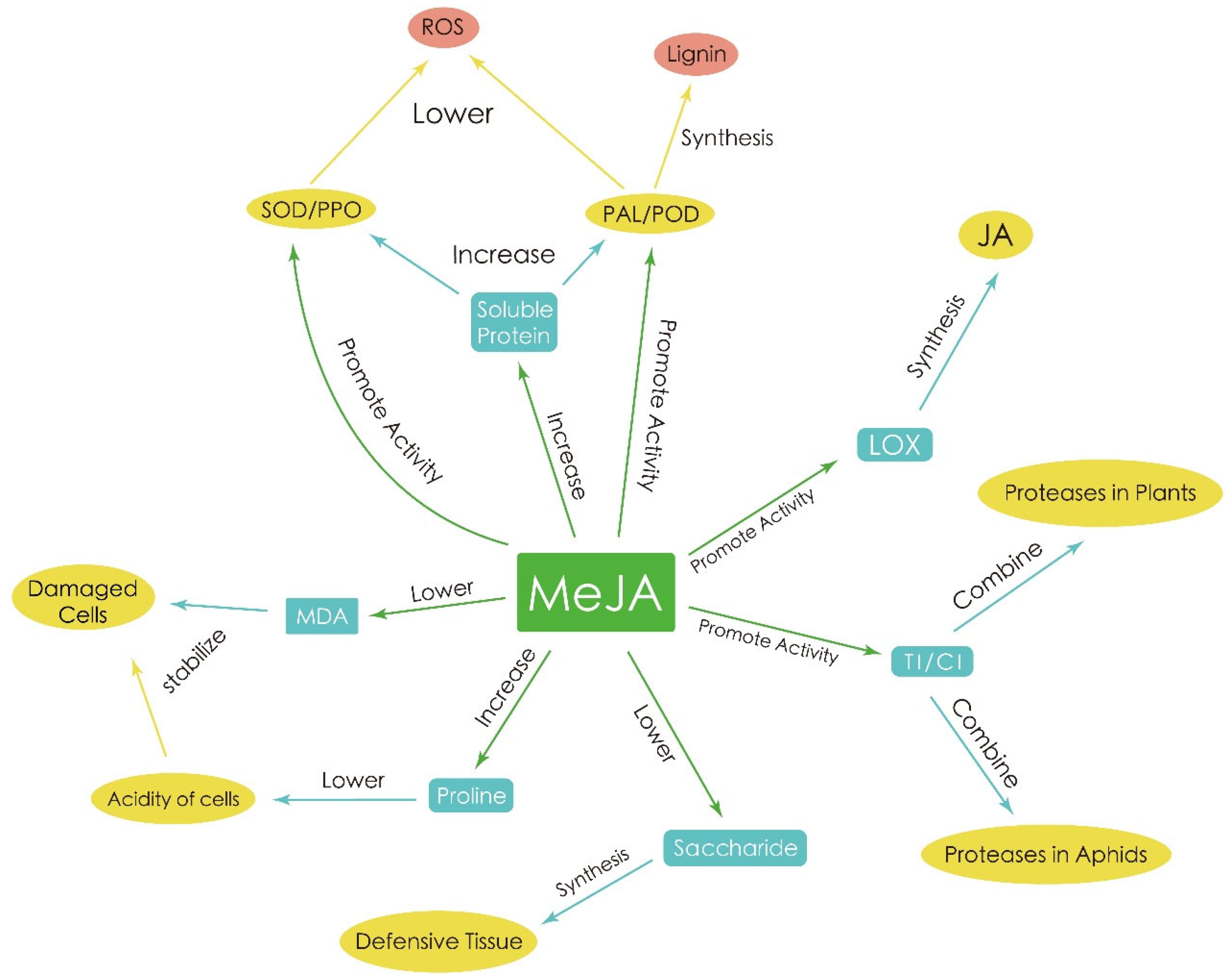
- Fan, J.; Zhang, X.Y.; Sun, X.Z.; Xu, B.Y. Effect of methyl jasmonate on aphid resistance of chrysanthemum. Ying Yong Sheng Tai Xue Bao 2020, 31, 4197–4205. [Google Scholar]
- Castro, A.M.; Clúa, A.; Giménez, D.O.; Tocho, É.; Tacaliti, M.S.; Collado, M.B.; Worland, A.J.; Bottini, R.; Snape, J.W. Genetic Resistance to Greenbug is Expressed with Higher Contents of Proteins and Non-Structural Carbohydrates in Wheat Substitution Lines. In Proceedings of the Wheat Production in Stressed Environments: 7th International Wheat Conference, Mar del Plata, Argentina, 27 November–2 December 2005; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar]
- Halliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Bi, J.L.; Felton, G.W. Foliar oxidative stress and insect herbivory: Primary compounds, secondary metabolites, and reactive oxygen species as components of induced resistance. J. Chem. Ecol. 1995, 21, 1511–1530. [Google Scholar] [CrossRef]
- Orozco-Cardenas, M.; Ryan, C.A. Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 6553–6557. [Google Scholar] [CrossRef]
- Moloi, M.J.; van der Westhuizen, A.J. The reactive oxygen species are involved in resistance responses of wheat to the Russian wheat aphid. J. Plant Physiol. 2006, 163, 1118–1125. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell 2005, 17, 1866–1875. [Google Scholar] [CrossRef] [PubMed]
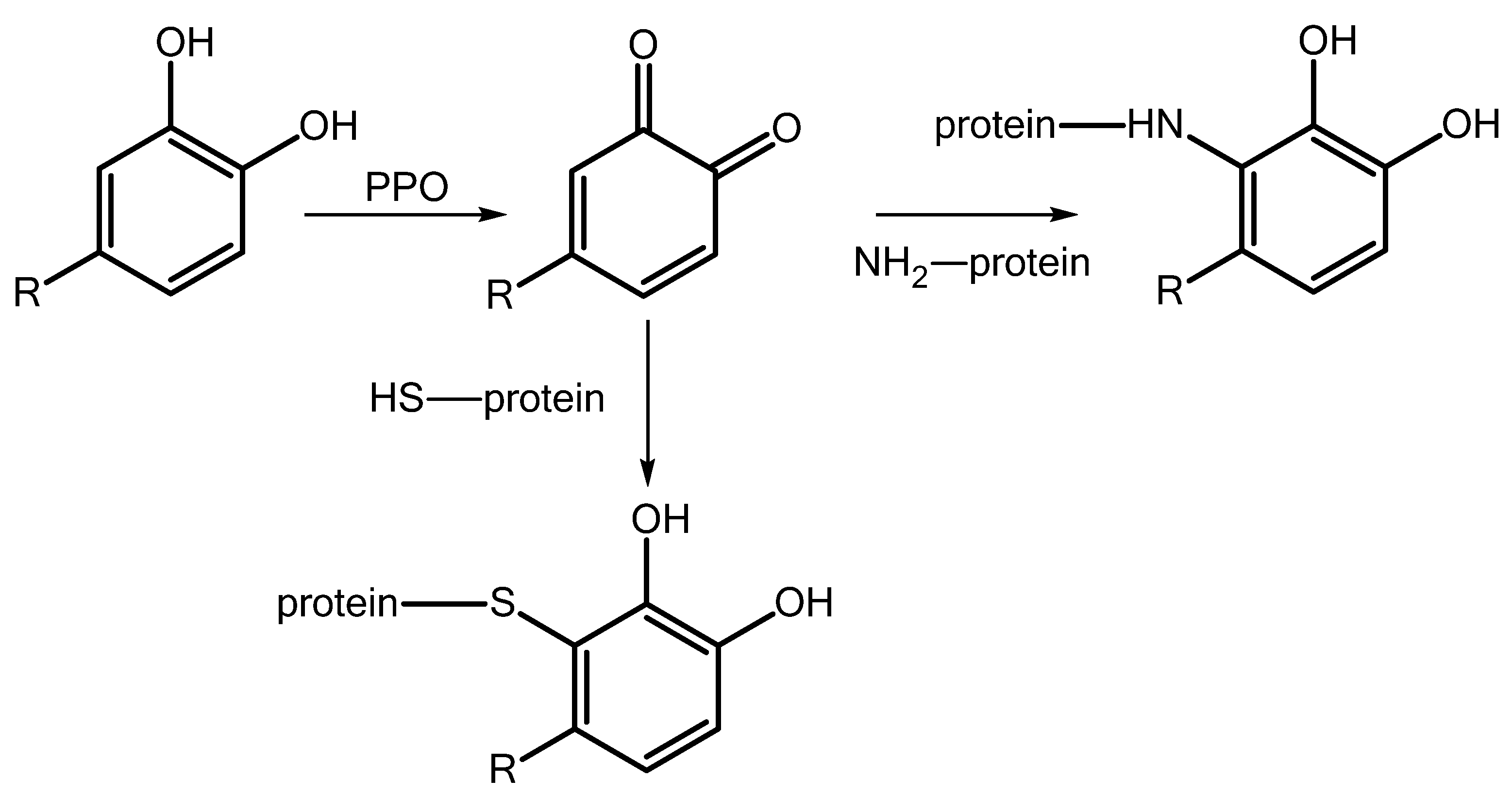
- Vámos-Vigyázó, L. Polyphenol oxidase and peroxidase in fruits and vegetables. Crit. Rev. Food Sci. Nutr. 1981, 15, 49–127. [Google Scholar] [CrossRef] [PubMed]
- Felton, G.W.; Donato, K.K.; Broadway, R.M.; Duffey, S.S. Impact of oxidized plant phenolics on the nutritional quality of dietar protein to a noctuid herbivore, Spodoptera exigua. J. Insect Physiol. 1992, 38, 277–285. [Google Scholar] [CrossRef]
- Mishra, B.B.; Gautam, S.; Sharma, A. Purification and characterisation of polyphenol oxidase (PPO) from eggplant (Solanum melongena). Food Chem. 2012, 134, 1855–1861. [Google Scholar] [CrossRef]
