Abstract
Genome editing has emerged as a powerful tool for accelerating crop improvement in horticultural crops by enabling precise modifications to their genetic makeup. This review provides an in-depth exploration of the applications, methodologies, and potential impacts of genome editing in horticulture. The review focuses on three major genome editing tools in horticulture, CRISPR-Cas9, TALENs, and ZFNs. The underlying mechanisms, applications, and potential challenges associated with each tool are discussed in detail. CRISPR-Cas9, being a versatile and widely used system, has the potential to enhance traits such as disease resistance, abiotic stress tolerance, nutritional content, and yield in horticultural crops. TALENs and ZFNs, although less commonly used, offer alternative options for targeted DNA modifications, and have demonstrated success in specific applications. We emphasize the potential benefits of genome editing in horticulture, including improved crop productivity, quality, and nutritional value. However, challenges such as off-target effects, delivery methods, and regulatory frameworks need to be addressed for the full realization of this technology’s potential. This review serves as a valuable resource for researchers, policymakers, and stakeholders, providing insights into the opportunities and complexities associated with harnessing genome editing for enhanced traits in horticultural crops. By navigating these challenges, genome editing can contribute to sustainable advancements in horticulture, benefiting both producers and consumers worldwide.
1. Introduction to Genome Editing in Agriculture
Genome editing is a powerful biotechnological tool with transformative potential for horticultural crops. It involves the precise modification of plant DNA to enhance desired traits, increase crop yield, and confer resistance to pests, diseases, and environmental stresses. Molecular tools like CRISPR-Cas9, TALENs, and ZFNs act as molecular scissors, allowing for the targeted alteration of specific DNA sequences with unprecedented accuracy. This technology allows scientists to introduce or enhance beneficial traits in crops, including disease resistance, improved nutrition, and drought tolerance.
Horticulture is a critical component of global food production and human well-being as it involves the cultivation and management of plants for food, aesthetics, and medicinal purposes [1]. Horticultural crops, such as fruits, vegetables, ornamental plants, and medicinal herbs, not only contribute to the nutritional needs of populations worldwide but also enhance the visual appeal of our surroundings. However, the productivity and quality of horticultural crops face significant constraints due to challenges such as biotic and abiotic stresses, limited genetic variation, and increasing demands for improved traits [2].
Plant breeders have employed various techniques, such as hybridization, selection, and genetic manipulation, to enhance horticultural crops [3]. These approaches have contributed to improved yield, disease resistance, and other desirable traits [4]. However, traditional breeding methods have limitations, including long breeding cycles, limited genetic variation, and complex genetic architectures [5]. Genome editing has emerged as a transformative technology with the potential to revolutionize crop improvement, including horticultural crops [6]. By enabling precise modifications in an organism’s DNA, genome editing offers an efficient method to manipulate specific genes and traits [7]. This technology holds immense promise for expediting crop improvement, overcoming genetic barriers, and addressing specific challenges in horticultural crops.
The CRISPR-Cas9 system is widely used as a genome editing tool, utilizing guide RNA to direct the Cas9 enzyme for precise DNA cleavage and modifications [8]. TALENs and ZFNs are alternative genome editing tools also utilized in horticultural crop research [9]. These tools enable the precise editing of plant genomes by targeting specific genes associated with desired traits. CRISPR-Cas9 has proven effective in improving important traits in various horticultural crops through targeted modifications [10]. By designing specific gRNAs, researchers can direct the Cas9 enzyme to target genes associated with traits of interest, including disease resistance, abiotic stress tolerance, nutritional content, and yield-related characteristics [11]. The precise nature of CRISPR-Cas9 allows for the introduction of beneficial mutations or targeted gene knockouts, simulating natural genetic variations and accelerating the breeding process [12].
TALENs and ZFNs, along with CRISPR-Cas9, have been employed in horticultural crop research as genome editing tools (Figure 1). TALENs and ZFNs utilize engineered DNA-binding proteins that can be customized to target specific genomic sequences [13]. Similar to CRISPR-Cas9, these tools induce targeted DNA cleavage and subsequent modifications at the desired genomic sites. TALENs employ DNA-binding domains derived from transcription activator-like effectors (TALEs), which are naturally occurring proteins in plant pathogenic bacteria [14]. Engineered TALE domains are utilized to bind specific DNA sequences and are fused with a nuclease domain to induce DNA cleavage [15]. In contrast, ZFNs are hybrid proteins that combine engineered zinc finger DNA-binding domains with the FokI nuclease domain derived from the FokI restriction enzyme [16]. The zinc finger domains are designed to recognize targeted DNA sequences, and the FokI domain cleaves the DNA at the desired site [17].
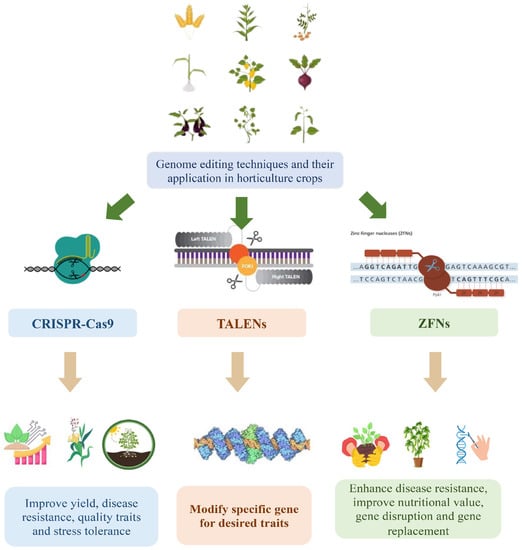
Figure 1.
Genome editing in horticulture crops.
These genome editing tools offer researchers the ability to precisely modify genes associated with desired traits in horticultural crops, facilitating the development of improved varieties [18]. By utilizing these tools, scientists can expedite the enhancement of traits such as disease resistance, abiotic stress tolerance, nutritional content, and yield potential. In this review, we examine the applications, methodologies, and potential impacts of genome editing tools, including CRISPR-Cas9, TALENs, and ZFNs, in horticultural crops (Table 1). We analyze the principles, advantages, and limitations of these tools, emphasizing their contributions to crop improvement.

Table 1.
Genome editing techniques and their applications in horticultural crops.
These genome editing techniques (Table 1) offer precise and efficient methods to modify the genetic makeup of horticultural crops, leading to desired improvements in various traits.
2. CRISPR-Cas9 in Horticultural Crops
The CRISPR-Cas9 system has gained prominence for its user-friendly nature, efficiency, and adaptability in genetic manipulation across organisms, including horticultural crops [21]. Based on bacteria’s defense mechanism against viral infections, CRISPR-Cas9 is a powerful tool [22] that enables precise genome editing in plants. This technology empowers researchers to target specific genes associated with desirable traits and introduce modifications to enhance agricultural characteristics [23]. This level of precision allows for the introduction of advantageous mutations, gene disruption, and the substitution of specific DNA sequences, leading to desired alterations in traits such as disease resistance, abiotic stress tolerance, nutritional composition, and yield-related attributes [24].
Table 2 presents examples of horticultural crops that have undergone genome editing using the CRISPR-Cas9 technique, along with the specific genes targeted and the resulting modified genetic traits. These modifications have led to significant improvements in fruit ripening, disease resistance, flowering time, tuberization, grain quality, and other desirable characteristics in the respective crops.

Table 2.
Horticultural crops subjected to genome editing techniques, modified genetic traits in various plant species.
The limited genetic diversity in horticultural crops poses challenges for developing improved varieties with enhanced traits [50]. CRISPR-Cas9 facilitates the accurate introduction of genetic variations, replicating the genetic diversity observed in wild relatives or closely related species [51]. Targeted modifications of specific genes or regulatory elements unlock untapped genetic potential and expand the available variation for crop enhancement [52], fostering the development of resilient, productive, and nutritionally valuable horticultural crops.
2.1. Structural Components and Mechanisms of CRISPR-Cas9Subsection Applications of CRISPR-Cas9 in Horticultural Crops
The CRISPR-Cas9 system comprises two essential components [53], the Cas9 nuclease and the guide RNA (gRNA). Together, they enable precise genome editing in horticultural crops. The Cas9 nuclease consists of a recognition domain, including two RNA-binding domains and a protospacer adjacent motif (PAM) recognition domain, responsible for binding to the target DNA sequence [54]. The nuclease domain of Cas9 possesses endonuclease activity, cleaving the DNA at the target site [55]. This activity is facilitated by the RuvC-like nuclease domain and the HNH nuclease domain within the Cas9 protein, leading to the generation of double-strand breaks (DSBs) at the targeted locus [56].
The guide RNA (gRNA) is a synthetic RNA molecule that guides the Cas9 nuclease to the target DNA sequence. It consists of two components: the CRISPR RNA (crRNA) derived from the bacterial genome’s CRISPR array, providing the complementary sequence information for the target DNA site, and the trans-activating CRISPR RNA (tracrRNA), which forms a complex with the crRNA, ensuring the assembly and stability of the Cas9-gRNA complex [7,56,57]. Alternatively, the crRNA and tracrRNA can be combined into a single gRNA molecule, simplifying the delivery and assembly of the Cas9-gRNA complex [58].
The gRNA guides the Cas9 nuclease to the target DNA sequence which induces a double-strand break (DSB) at the site complementary to the gRNA sequence, triggering cellular DNA repair mechanisms. The repair can occur through non-homologous end joining (NHEJ), resulting in indels that disrupt genes, or through homology-directed repair (HDR), which accurately repairs the DSB using a template DNA molecule [59,60,61,62].
2.2. Applications of CRISPR-Cas9 in Horticultural Crops
2.2.1. Trait Modification
The CRISPR-Cas9 system enables the precise modification of genes associated with desired traits in horticultural crops, including disease resistance, abiotic stress tolerance, fruit quality, nutritional composition, aroma, color, and post-harvest shelf life. Its application has successfully enhanced disease resistance in crops such as tomato, providing improved defense against pathogens like powdery mildew and bacterial spot [63].
CRISPR-Cas9 technology has been applied to combat powdery mildew, a destructive fungal disease that poses a significant threat to grapevines. Researchers targeted and modified the MLO (Mildew Resistance Locus O) gene in grapevines using CRISPR-Cas9, resulting in the development of powdery-mildew-resistant grape varieties. This study demonstrates the potential of CRISPR-Cas9 as a powerful tool for engineering disease-resistant grapes, highlighting its relevance in horticultural crop improvement [64].
Scientists utilized CRISPR-Cas9 technology to enhance resistance against Tomato Mosaic Virus (ToMV) in tomato crops. By targeting and modifying the eIF4E (eukaryotic translation initiation factor 4E) gene, a crucial player in ToMV infection, they successfully generated tomato plants with robust resistance to ToMV. This study highlights the efficacy of CRISPR-Cas9 as a transformative tool for engineering virus-resistant tomato varieties, showcasing its potential for advancing horticultural crop improvement [65].