- Kowalski, S.P.; Eannetta, N.T.; Hirzel, A.T.; Steffens, J.C. Purification and Characterization of Polyphenol Oxidase from Glandular Trichomes of Solanum berthaultii. Plant Physiol. 1992, 100, 677–684. [Google Scholar] [CrossRef] [PubMed]
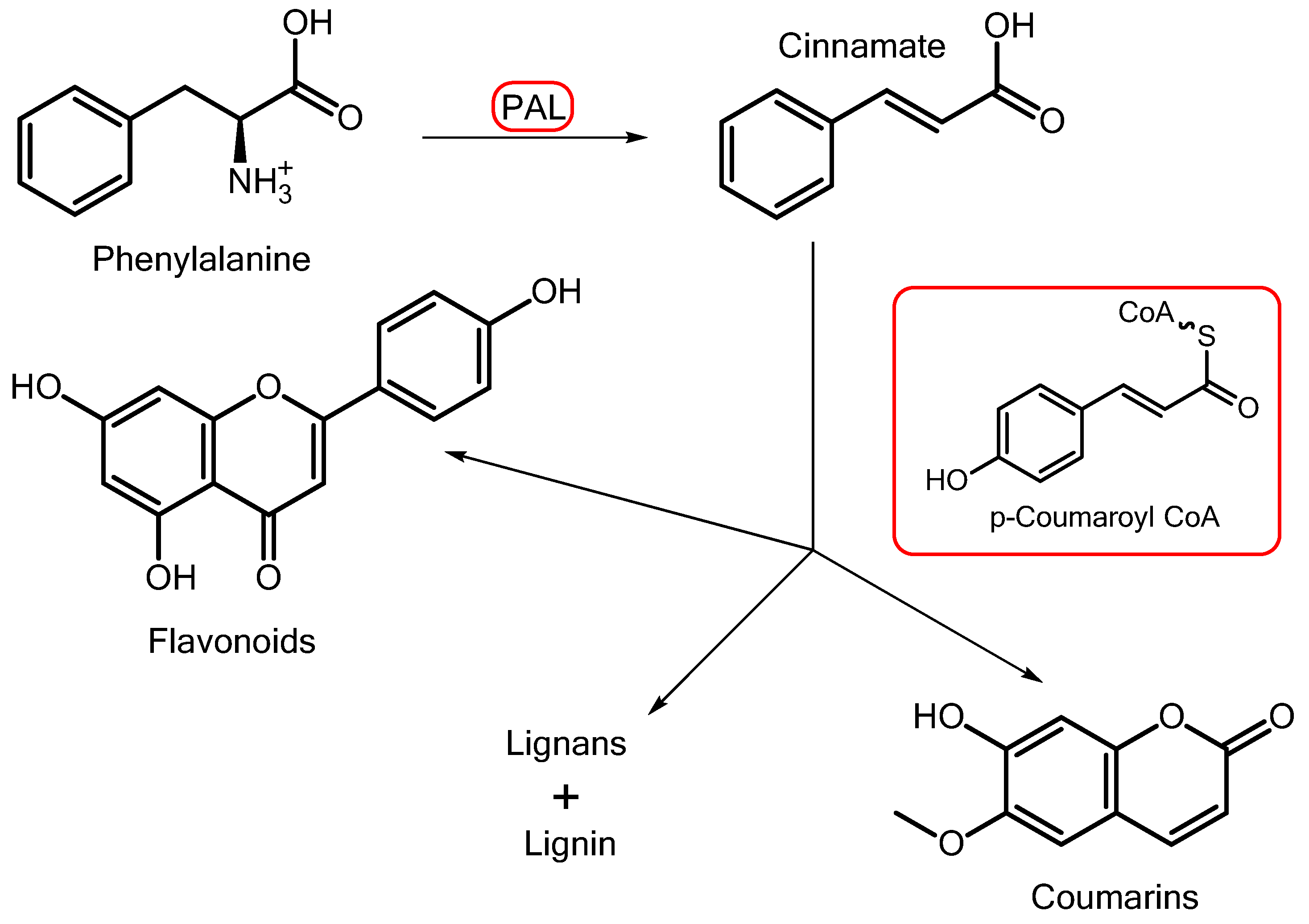
- Vogt, T. Phenylpropanoid Biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef]
- Dixon, R.A.; Achnine, L.; Kota, P.; Liu, C.J.; Reddy, M.S.; Wang, L. The phenylpropanoid pathway and plant defence–A genomics perspective. Mol. Plant Pathol. 2002, 3, 371–390. [Google Scholar] [CrossRef]
- Espelie, K.E.; Franceschi, V.R.; Kolattukudy, P.E. Immunocytochemical localization and time course of appearance of an anionic peroxidase associated with suberization in wound-healing potato tuber tissue. Plant Physiol. 1986, 81, 487–492. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Aispuro, E.; Vargas-Arispuro, I.; Martínez-Téllez, M. Plant Cell Wall Polymers: Function, Structure and Biological Activity of Their Derivatives. Polymerization 2012, 4, 63–86. [Google Scholar]
- Caffall, K.H.; Mohnen, D. The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Ridley, B.L.; O’Neill, M.A.; Mohnen, D. Pectins: Structure, biosynthesis, and oligogalacturonide-related signaling. Phytochemistry 2001, 57, 929–967. [Google Scholar] [CrossRef]
- Silva-Sanzana, C.; Estevez, J.M.; Blanco-Herrera, F. Influence of cell wall polymers and their modifying enzymes during plant-aphid interactions. J. Exp. Bot. 2020, 71, 3854–3864. [Google Scholar] [CrossRef] [PubMed]
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451. [Google Scholar] [CrossRef] [PubMed]
- Lionetti, V.; Fabri, E.; De Caroli, M.; Hansen, A.R.; Willats, W.G.; Piro, G.; Bellincampi, D. Three Pectin Methylesterase Inhibitors Protect Cell Wall Integrity for Arabidopsis Immunity to Botrytis. Plant Physiol. 2017, 173, 1844–1863. [Google Scholar] [CrossRef] [PubMed]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef] [PubMed]
- Kohorn, B.D. Cell wall-associated kinases and pectin perception. J. Exp. Bot. 2016, 67, 489–494. [Google Scholar] [CrossRef]
- Dorokhov, Y.L.; Komarova, T.V.; Petrunia, I.V.; Frolova, O.Y.; Pozdyshev, D.V.; Gleba, Y.Y. Airborne signals from a wounded leaf facilitate viral spreading and induce antibacterial resistance in neighboring plants. PLoS Pathog. 2012, 8, e1002640. [Google Scholar] [CrossRef]
- Von Dahl, C.C.; Hävecker, M.; Schlögl, R.; Baldwin, I.T. Caterpillar-elicited methanol emission: A new signal in plant-herbivore interactions? Plant J. 2006, 46, 948–960. [Google Scholar] [CrossRef]
- Hann, C.T.; Bequette, C.J.; Dombrowski, J.E.; Stratmann, J.W. Methanol and ethanol modulate responses to danger- and microbe-associated molecular patterns. Front. Plant Sci 2014, 5, 550. [Google Scholar] [CrossRef]
- Silva-Sanzana, C.; Celiz-Balboa, J.; Garzo, E.; Marcus, S.E.; Parra-Rojas, J.P.; Rojas, B.; Olmedo, P.; Rubilar, M.A.; Rios, I.; Chorbadjian, R.A.; et al. Pectin Methylesterases Modulate Plant Homogalacturonan Status in Defenses against the Aphid Myzus persicae. Plant Cell 2019, 31, 1913–1929. [Google Scholar] [CrossRef] [PubMed]
- An, S.H.; Sohn, K.H.; Choi, H.W.; Hwang, I.S.; Lee, S.C.; Hwang, B.K. Pepper pectin methylesterase inhibitor protein CaPMEI1 is required for antifungal activity, basal disease resistance and abiotic stress tolerance. Planta 2008, 228, 61–78. [Google Scholar] [CrossRef] [PubMed]
- Agüero, C.B.; Uratsu, S.L.; Greve, C.; Powell, A.L.; Labavitch, J.M.; Meredith, C.P.; Dandekar, A.M. Evaluation of tolerance to Pierce’s disease and Botrytis in transgenic plants of Vitis vinifera L. expressing the pear PGIP gene. Mol. Plant Pathol. 2005, 6, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Peñuelas, J.; Filella, I.; Stefanescu, C.; Llusià, J. Caterpillars of Euphydryas aurinia (Lepidoptera: Nymphalidae) feeding on Succisa pratensis leaves induce large foliar emissions of methanol. N. Phytol. 2005, 167, 851–857. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, F.; Zhang, W.; Wang, Y.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Cloning and expression characteristics of CmTPS1 like gene in Chrysanthemum morifolium. J. Nanjing Agric. Univ. 2020, 43, 58–64. [Google Scholar]