CRISPR-Cas9 system was used to enhance resistance against citrus canker, a destructive bacterial disease affecting citrus crops. By precisely targeting and modifying the susceptibility gene CsLOB1, they successfully generated citrus trees with increased resistance to citrus canker infection. This study demonstrates the transformative potential of CRISPR-Cas9 as a tool for developing disease-resistant citrus varieties, contributing to the advancement of horticultural crop protection strategies [66].
2.2.2. Gene Knockout
The CRISPR-Cas9 system enables precise disruption or knockout of target genes by inducing site-specific DNA double-strand breaks (DSBs), initiating error-prone DNA repair mechanisms. This technique has been extensively utilized to study gene function through the generation of loss-of-function mutants. Gene knockouts provide valuable insights into the roles of specific genes in various aspects of horticultural crop development, such as metabolism and responses to biotic and abiotic stresses [67].
The CRISPR-Cas9 system was employed to perform a gene knockout of CHS (chalcone synthase) in petunia plants. CHS is a crucial enzyme involved in flavonoid biosynthesis. The disruption of CHS led to significant alterations in pigment production, offering valuable insights into the functional importance of flavonoids in determining petunia flower coloration. This study contributes to a deeper comprehension of the regulatory mechanisms governing pigmentation in petunias and highlights the potential of CRISPR-Cas9 as a powerful tool for investigating specific gene functions related to horticultural crop traits [68].
In a study, CRISPR/Cas9 technology is employed to specifically target and modify the FaPG1 gene in strawberry plants. The FaPG1 gene encodes the polygalacturonase enzyme, which plays a critical role in pectin degradation and fruit softening. By utilizing Agrobacterium tumefaciens-mediated delivery, the researchers successfully generated FaPG1 knockout strawberry plants with modified gene function. This investigation showcases the significant potential of CRISPR/Cas9 as a potent tool for manipulating genes associated with fruit ripening, offering promising opportunities for enhancing fruit firmness in strawberry crops [69].
The CRISPR-Cas9 system was utilized to perform a gene knockout of the PSY1 gene, which is a vital enzyme involved in the biosynthesis of lycopene in tomatoes. This targeted disruption led to tomatoes with decreased lycopene content, offering valuable insights into the regulatory mechanisms governing carotenoid biosynthesis in tomatoes. The research highlights the efficacy of CRISPR-Cas9 in elucidating the molecular pathways associated with desirable traits in horticultural crops [70].
2.2.3. Gene Activation/Suppression
Through the CRISPR-Cas9 system, gene expression can be meticulously modulated by directing its action to gene promoters or regulatory elements. This capability allows for the selective activation or suppression of specific genes. This approach facilitates desired alterations in crop phenotypes by manipulating key genes associated with diverse biological processes. Researchers have effectively utilized CRISPR-Cas9 to enhance lettuce yield by activating crucial growth-related genes, highlighting the potential of this technique for precise gene regulation in horticultural crops [71].
Nitarska et al. [72] utilized CRISPR-Cas9 to activate endogenous F3’H gene expression in poinsettia (Euphorbia pulcherrima) plants. The introduction of a transcriptional activator through CRISPR-Cas9 led to enhanced F3’H gene expression, resulting in a change in bract color from vivid red to vivid reddish orange.
The study by Huang et al. [73] utilized CRISPR-Cas9 to suppress SlEIN2 gene expression in tomatoes by inducing small deletions in its promoter region. This resulted in reduced expression of SlEIN2 and delayed fruit ripening, highlighting the potential of CRISPR-Cas9 for gene suppression and extending tomato shelf life.
In a study, Huynh [74] employed CRISPR-Cas9 to activate the expression of the ZmDREB2A gene in maize plants. By introducing a transcriptional activator, they enhanced the expression of ZmDREB2A, a gene crucial for drought stress response. Consequently, the edited maize plants exhibited improved drought tolerance, as indicated by higher survival rates and enhanced growth in water-deficit conditions.
2.2.4. Genome Engineering for Crop Domestication
The utilization of CRISPR-Cas9 presents a valuable tool for the process of crop domestication, facilitating the rapid modification of wild or underutilized plant species with the aim of transforming them into potential horticultural crops. This innovative approach enables researchers to introduce precise genetic alterations associated with desirable agronomic traits, including but not limited to diminished bitterness, enhanced nutritional composition, and increased yield potential. Notably, the application of CRISPR/Cas9 technology has been investigated in crops such as watermelon, wherein targeted mutagenesis of the ClBG1 gene through CRISPR/Cas9 resulted in a reduction in seed size and an augmentation of seed germination [75].
In the context of crop domestication, seed dormancy, an innate mechanism that impedes germination under unfavorable conditions, has been subjected to negative selection. To investigate the regulation of seed dormancy in tomatoes, researchers utilized the CRISPR-Cas9 technology specifically to target Lycopen and modify the DELAY OF GERMINATION 1 (DOG1) gene, a key regulator in this process. By disrupting the DOG1 gene, they successfully induced the loss of seed dormancy in tomato plants, resulting in a phenotype resembling non-dormant characteristics observed in cultivated tomato varieties [76].
In the process of rice domestication, the acquisition of non-shattering seeds, a key trait favorably selected through human intervention, has significantly facilitated harvesting practices. To investigate the underlying molecular mechanism, researchers employed the CRISPR-Cas9 system to precisely target and modify the SH4 (SHATTERING4) gene in rice plants. By introducing specific mutations in the SH4 gene, they successfully achieved a reduction in seed shattering and enhanced the ease of harvesting, effectively recapitulating the non-shattering phenotype observed in domesticated rice varieties [77].
The MdERF3 (Ethylene Response Factor 3) gene, which encodes a crucial transcription factor involved in the regulation of fruit ripening and softening in apple plants, was specifically targeted by researchers. By employing CRISPR-Cas9 technology to introduce targeted disruptions in the MdERF3 gene, they achieved successful modulation of fruit ripening, resulting in delayed ripening and extended post-harvest shelf life of apples. This study highlights the potential of CRISPR-Cas9-mediated gene editing as a viable strategy for improving post-harvest characteristics in apple crops, providing insights for future advancements in apple breeding and cultivation [78].
2.2.5. Genome Editing for Quality Improvement
CRISPR-Cas9 offers the potential to enhance the quality attributes of horticultural crops by modifying genes related to flavor, nutritional content, texture, aroma, and color. Researchers have successfully utilized this technology to target genes associated with anthocyanin biosynthesis, resulting in the development of novel colors in flowers and fruits, thereby improving their sensory and nutritional qualities.
In a study, FAD2-1A and FAD2-1B genes were targeted in soybeans, which play a role in the conversion of oleic acid to linoleic acid, impacting oil composition and nutritional quality [79]. Through CRISPR-Cas9-mediated small insertions or deletions in these genes, they successfully enhanced the oleic acid content, thereby reducing levels of saturated fats and improving the nutritional quality of soybean oil [80].
Similarly, in a research study published in Nature Biotechnology [81], targeted vitamin C biosynthesis-related genes, including GDP-L-galactose phosphorylase (GGP) and L-galactono-1,4-lactone dehydrogenase (GLDH), in tomatoes. By employing CRISPR-Cas9, they successfully knocked out these genes, leading to tomato plants with increased vitamin C content and improved nutritional value. Likewise, Liu et al. [82] focused on the MaMADS1 gene, which governs fruit ripening and softening in bananas, to address the prevalent challenges of browning and deterioration in harvested bananas. Through the application of CRISPR-Cas9, they effectively disrupted this gene, resulting in delayed fruit ripening, prolonged shelf life, enhanced quality, and reduced post-harvest losses of bananas.
2.3. Potential Challenges Associated with CRISPR-Cas9 in Horticultural Crops
2.3.1. Off-Target Effects
The potential for off-target effects is a significant concern associated with CRISPR-Cas9, as described by Manghwar et al. [83]. These effects arise when the Cas9 nuclease unintentionally cleaves DNA sequences that bear similarity, but not exact identity, to the intended target site. Such off-target effects can result in unintended genetic modifications, potentially causing unpredictable consequences for the crop’s phenotype and genomic stability [84]. Current endeavors focus on improving gRNA design and enhancing the specificity of Cas9 to minimize off-target effects.
The occurrence of off-target effects during CRISPR-Cas9 gene editing can be influenced by multiple factors, as discussed by Manghwar et al. [83]. These factors include the similarity between the target site and off-target sites, the length and structure of the gRNA, the efficiency of the Cas9 enzyme, and the delivery method employed. To minimize off-target effects, researchers employ various strategies, including the careful selection of target sites with minimal genomic similarities, the design of highly specific gRNAs, optimization of Cas9 enzyme activity, and the utilization of advanced bioinformatics tools for predicting potential off-target sites [85].
Various methods, including whole-genome sequencing, targeted deep sequencing, and computational analysis, are utilized to detect and evaluate off-target effects of CRISPR-Cas9 gene editing [86]. These approaches aid researchers in assessing the specificity of CRISPR-Cas9 editing and identifying potential off-target modifications. Significantly, progress has been achieved in enhancing the specificity of CRISPR-Cas9 over the years. Development of Cas9 variants with improved fidelity, such as high-fidelity Cas9 (HiFi Cas9) and enhanced-specificity Cas9 (eSpCas9), has enabled the reduction of off-target effects while maintaining editing efficiency [87].
2.3.2. Delivery and Transformation Efficiency
The efficient delivery of CRISPR-Cas9 components into plant cells is essential for achieving successful genome editing. However, the transformation process for horticultural crops can be particularly challenging, especially in recalcitrant species or those with complex genomes [88]. Ongoing research focuses on improving delivery methods and enhancing transformation efficiency to facilitate broader applications of CRISPR-Cas9 across diverse horticultural crops [11].
Agrobacterium tumefaciens serves as a widely employed method for delivering CRISPR-Cas9 components into plant cells [89]. This approach involves introducing the CRISPR-Cas9 system into Agrobacterium, which subsequently infects plant tissues, facilitating the transfer of genetic material and enabling the delivery of CRISPR-Cas9 into the plant genome [90]. The efficiency of CRISPR-Cas9 delivery and transformation is influenced by several factors, including the choice of delivery method, plant species, tissue type, regeneration protocols, and the specific components of the CRISPR-Cas9 system. Optimization of these factors, along with the appropriate selection and design of target sequences, can enhance the transformation efficiency in horticultural crops [91].
Efficient transformation in horticultural crop species is influenced by species-specific requirements, including tissue culture protocols, regeneration capacity, and susceptibility to transformation methods, necessitating customized approaches for each species [92]. Furthermore, within a particular crop species, variations in transformation efficiency can be observed among different genotypes or cultivars [93]. While some genotypes exhibit higher amenability to transformation, others may present challenges, requiring further optimization and the utilization of tailored genetic transformation strategies [94].
Each delivery method for CRISPR-Cas9 exhibits specific limitations [95]. For example, Agrobacterium-mediated transformation may be ineffective in certain crops or tissue types, whereas particle bombardment can lead to random DNA integration and low transformation efficiency [96]. Certain horticultural crop genotypes inherently pose challenges for transformation due to their low regeneration capacities or high levels of tissue browning or necrosis [97].