- Zhang, Z.; Wang, C.B.; Fang, W.M.; Chen, F.D.; Jiang, J.F.; Guan, Z.Y.; Chen, S.M. ABA spraying enhanced aphids resistance in chrysanthemum and corresponding physiological mechanisms. Zhiwu Shengli Xuebao/Plant Physiol. J. 2014, 50, 1857–1862. [Google Scholar]
- Mishra, A.K.; Baek, K.H. Salicylic Acid Biosynthesis and Metabolism: A Divergent Pathway for Plants and Bacteria. Biomolecules 2021, 11, 705. [Google Scholar] [CrossRef] [PubMed]
- Avdiushko, S.A.; Brown, G.C.; Dahlman, D.L.; Hildebrand, D.F. Methyl Jasmonate Exposure Induces Insect Resistance in Cabbage and Tobacco. Environ. Entomol. 1997, 26, 642–654. [Google Scholar] [CrossRef]
- Liu, Q.; Luo, L.; Zheng, L. Lignins: Biosynthesis and Biological Functions in Plants. Int. J. Mol. Sci. 2018, 19, 335. [Google Scholar] [CrossRef]
- Li, M.; Wang, G. Effect of Drought Stress on Activities of Cell Defense Enzymes and Lipid Peroxidation in Glycyrrhiza uralensis Seedlings. Acta Ecol. Sin. 2002, 22, 503–507. [Google Scholar]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef]
- Bi, M.; Li, X.; Yan, X.; Liu, D.; Gao, G.; Zhu, P.; Mao, H. Chrysanthemum WRKY15-1 promotes resistance to Puccinia horiana Henn. via the salicylic acid signaling pathway. Hortic. Res. 2021, 8, 6. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Gao, T.; Li, P.; Tian, C.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; Chen, S. Chrysanthemum CmWRKY53 negatively regulates the resistance of chrysanthemum to the aphid Macrosiphoniella sanborni. Hortic. Res. 2020, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Carré, I.A.; Kim, J.Y. MYB transcription factors in the Arabidopsis circadian clock. J. Exp. Bot. 2002, 53, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Sheng, L.; Zhang, H.; Du, X.; An, C.; Xia, X.; Chen, F.; Jiang, J.; Chen, S. CmMYB19 Over-Expression Improves Aphid Tolerance in Chrysanthemum by Promoting Lignin Synthesis. Int. J. Mol. Sci. 2017, 18, 619. [Google Scholar] [CrossRef]