2.3.3. Off-Target Effects in Non-Coding Regions
Although protein-coding genes are the primary targets in many studies, non-coding regions of the genome are essential for gene regulation and plant development [98]. Off-target effects occurring in these regions have the potential to influence gene expression and regulatory networks, leading to unintended changes in plant physiology and development [99]. Therefore, conducting analysis and gaining a thorough understanding of potential off-target effects in non-coding regions is crucial to mitigate unintended alterations in gene regulation [100].
The identification of potential off-target sites in non-coding regions presents challenges due to the larger number of possible target sites compared to coding regions [101]. While bioinformatics tools are commonly used for predicting off-target sites, their accuracy may be lower for non-coding regions [102]. The presence of repetitive sequences and structural variations in the genome further complicates off-target prediction [103]. Unlike modifications in coding regions that directly impact gene function, the functional consequences of alterations in non-coding regions are often less evident [104]. Non-coding regions encompass regulatory elements, enhancers, promoters, and other essential regulatory sequences [105].
Non-coding regions play crucial roles in gene expression regulation and coordination of complex gene networks [106]. Modifying these regions can disrupt regulatory networks, impacting multiple genes and pathways beyond the intended target [107]. Understanding the full extent of these cascading effects is challenging and necessitates analyzing gene expression profiles and regulatory interactions. Off-target effects in non-coding regions may not always result in observable phenotypic changes, but they can contribute to phenotypic variability within edited plant populations [108]. Unintended consequences may manifest as altered growth patterns, changes in secondary metabolite profiles, or variations in stress responses [109]. Characterizing and comprehending these unintended effects can be intricate and requires extensive phenotypic analysis.
Various strategies have been utilized to mitigate off-target effects, such as selecting target sites with minimal similarity to non-coding regions, employing advanced bioinformatics tools for off-target prediction, and optimizing guide RNA design to enhance specificity [102]. Nevertheless, complete elimination of off-target effects remains challenging, necessitating ongoing enhancements in CRISPR-Cas9 technologies to minimize their occurrence.
2.3.4. Inheritance and Segregation of CRISPR-Edited Traits
Achieving the stable inheritance of CRISPR-edited traits through sexual reproduction in horticultural crops poses challenges [110]. Ensuring the presence of edited traits in germ cells and their reliable transmission to subsequent generations is essential [111]. To enhance the inheritance and segregation of CRISPR-edited traits, strategies such as screening and selection of edited lines, as well as the investigation of gene drive systems, are being explored [110].
Attaining extensive and efficient editing of all target sites in every plant cell poses challenges [112]. In certain cells, successful editing may not occur, leading to a mixture of edited and unedited cells within a single plant. This variation can result in diverse trait expression and inheritance patterns [113].
The CRISPR-Cas9 system utilizes DNA double-strand breaks (DSBs) to achieve targeted modifications [114]. Repair mechanisms are involved in mending the breaks, but errors can occur, leading to chromosomal rearrangements or translocations [115]. These genomic rearrangements can impact the inheritance patterns of edited traits and have unintended consequences for the stability and productivity of horticultural crops [116]. The genetic background of horticultural crops plays a role in the expression and inheritance of edited traits [117]. The presence of other alleles or genetic variations in the plant genome can modify the phenotypic outcomes and inheritance patterns of edited traits [118]. Understanding and considering these background effects are crucial for the accurate prediction of trait inheritance.
Researchers utilize backcrossing, selfing, and extensive genotyping to stabilize and validate the inheritance patterns of edited traits [119]. Advancements in genomics, molecular breeding, and genetic analysis methods aid in addressing these challenges and promoting the efficient inheritance and dissemination of desired traits in horticultural crops edited using CRISPR-Cas9.
2.4. Advancements in CRISPR-Cas9 to Overcome Initial Limitations
The initial implementation of CRISPR-Cas9 in horticultural crop genome editing encountered challenges related to off-target effects, resulting in unintended genetic modifications [120]. To address this limitation, significant progress has been made in developing improved Cas9 variants [121]. High-fidelity Cas9 and Cas9 nickase have been engineered to exhibit reduced off-target effects while maintaining efficient on-target editing [121]. Moreover, refined computational tools have been employed to enhance off-target prediction, enabling the better identification of potential off-target sites and guiding target selection for safer genome editing [122]. Enhancing specificity has been another crucial focus in CRISPR-Cas9 advancements for horticultural crop improvement [123]. Various research has explored alternatives to Cas proteins, such as Cas12a, which offer distinct protospacer adjacent motif (PAM) specificities, expanding the target range for precise editing [124]. Additionally, the development of base editors has allowed for the direct conversion of specific DNA bases without the need for double-strand breaks, further improving the precision of CRISPR-Cas9 editing and minimizing unintended mutations [125].
The utilization of viral vectors and nanoparticles has shown promise in enhancing the successful delivery of CRISPR-Cas9 machinery into different horticultural crops [126]. Furthermore, the optimization of tissue culture protocols and transformation techniques tailored to specific crop species has contributed to more effective editing outcomes in a wide range of horticultural crops [127]. These advancements in CRISPR-Cas9 technology have significantly addressed the initial disadvantages, enhancing its precision, specificity, and delivery capabilities for precise genome editing in horticultural crops. The continued progress in CRISPR-Cas9 holds tremendous potential for revolutionizing crop improvement and promoting sustainable agricultural practices.
3. TALENs (Transcription Activator-like Effector Nucleases)
TALENs, along with CRISPR-Cas9, are widely employed as genome editing tools for precise gene modifications in horticultural crops [114]. TALENs are engineered nucleases capable of inducing double-strand breaks (DSBs) at specific DNA sequences, enabling targeted gene editing [128]. Comprising a customizable DNA-binding domain derived from transcription activator-like effectors (TALEs) and a nuclease domain typically derived from the FokI endonuclease, TALENs offer a dual-component design [129].
The DNA-binding domain of TALENs is constructed using multiple repeats of TALEs, each recognizing a specific nucleotide in the target DNA sequence [130]. These TALEs consist of repeat units typically containing 33–35 amino acids in a central repeat region [131]. The specificity of TALENs is achieved through customizable repeat variable di-residues (RVDs) within each repeat unit, where different RVDs recognize different nucleotides, enabling the design of highly specific TALENs [132].
The nuclease domain of TALENs is derived from the FokI endonuclease, which requires dimerization for its DNA cleavage activity [133]. TALENs are designed as pairs, with each TALEN targeting one DNA strand [134]. Upon binding to their target sites, the FokI nuclease domains of the TALENs dimerize, forming a functional nuclease complex that induces double-strand breaks (DSBs) at the target site [135].
3.1. Applications of TALENs in Horticultural Crops
3.1.1. Gene Knockout
TALENs enable targeted disruption of genes through induced double-strand breaks (DSBs), allowing for gene knockout or loss-of-function mutations. This approach facilitates the investigation of gene function and the identification of genes associated with diverse horticultural traits. TALENs were utilized to knockout the SlAN2 gene in tomatoes, elucidating its functional role in fruit ripening. The generated SlAN2 knockout mutants exhibited delayed fruit ripening and modified fruit quality [136].
TALENs have been utilized for gene knockout in citrus crops, specifically targeting disease resistance-related genes. In a study on Xanthomonas citri, the disruption of the CsLOB1 gene using TALENs resulted in enhanced resistance to citrus canker disease [137]. Similarly, TALENs have been employed in grapevine to study gene function, with a specific focus on disease resistance. By targeting the VvWRKY52 gene, researchers successfully generated VvWRKY52 knockout mutants using TALENs, shedding light on its involvement in plant defense responses [138].
3.1.2. Gene Editing
TALENs enable precise gene editing through targeted DSB induction at specific sites, facilitating the insertion, deletion, or replacement of DNA sequences in horticultural crops, thereby enabling desired genetic modifications [139]. TALENs have been utilized to introduce targeted mutations in the StCDF1 gene of potato plants, resulting in altered tuberization patterns and flowering time [140].
3.2. Limitations of TALENs in Horticultural Crops
3.2.1. Design Complexity
The design and assembly of TALENs can be laborious and technically demanding, making them less scalable and limiting their widespread adoption in horticultural crop research, in contrast to the more accessible and versatile CRISPR-Cas9 system [13,141]. The process of TALENs involves identifying the specific DNA-binding domain and assembling the corresponding RVDs for sequence-specific recognition [142,143]. The assembly of TALEN constructs requires multiple cloning steps, which can be prone to errors and inefficiencies, resulting in lower transformation efficiency or difficulties in obtaining functional TALEN constructs [134,144].
The recognition mechanism of TALENs relies on their RVDs, which bind to specific nucleotides, limiting their flexibility in targeting certain DNA sequences [145]. Targeting repetitive or GC-rich regions poses challenges for TALENs, as designing specific RVDs for such sequences may be difficult [146]. Despite generally higher target specificity compared to previous genome editing tools, TALENs can still exhibit off-target effects attributed to partial complementarity between the TALEN and unintended DNA sequences [83,128].
3.2.2. Delivery and Transformation Efficiency
Efficient delivery and transformation of TALENs in horticultural crops, particularly recalcitrant species, pose challenges [110]. Attaining stable integration and high transformation efficiencies of TALEN constructs in the plant genome is crucial for successful genome editing [147]. Optimization of delivery methods and transformation protocols is essential to address these limitations.
TALEN delivery and transformation efficiency vary among plant species, with some exhibiting higher rates of success while others pose challenges [148,149]. Optimization and evaluation of TALEN delivery and transformation efficiency are necessary for each specific horticultural crop [150]. Genetic variation within a crop species and tissue specificity can also influence the efficiency of TALEN delivery and transformation [93,151,152].
TALEN-mediated transformations can result in off-target mutations and mosaicism, affecting the efficiency and reliability of the process [153,154]. Careful screening and selection of transformed plants are necessary to address these effects [155]. Ongoing efforts focus on enhancing TALEN delivery and transformation efficiency in horticultural crops through protocol optimization, tissue-specific methods, and technological advancements [156].
3.3. Advancements in TALENs to Overcome Limitations in Horticultural Crops
The initial development of TALENs encountered challenges related to the complex and time-consuming process of assembling custom-engineered TALE repeat arrays for recognizing specific DNA sequences. In response, researchers have made significant progress in developing modular and simplified TALE repeat architectures [145]. These advancements have streamlined the design process, enabling the more efficient construction of TALENs targeting various DNA sequences in horticultural crops [157].
In line with CRISPR-Cas9, improved delivery methods have been employed to enhance the efficiency of TALEN delivery into plant cells [114]. Viral vectors and nanoparticles have been explored as effective delivery tools, facilitating successful genome editing in a diverse range of horticultural crops [158]. These advancements in TALEN design and delivery have significantly addressed the initial limitations, making TALENs a more accessible and versatile genome editing tool for horticultural crop improvement. The continued progress in TALEN technology holds great potential for accelerating precision breeding and promoting sustainable agriculture.