- An, C.; Sheng, L.; Du, X.; Wang, Y.; Zhang, Y.; Song, A.; Jiang, J.; Guan, Z.; Fang, W.; Chen, F.; et al. Overexpression of CmMYB15 provides chrysanthemum resistance to aphids by regulating the biosynthesis of lignin. Hortic. Res. 2019, 6, 84. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Li, J.; Delatte, T.; Vervoort, J.; Gao, L.; Verstappen, F.; Xiong, W.; Gan, J.; Jongsma, M.A.; Wang, C. Modification of chrysanthemum odour and taste with chrysanthemol synthase induces strong dual resistance against cotton aphids. Plant Biotechnol. J. 2018, 16, 1434–1445. [Google Scholar] [CrossRef]
- Parry, A.D.; Horgan, R. Carotenoids and abscisic acid (ABA) biosynthesis in higher plants. Physiol. Plant. 1991, 82, 320–326. [Google Scholar] [CrossRef]
- Ren, H.; Gao, Z.; Chen, L.; Wei, K.; Liu, J.; Fan, Y.; Davies, W.J.; Jia, W.; Zhang, J. Dynamic analysis of ABA accumulation in relation to the rate of ABA catabolism in maize tissues under water deficit. J. Exp. Bot. 2007, 58, 211–219. [Google Scholar] [CrossRef]
- Li, F.; Zhang, H.; Zhao, H.; Gao, T.; Song, A.; Jiang, J.; Chen, F.; Chen, S. Chrysanthemum CmHSFA4 gene positively regulates salt stress tolerance in transgenic chrysanthemum. Plant Biotechnol. J. 2018, 16, 1311–1321. [Google Scholar] [CrossRef]
- Niu, L.; Pan, L.; Zeng, W.; Lu, Z.; Cui, G.; Fan, M.; Xu, Q.; Wang, Z.; Li, G. Dynamic transcriptomes of resistant and susceptible peach lines after infestation by green peach aphids (Myzus persicae Sülzer) reveal defence responses controlled by the Rm3 locus. BMC Genom. 2018, 19, 846. [Google Scholar] [CrossRef] [PubMed]
- Feinbaum, R.L.; Ausubel, F.M. Transcriptional regulation of the Arabidopsis thaliana chalcone synthase gene. Mol. Cell. Biol. 1988, 8, 1985–1992. [Google Scholar] [PubMed]
- Theologis, A.; Ecker, J.R.; Palm, C.J.; Federspiel, N.A.; Kaul, S.; White, O.; Alonso, J.; Altafi, H.; Araujo, R.; Bowman, C.L.; et al. Sequence and analysis of chromosome 1 of the plant Arabidopsis thaliana. Nature 2000, 408, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Tabata, S.; Kaneko, T.; Nakamura, Y.; Kotani, H.; Kato, T.; Asamizu, E.; Miyajima, N.; Sasamoto, S.; Kimura, T.; Hosouchi, T.; et al. Sequence and analysis of chromosome 5 of the plant Arabidopsis thaliana. Nature 2000, 408, 823–826. [Google Scholar]
- Cho, A.; Jang, H.; Baek, S.; Kim, M.-J.; Yim, B.; Huh, S.M.; Kwon, S.-H.; Yu, H.-J.; Mun, J.H. An improved Raphanus sativus cv. WK10039 genome localizes centromeres, uncovers variation of DNA methylation and resolves arrangement of the ancestral Brassica genome blocks in radish chromosomes. Theor. Appl. Genet. 2022, 135, 1731–1750. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, J.; Luo, L.; Ye, S.F.; Yang, Y.; Zhang, G.; Wang, X.; Zhang, J. The Cotton GhRac6 Gene Encoding a Plant ROP/RAC Protein Improves the Plant Defense Response to Aphid Feeding. Plant Mol. Biol. Rep. 2018, 36, 888–896. [Google Scholar] [CrossRef]
- Pitrat, M.; Lecoq, H. Inheritance of resistance to cucumber mosaic virus transmission by Aphis gossypii in Cucumis melo. Phytopathology 1980, 70, 958–961. [Google Scholar] [CrossRef]
- Laila, R.; Robin, A.H.; Yang, K.; Park, J.I.; Suh, M.C.; Kim, J.; Nou, I.S. Developmental and Genotypic Variation in Leaf Wax Content and Composition, and in Expression of Wax Biosynthetic Genes in Brassica oleracea var. capitata. Front. Plant Sci. 2016, 7, 1972. [Google Scholar] [CrossRef]
- Cheng, C.-Y.; Krishnakumar, V.; Chan, A.P.; Thibaud-Nissen, F.; Schobel, S.; Town, C.D. Araport11: A complete reannotation of the Arabidopsis thaliana reference genome. Plant J. 2017, 89, 789–804. [Google Scholar] [CrossRef]
- Patwari, P.; Salewski, V.; Gutbrod, K.; Kreszies, T.; Dresen-Scholz, B.; Peisker, H.; Steiner, U.; Meyer, A.J.; Schreiber, L.; Dörmann, P. Surface wax esters contribute to drought tolerance in Arabidopsis. Plant J. 2019, 98, 727–744. [Google Scholar] [CrossRef]
- Takeuchi, M.; Kegasa, T.; Watanabe, A.; Tamura, M.; Tsutsumi, Y. Expression analysis of transporter genes for screening candidate monolignol transporters using Arabidopsis thaliana cell suspensions during tracheary element differentiation. J. Plant Res. 2018, 131, 297–305. [Google Scholar] [CrossRef]
| Gene Abbreviation | Accession Number | Source | Function | Reference | Predictive Protein | % Identity | Alignment Length | E-Value |