4. ZFNs (Zinc Finger Nucleases)
ZFNs, an engineered nuclease class, have been utilized for genome editing in horticultural crops [159]. Comprising two main components, ZFPs and a nuclease domain from FokI endonuclease, ZFNs exhibit sequence-specific DNA recognition [160,161]. Each zinc finger module, approximately 30 amino acids in length, targets three DNA bases, and multiple modules enable precise targeting of specific DNA sequences [162,163]. ZFNs are employed in pairs, with each ZFN targeting one DNA strand [164]. The binding of the ZFNs to their target sites leads to FokI nuclease domain dimerization, generating a functional nuclease complex that induces double-strand breaks (DSBs) at the target site [135].
4.1. ZFNs and Their Applications
Zinc finger nucleases (ZFNs) enable targeted mutations through ZFN-mediated site-specific mutagenesis, leveraging non-homologous end joining (NHEJ) repair mechanisms for introducing mutations [165,166]. The successful targeting of both transgene and native sequences has been demonstrated in plant species like Arabidopsis and tobacco [165,166]. ZFNs have also been applied to remove transgenes in tobacco plants via NHEJ-mediated repairs, leading to truncated modifications at the targeted sites and transgene elimination [167].
ZFNs, when co-delivered with donor DNA molecules, have shown the capacity to induce site-specific homology-directed repair (HDR) in tobacco and corn (Zea mays) plants, facilitating accurate integration of the donor DNA into their genomes [168,169]. Zinc finger nucleases (ZFNs) have demonstrated their potential as powerful tools for site-specific mutagenesis in tobacco and corn cells, inducing targeted mutations. This feature enhances gene discovery and facilitates crop plant development. Efficient expression of ZFNs in regenerating cells or tissues is essential for successful site-specific mutagenesis. In Arabidopsis, transgenic strategies have been utilized to achieve high ZFN expression levels in shoot apical meristem L2 cells, leading to the generation of mutated seeds.
Various strategies have been employed to ensure efficient expression of ZFNs in transgenic Arabidopsis plants. Inducible stable expression systems, such as those activated by heat shock or estrogen, have been utilized [165,170,171]. These systems enable controlled induction of ZFN expression at specific time points. Additionally, constitutive expression of ZFNs, where ZFNs are continuously expressed, has been implemented [172]. Both approaches have proven successful in driving the expression of ZFNs in transgenic Arabidopsis plants, facilitating efficient site-specific mutagenesis.
4.2. Challenges Associated with ZFNs in Horticultural Crops
Designing ZFNs entails the custom engineering of DNA sequence-specific zinc finger proteins (ZFPs), a process that demands expertise in protein engineering and DNA binding specificity [173]. This design complexity hampers the widespread adoption of ZFNs and restricts their applicability to a broader spectrum of target sequences in horticultural crops [174].
The complex genomes of horticultural crops, characterized by repetitive sequences and high heterozygosity, pose challenges in identifying unique and suitable target sites for ZFN binding [175]. Repetitive sequences increase the risk of off-target effects, necessitating meticulous design and validation of ZFNs [176]. Furthermore, horticultural crops often possess gene families with closely related members, requiring careful selection of ZFNs to target specific members without affecting others. Sequence analysis and bioinformatics tools aid in identifying unique target sites within gene family members [177].
The effective delivery of ZFNs into plant cells is essential for achieving successful mutagenesis [178]. However, the diverse cell types, tissue structures, and cell wall compositions in horticultural crops can present challenges in delivering ZFNs to specific cells or tissues [179]. Tailoring delivery methods, such as Agrobacterium-mediated transformation or particle bombardment, to each crop is necessary for efficient delivery [180].
4.3. Advancements in ZFNs Overcoming Challenges in Horticultural Crop Genome Editing
The early development of ZFNs faced challenges due to the custom engineering required for recognizing specific DNA sequences, leading to complexities in the design process [181]. To address this limitation, researchers have made significant strides in the advancement of modular ZFN platforms [182]. These platforms offer increased design flexibility, allowing for the targeting of various DNA sequences with reduced design complexity [145]. By adopting modular ZFN architectures, the design process has been streamlined, facilitating the construction of ZFNs tailored to diverse target sites in horticultural crops [183].
Similar to CRISPR-Cas9 and TALENs, enhanced specificity has been a primary focus in the advancements of ZFNs for horticultural crop genome editing. Researchers have made improvements in target site selection algorithms and ZFN architecture modifications to achieve greater specificity [184]. These developments have resulted in reduced off-target effects, enhancing the precision and safety of ZFN-mediated genome editing in horticultural crops. The continuous progress in ZFN technology has addressed the initial challenges encountered during its development, making ZFNs a valuable and versatile tool for precise genome editing in horticultural crop improvement [185]. The refined design strategies and improved specificity hold promising potential for accelerating genetic improvement in crops and contributing to sustainable agriculture.
5. Regulatory and Ethical Considerations
Genome editing has emerged as a promising approach for sustainable agriculture and crop enhancement, aiming to create transgene-free plants [186]. However, to ensure responsible and safe utilization of this technology, it is imperative to address key regulatory and ethical aspects. As genome editing advances, a thorough understanding of international regulations, regional policies, and ethical implications becomes essential in promoting its widespread adoption for crop improvement in agriculture.
5.1. Regulatory Frameworks for Genome-Edited Crops
Genome editing techniques like CRISPR-Cas9, TALENs, and ZFNs have sparked inquiries regarding the regulatory status of genome-edited crops and the definition of Genetically Modified Organisms (GMOs) [187]. Different regions employ varied approaches to govern these crops, impacting their commercialization, import/export, and release. The challenges of classifying genome-edited crops under existing GMO regulations necessitate harmonization efforts for consistent global oversight [188].
In Europe, stringent GMO regulations are implemented, following a precautionary approach to assess the potential risks associated with genetically modified crops [189]. Genome-edited crops generally face the same regulations as transgenic GMOs, irrespective of the absence of foreign DNA [190]. Approval and commercialization of genome-edited crops necessitate thorough safety assessments and strict regulatory adherence [191]. Conversely, the United States adopts a product-focused regulatory approach rather than a process-based one for genome-edited crops [192]. If the final product lacks foreign DNA or if the introduced changes could have occurred through conventional breeding, the crop may not be categorized as a GMO [193]. However, regulatory status may vary depending on the specific genetic modifications introduced [194]. Regions such as Asia, Africa, and Latin America exhibit diverse GMO regulations and stances on genome-edited crops [195,196]. Some countries align with Europe’s strict regulations, while others follow the product-based approach of the United States [197].
5.2. Safety Assessment and Risk Analysis
The safety evaluation of genome-edited crops constitutes a pivotal aspect of regulatory deliberations [198]. It is imperative to conduct thorough assessments to examine off-target effects and unintended consequences arising from genome editing, ensuring the absence of unforeseen alterations in the plant’s genome [199]. Environmental and ecological impact evaluations are indispensable in gauging potential ecological risks linked to the release of genome-edited crops into the environment [200]. Furthermore, detailed studies focusing on the potential allergenicity and toxicity of edited crops are essential to ensure consumer safety.
5.3. Public Perception and Ethical Considerations
The public perception of genome-edited crops exerts a considerable influence on their acceptance and widespread adoption [201]. Ethical considerations surrounding genetic modification and the perceived risks associated with genome editing can significantly impact societal acceptance [202]. To address these concerns effectively, transparent communication and active engagement with the public, stakeholders, and policymakers are crucial [203]. Establishing trust through open dialogue is essential for addressing ethical issues related to genome editing in plants.
5.4. International Harmonization and Collaboration
The international harmonization of regulations and inter-country collaboration are pivotal for enabling the smooth movement and trade of genome-edited crops across borders [204]. Promoting open dialogue and data sharing among regulatory entities facilitates efficient risk assessments and well-informed decision-making processes [205]. Standardizing protocols for the detection and identification of genome-edited crops is beneficial in ensuring that accurate labeling and traceability measures are in place [206].
6. Future Prospects of Genome-Edited Crops in Horticulture
Genome editing holds promise for developing disease-resistant horticultural crops, reducing reliance on pesticides, and mitigating crop losses [207,208]. By targeting and modifying genes associated with disease susceptibility, crops can be engineered to enhance resilience against pathogens [208]. Climate change impacts horticultural crops through increased temperatures, droughts, and extreme weather events, affecting productivity [209]. Genome editing enables the enhancement of abiotic stress tolerance by modifying stress response genes, fostering crop adaptation to adverse environmental conditions, and ensuring food security [210,211].
Genome editing offers the potential for enhancing the nutritional content of horticultural crops by precisely modifying genes involved in nutrient uptake, synthesis, and metabolism [212]. This can result in crops with elevated levels of essential vitamins, minerals, and beneficial compounds, addressing malnutrition and promoting human health [180,213]. Targeting genes associated with plant architecture, flowering time, and fruit development can improve yield and productivity in horticultural crops [214]. The optimization of these traits can lead to crops with increased yields, extended shelf life, and improved quality, meeting the global demand for food [215].
Genome editing enables the creation of novel horticultural crop varieties with improved traits that surpass traditional breeding methods [216]. Precise genetic modifications result in unique plant characteristics, enhancing flavor, color, aroma, and other desirable attributes, appealing to consumer preferences and expanding market opportunities [217,218]. Genome-edited crops contribute to sustainable agriculture by reducing chemical inputs, improving resource-use efficiency, and enhancing stress tolerance, thus minimizing environmental impacts associated with conventional practices [219]. This fosters sustainable and environmentally friendly horticultural production systems.
7. Conclusions
In this review, we investigated the applications, methodologies, and potential impacts of genome editing in horticulture, focusing on CRISPR-Cas9, TALENs, and ZFNs as promising tools. CRISPR-Cas9, with its versatility and efficiency, has revolutionized horticultural crop improvement by enabling precise modifications targeting genes associated with disease resistance, abiotic stress tolerance, nutrition, and yield. Challenges such as off-target effects, delivery methods, and regulatory considerations need attention to fully exploit CRISPR-Cas9’s potential. TALENs and ZFNs offer alternative options with successful applications in specific contexts. The benefits of genome editing in horticultural crops are substantial, encompassing disease protection, stress resilience, nutrition enhancement, and increased yield for food security. Addressing limitations, challenges, and ethical considerations will facilitate the sustainable and impactful implementation of genome editing in horticulture, benefiting stakeholders at various levels.
Author Contributions
Conceptualization and methodology, M.A.D.; software and validation, R.S.; formal analysis and investigation, M.P.S. and J.-K.Y.; writing—original draft preparation, M.A.D.; writing—review and editing, R.S.; supervision and project administration, M.P.S. and J.W.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Acknowledgments
The authors wish to thank the Department of Industrial Plant Science and Technology, Chungbuk National University, Cheongju 28644, Republic of Korea, and The Entomology Research Institute, Loyola College Chennai—34, Tamil Nadu, India, for extending the necessary support and guidance.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Okunlola, A.I.; Adepoju, A.O.; Akinpetide, E.O. The significant role of horticulture in environmental aesthetics and management. Int. J. Hortic. 2016, 6, 17. [Google Scholar] [CrossRef]
- Salgotra, R.K.; Chauhan, B.S. Genetic Diversity, Conservation, and Utilization of Plant Genetic Resources. Genes 2023, 14, 174. [Google Scholar] [CrossRef]
- Borlaug, N.E. Contributions of Conventional Plant-Breeding to Food-Production. Science 1983, 219, 689–693. [Google Scholar] [CrossRef] [PubMed]
- Sharma, H.C.; Crouch, J.H.; Sharma, K.K.; Seetharama, N.; Hash, C.T. Applications of biotechnology for crop improvement: Prospects and constraints. Plant Sci. 2002, 163, 381–395. [Google Scholar] [CrossRef]
- Beaver, J.S.; Osorno, J.M. Achievements and limitations of contemporary common bean breeding using conventional and molecular approaches. Euphytica 2009, 168, 145–175. [Google Scholar] [CrossRef]
- Xiong, J.S.; Ding, J.; Li, Y. Genome-editing technologies and their potential application in horticultural crop breeding. Hortic. Res. 2015, 2, 15019. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Thurtle-Schmidt, D.M.; Lo, T.W. Molecular biology at the cutting edge: A review on CRISPR/CAS9 gene editing for undergraduates. Biochem. Mol. Biol. Edu. 2018, 46, 195–205. [Google Scholar] [CrossRef]
- Xu, J.M.; Hua, K.; Lang, Z.B. Genome editing for horticultural crop improvement. Hortic. Res. 2019, 6, 113. [Google Scholar] [CrossRef]
- Erpen-Dalla Corte, L.; Mahmoud, L.M.; Moraes, T.S.; Mou, Z.L.; Grosser, J.W.; Dutt, M. Development of Improved Fruit, Vegetable, and Ornamental Crops Using the CRISPR/Cas9 Genome Editing Technique. Plants 2019, 8, 601. [Google Scholar] [CrossRef]
- Zhang, D.Q.; Zhang, Z.Y.; Unver, T.; Zhang, B.H. CRISPR/Cas: A powerful tool for gene function study and crop improvement. J. Adv. Res. 2021, 29, 207–221. [Google Scholar] [CrossRef] [PubMed]
- Rani, R.; Yadav, P.; Barbadikar, K.M.; Baliyan, N.; Malhotra, E.V.; Singh, B.K.; Kumar, A.; Singh, D. CRISPR/Cas9: A promising way to exploit genetic variation in plants. Biotechnol. Lett. 2016, 38, 1991–2006. [Google Scholar] [CrossRef] [PubMed]
- Gaj, T.; Gersbach, C.A.; Barbas, C.F. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013, 31, 397–405. [Google Scholar] [CrossRef] [PubMed]
- Sun, N.; Zhao, H.M. Transcription activator-like effector nucleases (TALENs): A highly efficient and versatile tool for genome editing. Biotechnol. Bioeng. 2013, 110, 1811–1821. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. INNOVATION TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Bio. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef]
- Pattanayak, V.; Ramirez, C.L.; Joung, J.K.; Liu, D.R. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat. Methods 2011, 8, 765–770. [Google Scholar] [CrossRef]
- Osakabe, Y.; Osakabe, K. Genome Editing with Engineered Nucleases in Plants. Plant Cell Physiol. 2015, 56, 389–400. [Google Scholar] [CrossRef]
- Bhagwat, A.C.; Patil, A.M.; Saroj, S.D. CRISPR/Cas 9-Based Editing in the Production of Bioactive Molecules. Mol. Biotechnol. 2022, 64, 245–251. [Google Scholar] [CrossRef]
- Khanzadi, M.N.; Khan, A.A. CRISPR/Cas9: Nature’s gift to prokaryotes and an auspicious tool in genome editing. J. Basic Microb. 2020, 60, 91–102. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; He, S.L. CRISPR-Cas9: Tool for Qualitative and Quantitative Plant Genome Editing. Front. Plant Sci. 2016, 7, 1740. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.J.; Wang, L.Q. CRISPR/Cas9 technology for improving agronomic traits and future prospective in agriculture. Planta 2021, 254, 68. [Google Scholar] [CrossRef]
- Rasheed, A.; Barqawi, A.A.; Mahmood, A.; Nawaz, M.; Shah, A.N.; Bay, D.H.; Alahdal, M.A.; Hassan, M.U.; Qari, S.H. CRISPR/Cas9 is a powerful tool for precise genome editing of legume crops: A review. Mol. Biol. Rep. 2022, 49, 5595–5609. [Google Scholar] [CrossRef] [PubMed]
- Urnov, F.D.; Rebar, E.J.; Holmes, M.C.; Zhang, H.S.; Gregory, P.D. Genome editing with engineered zinc finger nucleases. Nat. Rev. Genet. 2010, 11, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Wai, A.H.; Naing, A.H.; Lee, D.J.; Kim, C.K.; Chung, M.Y. Molecular genetic approaches for enhancing stress tolerance and fruit quality of tomato. Plant Biotechnol. Rep. 2020, 14, 515–537. [Google Scholar] [CrossRef]
- Gonzales, L.R.; Shi, L.; Bergonzi, S.B.; Oortwijn, M.; Franco-Zorrilla, J.M.; Solano-Tavira, R.; Visser, R.G.F.; Abelenda, J.A.; Bachem, C.W.B. Potato CYCLING DOF FACTOR 1 and its lncRNA counterpart StFLORE link tuber development and drought response. Plant J. 2021, 105, 855–869. [Google Scholar] [CrossRef]
- Henry, R.J.; Furtado, A.; Rangan, P. Wheat seed transcriptome reveals genes controlling key traits for human preference and crop adaptation. Curr. Opin. Plant Biol. 2018, 45, 231–236. [Google Scholar] [CrossRef]
- Yang, W.; Ren, J.; Liu, W.; Liu, D.; Xie, K.; Zhang, F.; Wang, P.; Guo, W.; Wu, X. An efficient transient gene expression system for protein subcellular localization assay and genome editing in citrus protoplasts. Hortic. Plant J. 2023, 9, 425–436. [Google Scholar] [CrossRef]
- Martin-Pizarro, C.; Trivino, J.C.; Pose, D. Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 2019, 70, 885–895. [Google Scholar] [CrossRef]
- Capriotti, L.; Baraldi, E.; Mezzetti, B.; Limera, C.; Sabbadini, S. Biotechnological Approaches: Gene Overexpression, Gene Silencing, and Genome Editing to Control Fungal and Oomycete Diseases in Grapevine. Int. J. Mol. Sci. 2020, 21, 5701. [Google Scholar] [CrossRef]
- Afrin, K.S.; Rahim, M.A.; Jung, H.J.; Park, J.I.; Kim, H.T.; Nou, I.S. Development of Molecular Marker through Genome Realignment for Specific Detection of Xanthomonas campestris pv. campestris Race 5, a Pathogen of Black Rot Disease. J. Microbiol. Biotechnol. 2019, 29, 785–793. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. CRISPR-edited crops free to enter market, skip regulation. Nat. Biotechnol. 2016, 34, 582. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.H.; Sheng, O.; Deng, G.M.; He, W.D.; Dong, T.; Yang, Q.S.; Dou, T.X.; Li, C.Y.; Gao, H.J.; Liu, S.W.; et al. CRISPR/Cas9-mediated genome editing of MaACO1 (aminocyclopropane-1-carboxylate oxidase 1) promotes the shelf life of banana fruit. Plant Biotechnol. J. 2021, 19, 654–656. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Deng, Y.J.; Liu, J.X.; Duan, A.Q.; Liu, H.; Xiong, A.S. DcCCD4 catalyzes the degradation of alpha-carotene and beta-carotene to affect carotenoid accumulation and taproot color in carrot. Plant J. 2021, 108, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Abdullah; Faraji, S.; Mehmood, F.; Malik, H.M.T.; Ahmed, I.; Heidari, P.; Poczai, P. The GASA Gene Family in Cacao (Theobroma cacao, Malvaceae): Genome Wide Identification and Expression Analysis. Agronomy 2021, 11, 1425. [Google Scholar] [CrossRef]
- Nonaka, S.; Ito, M.; Ezura, H. Targeted modification of CmACO1 by CRISPR/Cas9 extends the shelf-life of Cucumis melo var. reticulatus melon. Front. Genome Ed. 2023, 5, 1176125. [Google Scholar] [CrossRef]
- Mishra, R.; Mohanty, J.N.; Mahanty, B.; Joshi, R.K. A single transcript CRISPR/Cas9 mediated mutagenesis of CaERF28 confers anthracnose resistance in chilli pepper (Capsicum annuum L.). Planta 2021, 254, 5. [Google Scholar] [CrossRef]
- Wang, C.P.; Li, Y.; Wang, N.; Yu, Q.; Li, Y.H.; Gao, J.P.; Zhou, X.F.; Ma, N. An efficient CRISPR/Cas9 platform for targeted genome editing in rose (Rosa hybrida). J. Integr. Plant Biol. 2023, 65, 895–899. [Google Scholar] [CrossRef]
- Pechar, G.S.; Donaire, L.; Gosalvez, B.; Garcia-Almodovar, C.; Sanchez-Pina, M.A.; Truniger, V.; Aranda, M.A. Editing melon eIF4E associates with virus resistance and male sterility. Plant Biotechnol. J. 2022, 20, 2006–2022. [Google Scholar] [CrossRef]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.R.; Yang, M.J.; Sheng, J.P.; Shen, L. Reduced Drought Tolerance by CRISPR/Cas9-Mediated SlMAPK3 Mutagenesis in Tomato Plants. J. Agr. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef]