|---|---|---|---|---|---|---|---|---|
| Rm | XP_007225245.1 | Prunus persica | beta-xylosidase/alpha-L-arabinofuranosidase 2 | [96] | CHR00089971-RA | 77.242 | 747 | 0.0 |
| CHS | AAA32771.1 | Arabidopsis thaliana | Chalcone synthase | [97] | CHR00028844-RA | 85.751 | 386 | 0.0 |
| CHR00062418-RA | 85.459 | 392 | 0.0 | |||||
| CHR00019175-RA | 84.949 | 392 | 0.0 | |||||
| Cu/ZnSOD | NP_001077494.1 | Arabidopsis thaliana | copper/zinc superoxide dismutase | [98] | CHR00045542-RA | 82.590 | 659 | 0.0 |
| CHR00083274-RA | 80.980 | 694 | 0.0 | |||||
| CHR00058326-RA | 78.242 | 694 | 0.0 | |||||
| CHR00001194-RA | 76.871 | 147 | 5.26 × 10−78 | |||||
| CHR00061843-RA | 76.712 | 146 | 5.72 × 10−78 | |||||
| PRX7 | NP_199033.1 | Arabidopsis thaliana | Peroxidase superfamily protein | [99] | CHR00037301-RA | 73.810 | 294 | 3.63 × 10−166 |
| TCH4 | KAJ4877013.1 | Raphanus sativus | Tyrosine decarboxylase 1 | [100] | CHR00084602-RA | 73.222 | 478 | 0.0 |
| GhRac | NP_001314362.1 | Gossypium hirsutum | ROP/Rac protein | [101] | CHR00004359-RA | 93.333 | 180 | 8.85 × 10−127 |
| Vat | XP_008465871 | Cucumis melo | Heavy metal-associated isoprenylated plant protein 39 | [102] | CHR00066820-RA | 93.056 | 72 | 4.58 × 10−30 |
| CHR00086923-RA | 93.056 | 72 | 4.58 × 10−30 |
| Gene Abbreviation | Accession Number | Source | Recname | Reference | Predictive Protein | % Identity | Alignment Length | E-Value |
|---|---|---|---|---|---|---|---|---|
| LTP2 | Q9S7I3.1 | Arabidopsis thaliana | Non-specific lipid-transfer protein 2 | [104] | CHR00049734-RA CHR00038208-RA | 50.847 48.246 | 118 114 | 5.15 × 10−33 1.17 × 10−30 |
| FAR3 | Q93ZB9.1 | Arabidopsis thaliana | Fatty acyl-CoA reductase 3 | [104] | CHR00057591-RA CHR00092239-RA | 57.407 51.120 | 486 491 | 0.0 2.16 × 10−170 |
| WSD1 | Q93ZR6.1 | Arabidopsis thaliana | Wax ester synthase/ diacylglycerol acyltransferase 1 | [105] | CHR00049351-RA CHR00071533-RA | 39.738 41.304 | 458 368 | 3.81 × 10−119 7.34 × 10−85 |
| ABCG11 | Q8RXN0.1 | Arabidopsis thaliana | ABC transporter G family member 11 | [106] | CHR00028885-RA CHR00053055-RA | 77.874 57.803 | 696 628 | 0.0 0.0 |
| LACS1 | O22898.1 | Arabidopsis thaliana | Long chain acyl-CoA synthetase 1 | [104] | CHR00023806-RA CHR00075383-RA | 62.367 64.791 | 659 622 | 0.0 0.0 |
| KCS1 | Q9MAM3.1 | Arabidopsis thaliana | 3-ketoacyl-CoA synthase 1 | [104] | CHR00017132-RA | 71.734 | 467 | 0.0 |
| CHR00049310-RA | 62.475 | 493 | 0.0 | |||||
| CHR00035436-RA | 62.348 | 494 | 0.0 | |||||
| KCR1 | Q8L9C4.1 | Arabidopsis thaliana | Very-long-chain 3-oxoacyl-CoA reductase 1 | [104] | CHR00038450-RA CHR00022927-RA | 64.762 61.538 | 315 312 | 9.70 × 10−159 1.42 × 10−140 |
| ECR | Q9M2U2.1 | Arabidopsis thaliana | Very-long-chain enoyl-CoA reductase | [104] | CHR00089558-RA CHR00031281-RA | 78.333 66.304 | 300 276 | 0.0 2.50 × 10−130 |
| CER3 | Q8H1Z0.1 | Arabidopsis thaliana | Very-long-chain aldehyde decarbonylase CER3 | [104] | CHR00091218-RA CHR00066836-RA | 66.341 61.280 | 615 625 | 0.0 0.0 |
| MAH1 | Q9FVS9.1 | Arabidopsis thaliana | Alkane hydroxylase MAH1 | [104] | CHR00033702-RA CHR00073663-RA | 45.669 46.220 | 508 463 | 4.13 × 10−167 6.45 × 10−152 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, C.; Xue, W.; Li, Z.; Shi, J.; Yu, G.; Zhang, Y. Presenting the Secrets: Exploring Endogenous Defense Mechanisms in Chrysanthemums against Aphids. Horticulturae 2023, 9, 937. https://doi.org/10.3390/horticulturae9080937
Xia C, Xue W, Li Z, Shi J, Yu G, Zhang Y. Presenting the Secrets: Exploring Endogenous Defense Mechanisms in Chrysanthemums against Aphids. Horticulturae. 2023; 9(8):937. https://doi.org/10.3390/horticulturae9080937
Chicago/Turabian StyleXia, Changchen, Wanjie Xue, Zhuozheng Li, Jiaxu Shi, Guofu Yu, and Yang Zhang. 2023. "Presenting the Secrets: Exploring Endogenous Defense Mechanisms in Chrysanthemums against Aphids" Horticulturae 9, no. 8: 937. https://doi.org/10.3390/horticulturae9080937
APA StyleXia, C., Xue, W., Li, Z., Shi, J., Yu, G., & Zhang, Y. (2023). Presenting the Secrets: Exploring Endogenous Defense Mechanisms in Chrysanthemums against Aphids. Horticulturae, 9(8), 937. https://doi.org/10.3390/horticulturae9080937