- Okuzaki, A.; Ogawa, T.; Koizuka, C.; Kaneko, K.; Inaba, M.; Imamura, J.; Koizuka, N. CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 2018, 131, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Herath, D.; Voogd, C.; Mayo-Smith, M.; Yang, B.; Allan, A.C.; Putterill, J.; Varkonyi-Gasic, E. CRISPR-Cas9-mediated mutagenesis of kiwifruit BFT genes results in an evergrowing but not early flowering phenotype. Plant Biotechnol. J. 2022, 20, 2064–2076. [Google Scholar] [CrossRef]
- Shu, P.; Li, Z.Y.; Min, D.D.; Zhang, X.H.; Ai, W.; Li, J.Z.; Zhou, J.X.; Li, Z.L.; Li, F.J.; Li, X.A. CRISPR/Cas9-Mediated SlMYC2 Mutagenesis Adverse to Tomato Plant Growth and MeJA-Induced Fruit Resistance to Botrytis cinerea. J. Agr. Food Chem. 2020, 68, 5529–5538. [Google Scholar] [CrossRef]
- Ma, J.; Sun, S.; Whelan, J.; Shou, H.X. CRISPR/Cas9-Mediated Knockout of GmFATB1 Significantly Reduced the Amount of Saturated Fatty Acids in Soybean Seeds. Int. J. Mol. Sci. 2021, 22, 3877. [Google Scholar] [CrossRef]
- Wang, R. What Makes ‘Hayward’ Kiwifruit Store so Well? The Biological Basis for the Postharvest Behaviour of ‘Hayward’ Kiwifruit. Ph.D. Thesis, The University of Auckland, Auckland, New Zealand, 2021. [Google Scholar]
- Wang, H.X.; Wu, Y.L.; Zhang, Y.D.; Yang, J.; Fan, W.J.; Zhang, H.; Zhao, S.S.; Yuan, L.; Zhang, P. CRISPR/Cas9-Based Mutagenesis of Starch Biosynthetic Genes in Sweet Potato (Ipomoea Batatas) for the Improvement of Starch Quality. Int. J. Mol. Sci. 2019, 20, 4702. [Google Scholar] [CrossRef]
- Brewer, S.E.; Chambers, A.H. CRISPR/Cas9-mediated genome editing of phytoene desaturase in Carica papaya L. J. Hortic. Sci. Biotechnol. 2022, 97, 580–592. [Google Scholar] [CrossRef]
- Maioli, A.; Gianoglio, S.; Moglia, A.; Acquadro, A.; Valentino, D.; Milani, A.M.; Prohens, J.; Orzaez, D.; Granell, A.; Lanteri, S.; et al. Simultaneous CRISPR/Cas9 Editing of Three PPO Genes Reduces Fruit Flesh Browning in Solanum melongena L. Front. Plant Sci. 2020, 11, 607161. [Google Scholar] [CrossRef]
- Gomez, M.A.; Lin, Z.D.; Moll, T.; Chauhan, R.D.; Hayden, L.; Renninger, K.; Beyene, G.; Taylor, N.J.; Carrington, J.C.; Staskawicz, B.J.; et al. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 2019, 17, 421–434. [Google Scholar] [CrossRef]
- Krishna, H.; Alizadeh, M.; Singh, D.; Singh, U.; Chauhan, N.; Eftekhari, M.; Sadh, R.K. Somaclonal variations and their applications in horticultural crops improvement. 3 Biotech 2016, 6, 54. [Google Scholar] [CrossRef]
- Søren, K.; Toni, W.; Christoph, D.; Hanne, C.T.; Magnus, R.; Morten Egevang, J.; Qiongxian, L.; Cynthia, V.; Emiko, M.; Jeppe Thulin, Ø.; et al. FIND-IT: Ultrafast mining of genome diversity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Wagh, S.G.; Pohare, M.B. Current and Future Prospects of Plant Breeding with CRISPR/Cas. Current. J. Appl. Sci. Technol. 2019, 38, 1–17. [Google Scholar] [CrossRef]
- Zhu, L.H.J.; Holmes, B.R.; Aronin, N.; Brodsky, M.H. CRISPRseek: A Bioconductor Package to Identify Target-Specific Guide RNAs for CRISPR-Cas9 Genome-Editing Systems. PLoS ONE 2014, 9, e108424. [Google Scholar] [CrossRef]
- Jinek, M.; Jiang, F.G.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.B.; Anders, C.; Hauer, M.; Zhou, K.H.; Lin, S.; et al. Structures of Cas9 Endonucleases Reveal RNA-Mediated Conformational Activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef]
- Anders, C.; Niewoehner, O.; Duerst, A.; Jinek, M. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 2014, 513, 569–573. [Google Scholar] [CrossRef]
- Gasiunas, G.; Siksnys, V. RNA-dependent DNA endonuclease Cas9 of the CRISPR system: Holy Grail of genome editing? Trends Microbiol. 2013, 21, 562–567. [Google Scholar] [CrossRef]
- Jiang, F.G.; Doudna, J.A. CRISPR-Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 2017, 46, 505–529. [Google Scholar] [CrossRef]
- Al Abdallah, Q.; Ge, W.B.; Fortwendel, J.R. A Simple and Universal System for Gene Manipulation in Aspergillus fumigatus: In Vitro-Assembled Cas9-Guide RNA Ribonucleoproteins Coupled with Microhomology Repair Templates. Msphere 2017, 2, e00446-17. [Google Scholar] [CrossRef]
- Lemos, B.R.; Kaplan, A.C.; Bae, J.E.; Ferrazzoli, A.E.; Kuo, J.; Anand, R.P.; Waterman, D.P.; Haber, J.E. CRISPR/Cas9 cleavages in budding yeast reveal templated insertions and strand-specific insertion/deletion profiles. Proc. Natl. Acad. Sci. USA 2018, 115, E2040–E2047. [Google Scholar] [CrossRef]
- Song, F.; Stieger, K. Optimizing the DNA Donor Template for Homology-Directed Repair of Double-Strand Breaks. Mol. Ther. Nucleic Acids 2017, 7, 53–60. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Platt, R.J.; Zhang, F. Therapeutic genome editing: Prospects and challenges. Nat. Med. 2015, 21, 121–131. [Google Scholar] [CrossRef]
- Danner, E.; Bashir, S.; Yumlu, S.; Wurst, W.; Wefers, B.; Kuhn, R. Control of gene editing by manipulation of DNA repair mechanisms. Mamm. Genome 2017, 28, 262–274. [Google Scholar] [CrossRef]
- Boubakri, H. Recent progress in CRISPR/Cas9-based genome editing for enhancing plant disease resistance. Gene 2023, 866, 147334. [Google Scholar] [CrossRef]
- Wan, D.Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.Y.; Wang, Y.J.; Wen, Y.Q. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116. [Google Scholar] [CrossRef]
- Atarashi, H.; Jayasinghe, W.H.; Kwon, J.; Kim, H.; Taninaka, Y.; Igarashi, M.; Ito, K.; Yamada, T.; Masuta, C.; Nakahara, K.S. Artificially Edited Alleles of the Eukaryotic Translation Initiation Factor 4E1 Gene Differentially Reduce Susceptibility to Cucumber Mosaic Virus and Potato Virus Y in Tomato. Front. Microbiol. 2020, 11, 564310. [Google Scholar] [CrossRef]
- Peng, A.; Chen, S.; Lei, T.; Xu, L.; He, Y.; Wu, L.; Yao, L.; Zou, X. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 2017, 15, 1509–1519. [Google Scholar] [CrossRef]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell 2017, 169, 1142–1155.e12. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, X.J.; Huang, R.W.; Yang, S.; Li, M.Y.; Guo, Y.L. CRISPR/Cas9-mediated targeted mutation reveals a role for AN4 rather than DPL in regulating venation formation in the corolla tube of Petunia hybrida. Hortic. Res. 2021, 8, 116. [Google Scholar] [CrossRef]
- Lopez-Casado, G.; Sanchez-Raya, C.; Ric-Varas, P.D.; Paniagua, C.; Blanco-Portales, R.; Munoz-Blanco, J.; Pose, S.; Matas, A.J.; Mercado, J.A. CRISPR/Cas9 editing of the polygalacturonase FaPG1 gene improves strawberry fruit firmness. Hortic. Res. 2023, 10, uhad011. [Google Scholar] [CrossRef]
- Li, X.D.; Wang, Y.N.; Chen, S.; Tian, H.Q.; Fu, D.Q.; Zhu, B.Z.; Luo, Y.B.; Zhu, H.L. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 2018, 9, 559. [Google Scholar] [CrossRef]
- Beracochea, V.; Stritzler, M.; Radonic, L.; Bottero, E.; Jozefkowicz, C.; Darqui, F.; Ayub, N.; Bilbao, M.L.; Soto, G. CRISPR/Cas9-mediated knockout of SPL13 radically increases lettuce yield. Plant Cell Rep. 2023, 42, 645–647. [Google Scholar] [CrossRef]
- Nitarska, D.; Boehm, R.; Debener, T.; Lucaciu, R.C.; Halbwirth, H. First genome edited poinsettias: Targeted mutagenesis of flavonoid 3′-hydroxylase using CRISPR/Cas9 results in a colour shift. Plant Cell Tissue Organ Cult. 2021, 147, 49–60. [Google Scholar] [CrossRef]
- Huang, W.; Hu, N.; Xiao, Z.N.; Qiu, Y.P.; Yang, Y.; Yang, J.; Mao, X.; Wang, Y.C.; Li, Z.G.; Guo, H.W. A molecular framework of ethylene-mediated fruit growth and ripening processes in tomato. Plant Cell 2022, 34, 3280–3300. [Google Scholar] [CrossRef]
- Huynh, T.T.H.; Nguyen, T.L.; Luu, H.L.; Nguyen, H.H.; Le, H.D.; Bui, M.M.; Pham, T.H.; Doan, T.B.T.; Le, T.T.H.; Ha, H.H.; et al. Isolation and Characterization of a Dreb Homolog Gene from a Local Drought-Tolerant Maize Cultivar. Acta Biol. Cracoviensia Bot. 2019, 61, 13–24. [Google Scholar] [CrossRef]
- Wang, Y.P.; Wang, J.F.; Guo, S.G.; Tian, S.W.; Zhang, J.; Ren, Y.; Li, M.Y.; Gong, G.Y.; Zhang, H.Y.; Xu, Y. CRISPR/Cas9-mediated mutagenesis of ClBG1 decreased seed size and promoted seed germination in watermelon. Hortic. Res. 2021, 8, 70. [Google Scholar] [CrossRef]
- Kishchenko, O.; Zhou, Y.Z.; Jatayev, S.; Shavrukov, Y.; Borisjuk, N. Gene editing applications to modulate crop flowering time and seed dormancy. Abiotech 2020, 1, 233–245. [Google Scholar] [CrossRef]
- Lv, S.W.; Wu, W.G.; Wang, M.H.; Meyer, R.S.; Ndjiondjop, M.N.; Tan, L.B.; Zhou, H.Y.; Zhang, J.W.; Fu, Y.C.; Cai, H.W.; et al. Genetic control of seed shattering during African rice domestication. Nat. Plants 2018, 4, 331–337. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.X.; Zhang, L.C.; Ji, Y.L.; Tan, D.M.; Yuan, H.; Wang, A.D. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef]
- Jo, H.; Woo, C.; Norah, N.; Song, J.T.; Lee, J.D. Novel Allele of FAD2-1A from an EMS-Induced Mutant Soybean Line (PE529) Produces Elevated Levels of Oleic Acid in Soybean Oil. Agronomy 2022, 12, 2115. [Google Scholar] [CrossRef]
- Fu, M.X.; Chen, L.; Cai, Y.P.; Su, Q.; Chen, Y.Y.; Hou, W.S. CRISPR/Cas9-Mediated Mutagenesis of GmFAD2-1A and/or GmFAD2-1B to Create High-Oleic-Acid Soybean. Agronomy 2022, 12, 3218. [Google Scholar] [CrossRef]
- Mellidou, I.; Koukounaras, A.; Kostas, S.; Patelou, E.; Kanellis, A.K. Regulation of Vitamin C Accumulation for Improved Tomato Fruit Quality and Alleviation of Abiotic Stress. Genes 2021, 12, 694. [Google Scholar] [CrossRef]
- Liu, J.H.; Liu, M.T.; Wang, J.Y.; Zhang, J.; Miao, H.X.; Wang, Z.; Jia, C.H.; Zhang, J.B.; Xu, B.Y.; Jin, Z.Q. Transcription factor MaMADS36 plays a central role in regulating banana fruit ripening. J. Exp. Bot. 2021, 72, 7078–7091. [Google Scholar] [CrossRef]
- Manghwar, H.; Li, B.; Ding, X.; Hussain, A.; Lindsey, K.; Zhang, X.L.; Jin, S.X. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 2020, 7, 1902312. [Google Scholar] [CrossRef]
- Zhao, H.; Wolt, J.D. Risk associated with off-target plant genome editing and methods for its limitation. Emerg. Top. Life Sci. 2017, 1, 231–240. [Google Scholar] [CrossRef]
- Peng, R.X.; Lin, G.G.; Li, J.M. Potential pitfalls of CRISPR/Cas9-mediated genome editing. Febs. J. 2016, 283, 1218–1231. [Google Scholar] [CrossRef]
- Kim, D.; Bae, S.; Park, J.; Kim, E.; Kim, S.; Yu, H.R.; Hwang, J.; Kim, J.I.; Kim, J.S. Digenome-seq: Genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat. Methods 2015, 12, 237–243. [Google Scholar] [CrossRef]
- Vakulskas, C.A.; Dever, D.P.; Rettig, G.R.; Turk, R.; Jacobi, A.M.; Collingwood, M.A.; Bode, N.M.; McNeill, M.S.; Yan, S.Q.; Camarena, J.; et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018, 24, 1216–1224. [Google Scholar] [CrossRef]
- Anjanappa, R.B.; Gruissem, W. Current progress and challenges in crop genetic transformation. J. Plant Physiol. 2021, 261, 153411. [Google Scholar] [CrossRef]
- Gonzalez, M.N.; Massa, G.A.; Andersson, M.; Oneto, C.A.D.; Turesson, H.; Storani, L.; Olsson, N.; Falt, A.S.; Hofvander, P.; Feingold, S.E. Comparative potato genome editing: Agrobacterium tumefaciens-mediated transformation and protoplasts transfection delivery of CRISPR/Cas9 components directed to StPPO2 gene. Plant Cell Tissue Organ Cult. 2021, 145, 291–305. [Google Scholar] [CrossRef]
- Zlobin, N.E.; Lebedeva, M.V.; Taranov, V.V. CRISPR/Cas9 genome editing through in planta transformation. Crit. Rev. Biotechnol. 2020, 40, 153–168. [Google Scholar] [CrossRef]
- Bhowmik, P.; Konkin, D.; Polowick, P.; Hodgins, C.L.; Subedi, M.; Xiang, D.; Yu, B.; Patterson, N.; Rajagopalan, N.; Babic, V.; et al. CRISPR/Cas9 gene editing in legume crops: Opportunities and challenges. Legume Sci. 2021, 3, e96. [Google Scholar] [CrossRef]
- Cardi, T.; D’Agostino, N.; Tripodi, P. Genetic Transformation and Genomic Resources for Next-Generation Precise Genome Engineering in Vegetable Crops. Front. Plant Sci. 2017, 8, 241. [Google Scholar] [CrossRef]
- Kausch, A.P.; Nelson-Vasilchik, K.; Hague, J.; Mookkan, M.; Quemada, H.; Dellaporta, S.; Fragoso, C.; Zhang, Z.Y.J. Edit at will: Genotype independent plant transformation in the era of advanced genomics and genome editing. Plant Sci. 2019, 281, 186–205. [Google Scholar] [CrossRef]
- Lee, K.S.; Wang, K. Strategies for genotype-flexible plant transformation. Curr. Opin. Biotechnol. 2023, 79, 102848. [Google Scholar] [CrossRef]
- Son, S.; Park, S.R. Challenges Facing CRISPR/Cas9-Based Genome Editing in Plants. Front. Plant Sci. 2022, 13, 902413. [Google Scholar] [CrossRef]
- Jackson, M.A.; Anderson, D.J.; Birch, R.G. Comparison of Agrobacterium and particle bombardment using whole plasmid or minimal cassette for production of high-expressing, low-copy transgenic plants. Transgenic Res. 2013, 22, 143–151. [Google Scholar] [CrossRef]
- Vats, S.; Kumawat, S.; Kumar, V.; Patil, G.B.; Joshi, T.; Sonah, H.; Sharma, T.R.; Deshmukh, R. Genome Editing in Plants: Exploration of Technological Advancements and Challenges. Cells 2019, 8, 1386. [Google Scholar] [CrossRef]
- Basak, J.; Nithin, C. Targeting Non-Coding RNAs in Plants with the CRISPR-Cas Technology is a Challenge yet Worth Accepting. Front. Plant Sci. 2015, 6, 1001. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-Guided Platform for Sequence-Specific Control of Gene Expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Liu, S.J.; Lim, D.A. Modulating the expression of long non-coding RNAs for functional studies. EMBO Rep. 2018, 19, e46955. [Google Scholar] [CrossRef]
- Tycko, J.; Wainberg, M.; Marinov, G.K.; Ursu, O.; Hess, G.T.; Ego, B.K.; Aradhana; Li, A.; Truong, A.; Trevino, A.E.; et al. Mitigation of off-target toxicity in CRISPR-Cas9 screens for essential non-coding elements. Nat. Commun. 2019, 10, 4063. [Google Scholar] [CrossRef]
- Zischewski, J.; Fischer, R.; Bortesi, L. Detection of on-target and off-target mutations generated by CRISPR/Cas9 and other sequence-specific nucleases. Biotechnol. Adv. 2017, 35, 95–104. [Google Scholar] [CrossRef]
- Hoijer, I.; Johansson, J.; Gudmundsson, S.; Chin, C.S.; Bunikis, I.; Haggqvist, S.; Emmanouilidou, A.; Wilbe, M.; den Hoed, M.; Bondeson, M.L.; et al. Amplification-free long-read sequencing reveals unforeseen CRISPR-Cas9 off-target activity. Genome Biol. 2020, 21, 290. [Google Scholar] [CrossRef]
- Teotia, S.; Singh, D.; Tang, X.Q.; Tang, G.L. Essential RNA-Based Technologies and Their Applications in Plant Functional Genomics. Trends Biotechnol. 2016, 34, 106–123. [Google Scholar] [CrossRef]
- Durr, J.; Papareddy, R.; Nakajima, K.; Gutierrez-Marcos, J. Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci. Rep. 2018, 8, 4443. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L.H. The Critical Role of miRNAs in Regulation of Flowering Time and Flower Development. Genes 2020, 11, 319. [Google Scholar] [CrossRef]
- Shin, S.Y.; Shin, C. Regulatory non-coding RNAs in plants: Potential gene resources for the improvement of agricultural traits. Plant Biotechnol. Rep. 2016, 10, 35–47. [Google Scholar] [CrossRef]
- Han, H.A.; Pang, J.K.S.; Soh, B.S. Mitigating off-target effects in CRISPR/Cas9-mediated in vivo gene editing. J. Mol. Med. 2020, 98, 615–632. [Google Scholar] [CrossRef]
- Salika, R.; Riffat, J. Abiotic stress responses in maize: A review. Acta Physiol. Plant. 2021, 43, 130. [Google Scholar] [CrossRef]
- Mao, Y.F.; Botella, J.R.; Liu, Y.G.; Zhu, J.K. Gene editing in plants: Progress and challenges. Natl. Sci. Rev. 2019, 6, 421–437. [Google Scholar] [CrossRef]
- Xu, R.F.; Li, H.; Qin, R.Y.; Li, J.; Qiu, C.H.; Yang, Y.C.; Ma, H.; Li, L.; Wei, P.C.; Yang, J.B. Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci. Rep. 2015, 5, 11491. [Google Scholar] [CrossRef]
- Cermak, T.; Baltes, N.J.; Cegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef]
- Zhu, H.C.; Li, C.; Gao, C.X. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 782. [Google Scholar] [CrossRef]
- Mahfouz, M.M.; Piatek, A.; Stewart, C.N. Genome engineering via TALENs and CRISPR/Cas9 systems: Challenges and perspectives. Plant Biotechnol. J. 2014, 12, 1006–1014. [Google Scholar] [CrossRef]
- Eki, R.; She, J.; Parlak, M.; Benamar, M.; Du, K.P.; Kumar, P.; Abbas, T. A robust CRISPR-Cas9-based fluorescent reporter assay for the detection and quantification of DNA double-strand break repair. Nucleic Acids Res. 2020, 48, e126. [Google Scholar] [CrossRef]
- Bradford, K.J.; Van Deynze, A.; Gutterson, N.; Parrott, W.; Strauss, S.H. Regulating transgenic crops sensibly: Lessons from plant breeding, biotechnology and genomics. Nat. Biotechnol. 2005, 23, 439–444. [Google Scholar] [CrossRef]
- Alonge, M.; Wang, X.G.; Benoit, M.; Soyk, S.; Pereira, L.; Zhang, L.; Suresh, H.; Ramakrishnan, S.; Maumus, F.; Ciren, D.; et al. Major Impacts of Widespread Structural Variation on Gene Expression and Crop Improvement in Tomato. Cell 2020, 182, 145–161. [Google Scholar] [CrossRef]
- Zhou, H.B.; Liu, B.; Weeks, D.P.; Spalding, M.H.; Yang, B. Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res. 2014, 42, 10903–10914. [Google Scholar] [CrossRef]
- Lorenzo, C.D.; Debray, K.; Herwegh, D.; Develtere, W.; Impens, L.; Schaumont, D.; Vandeputte, W.; Aesaert, S.; Coussens, G.; De Boe, Y.; et al. BREEDIT: A multiplex genome editing strategy to improve complex quantitative traits in maize. Plant Cell 2023, 35, 1160. [Google Scholar] [CrossRef]
- Modrzejewski, D.; Hartung, F.; Sprink, T.; Krause, D.; Kohl, C.; Schiemann, J.; Wilhelm, R. What is the available evidence for the application of genome editing as a new tool for plant trait modification and the potential occurrence of associated off-target effects: A systematic map protocol. Environ. Evid. 2018, 7, 18. [Google Scholar] [CrossRef]
- Zhang, D.; Zhang, H.; Li, T.; Chen, K.; Qiu, J.-L.; Gao, C. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017, 18, 191. [Google Scholar] [CrossRef]
- Zhao, Z.; Shang, P.; Mohanraju, P.; Geijsen, N. Prime editing: Advances and therapeutic applications. Trends Biotechnol. 2023, 41, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, A.; Gill, R.A.; Hassan, M.U.; Mahmood, A.; Qari, S.; Zaman, Q.U.; Ilyas, M.; Aamer, M.; Batool, M.; Li, H. A critical review: Recent advancements in the use of CRISPR/Cas9 technology to enhance crops and alleviate global food crises. Curr. Issues Mol. Biol. 2021, 43, 1950–1976. [Google Scholar] [CrossRef] [PubMed]
- Wada, N.; Osakabe, K.; Osakabe, Y. Expanding the plant genome editing toolbox with recently developed CRISPR–Cas systems. Plant Physiol. 2022, 188, 1825–1837. [Google Scholar] [CrossRef] [PubMed]
- Slesarenko, Y.S.; Lavrov, A.V.; Smirnikhina, S.A. Off-target effects of base editors: What we know and how we can reduce it. Curr. Genet. 2022, 68, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Ghogare, R.; Ludwig, Y.; Bueno, G.M.; Slamet-Loedin, I.H.; Dhingra, A. Genome editing reagent delivery in plants. Transgenic Res. 2021, 30, 321–335. [Google Scholar] [CrossRef]
- Wijerathna-Yapa, A.; Ramtekey, V.; Ranawaka, B.; Basnet, B.R. Applications of in vitro tissue culture technologies in breeding and genetic improvement of wheat. Plants 2022, 11, 2273. [Google Scholar] [CrossRef]
- Lee, J.; Chung, J.H.; Kim, H.M.; Kim, D.W.; Kim, H. Designed nucleases for targeted genome editing. Plant Biotechnol. J. 2016, 14, 448–462. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Cong, L.; Zhou, Y.; Cunniff, M.M.; Feng, G.P.; Zhang, F. A transcription activator-like effector toolbox for genome engineering. Nat. Protoc. 2012, 7, 171–192. [Google Scholar] [CrossRef]
- Morbitzer, R.; Romer, P.; Boch, J.; Lahaye, T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 21617–21622. [Google Scholar] [CrossRef]
- Miller, J.C.; Tan, S.Y.; Qiao, G.J.; Barlow, K.A.; Wang, J.B.; Xia, D.F.; Meng, X.D.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–148. [Google Scholar] [CrossRef]
- Christian, M.L.; Demorest, Z.L.; Starker, C.G.; Osborn, M.J.; Nyquist, M.D.; Zhang, Y.; Carlson, D.F.; Bradley, P.; Bogdanove, A.J.; Voytas, D.F. Targeting G with TAL Effectors: A Comparison of Activities of TALENs Constructed with NN and NK Repeat Variable Di-Residues. PLoS ONE 2012, 7, e45383. [Google Scholar] [CrossRef]
- Xiao, A.; Wu, Y.D.; Yang, Z.P.; Hu, Y.Y.; Wang, W.Y.; Zhang, Y.T.; Kong, L.; Gao, G.; Zhu, Z.Y.; Lin, S.; et al. EENdb: A database and knowledge base of ZFNs and TALENs for endonuclease engineering. Nucleic Acids Res. 2013, 41, D415–D422. [Google Scholar] [CrossRef][Green Version]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, 7879. [Google Scholar] [CrossRef]
- Puchta, H.; Fauser, F. Synthetic nucleases for genome engineering in plants: Prospects for a bright future. Plant J. 2014, 78, 727–741. [Google Scholar] [CrossRef] [PubMed]
- Mackon, E.; Jeazet Dongho Epse Mackon, G.C.; Guo, Y.; Ma, Y.; Yao, Y.; Liu, P. Development and application of CRISPR/Cas9 to improve anthocyanin pigmentation in plants: Opportunities and perspectives. Plant Sci. 2023, 333, 111746. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.; Orbovic, V.; Jones, J.B.; Wang, N. Modification of the PthA4 effector binding elements in Type I CsLOB1 promoter using Cas9/sgRNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4:dCsLOB1.3 infection. Plant Biotechnol. J. 2016, 14, 1291–1301. [Google Scholar] [CrossRef]
- Jaganathan, D.; Ramasamy, K.; Sellamuthu, G.; Jayabalan, S.; Venkataraman, G. CRISPR for Crop Improvement: An Update Review. Front. Plant Sci. 2018, 9, 985. [Google Scholar] [CrossRef]
- Holkers, M.; Maggio, I.; Henriques, S.F.D.; Janssen, J.M.; Cathomen, T.; Goncalves, M.A.F.V. Adenoviral vector DNA for accurate genome editing with engineered nucleases. Nat. Methods 2014, 11, 1051–1057. [Google Scholar] [CrossRef]
- Tiwari, J.K.; Challam, C.; Chakrabarti, S.K.; Feingold, S.E. Climate-Smart Potato: An Integrated Breeding, Genomics, and Phenomics Approach. In Genomic Designing of Climate-Smart Vegetable Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 1–46. [Google Scholar]
- Wei, C.; Liu, J.; Yu, Z.; Zhang, B.; Gao, G.; Jiao, R. TALEN or Cas9—Rapid, efficient and specific choices for genome modifications. J. Genet. Genom. 2013, 40, 281–289. [Google Scholar] [CrossRef]
- Doyle, E.L.; Stoddard, B.L.; Voytas, D.F.; Bogdanove, A.J. TAL effectors: Highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 2013, 23, 390–398. [Google Scholar] [CrossRef][Green Version]
- Lee, C.M.; Cradick, T.J.; Fine, E.J.; Bao, G. Nuclease Target Site Selection for Maximizing On-target Activity and Minimizing Off-target Effects in Genome Editing. Mol. Ther. 2016, 24, 475–487. [Google Scholar] [CrossRef]
- Wright, D.A.; Li, T.; Yang, B.; Spalding, M.H. TALEN-mediated genome editing: Prospects and perspectives. Biochem. J. 2014, 462, 15–24. [Google Scholar] [CrossRef]
- Li, H.Y.; Yang, Y.; Hong, W.Q.; Huang, M.Y.; Wu, M.; Zhao, X. Applications of genome editing technology in the targeted therapy of human diseases: Mechanisms, advances and prospects. Signal Transduct. Target. Ther. 2020, 5, 1. [Google Scholar] [CrossRef]
- Castro, N.G.; Bjelic, J.; Malhotra, G.; Huang, C.; Alsaffar, S.H. Comparison of the Feasibility, Efficiency, and Safety of Genome Editing Technologies. Int. J. Mol. Sci. 2021, 22, 10355. [Google Scholar] [CrossRef]
- Zhu, C.F.; Bortesi, L.; Baysal, C.; Twyman, R.M.; Fischer, R.; Capell, T.; Schillberg, S.; Christou, P. Characteristics of Genome Editing Mutations in Cereal Crops. Trends Plant Sci. 2017, 22, 38–52. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, F.; Li, X.H.; Baller, J.A.; Qi, Y.P.; Starker, C.G.; Bogdanove, A.J.; Voytas, D.F. Transcription Activator-Like Effector Nucleases Enable Efficient Plant Genome Engineering. Plant Physiol. 2013, 161, 20–27. [Google Scholar] [CrossRef]
- Malzahn, A.; Lowder, L.; Qi, Y.P. Plant genome editing with TALEN and CRISPR. Cell Biosci. 2017, 7, 21. [Google Scholar] [CrossRef]
- Ansari, W.A.; Chandanshive, S.U.; Bhatt, V.; Nadaf, A.B.; Vats, S.; Katara, J.L.; Sonah, H.; Deshmukh, R. Genome Editing in Cereals: Approaches, Applications and Challenges. Int. J. Mol. Sci. 2020, 21, 4040. [Google Scholar] [CrossRef]
- Voytas, D.F.; Gao, C.X. Precision Genome Engineering and Agriculture: Opportunities and Regulatory Challenges. PLoS Biol. 2014, 12, e1001877. [Google Scholar] [CrossRef]
- Yau, Y.Y.; Stewart, C.N. Less is more: Strategies to remove marker genes from transgenic plants. BMC Biotechnol. 2013, 13, 36. [Google Scholar] [CrossRef]
- Yee, J.K. Off-target effects of engineered nucleases. FEBS J. 2016, 283, 3239–3248. [Google Scholar] [CrossRef]
- Gurushidze, M.; Hensel, G.; Hiekel, S.; Schedel, S.; Valkov, V.; Kumlehn, J. True-Breeding Targeted Gene Knock-Out in Barley Using Designer TALE-Nuclease in Haploid Cells. PLoS ONE 2014, 9, e92046. [Google Scholar] [CrossRef]
- Kopischke, S.; Schussler, E.; Althoff, F.; Zachgo, S. TALEN-mediated genome-editing approaches in the liverwort Marchantia polymorpha yield high efficiencies for targeted mutagenesis. Plant Methods 2017, 13, 20. [Google Scholar] [CrossRef]
- Singh, R.K.; Prasad, M. Advances in Agrobacterium tumefaciens-mediated genetic transformation of graminaceous crops. Protoplasma 2016, 253, 691–707. [Google Scholar] [CrossRef]
- Čermák, T.; Curtin, S.J.; Gil-Humanes, J.; Čegan, R.; Kono, T.J.; Konečná, E.; Belanto, J.J.; Starker, C.G.; Mathre, J.W.; Greenstein, R.L. A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 2017, 29, 1196–1217. [Google Scholar]
- Yan, Y.; Zhu, X.; Yu, Y.; Li, C.; Zhang, Z.; Wang, F. Nanotechnology strategies for plant genetic engineering. Adv. Mater. 2022, 34, 2106945. [Google Scholar]
- Govindan, G.; Ramalingam, S. Programmable Site-Specific Nucleases for Targeted Genome Engineering in Higher Eukaryotes. J. Cell Physiol. 2016, 231, 2380–2392. [Google Scholar] [CrossRef]
- Durai, S.; Mani, M.; Kandavelou, K.; Wu, J.; Porteus, M.H.; Chandrasegaran, S. Zinc finger nucleases: Custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005, 33, 5978–5990. [Google Scholar] [CrossRef]
- Mandell, J.G.; Barbas, C.F. Zinc finger tools: Custom DNA-binding domains for transcription factors and nucleases. Nucleic Acids Res. 2006, 34, W516–W523. [Google Scholar] [CrossRef]
- Hall, T.M.T. Multiple modes of RNA recognition by zinc finger proteins. Curr. Opin. Struct. Biol. 2005, 15, 367–373. [Google Scholar] [CrossRef]
- Cathomen, T.; Joung, J.K. Zinc-finger nucleases: The next generation emerges. Mol. Ther. 2008, 16, 1200–1207. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Kim, J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, A.; Plaisier, C.L.; Carroll, D.; Drews, G.N. Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc. Natl. Acad. Sci. USA 2005, 102, 2232–2237. [Google Scholar] [CrossRef]
- Maeder, M.L.; Thibodeau-Beganny, S.; Osiak, A.; Wright, D.A.; Anthony, R.M.; Eichtinger, M.; Jiang, T.; Foley, J.E.; Winfrey, R.J.; Townsend, J.A.; et al. Rapid “Open-Source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 2008, 31, 294–301. [Google Scholar] [CrossRef]
- Marton, I.; Zuker, A.; Shklarman, E.; Zeevi, V.; Tovkach, A.; Roffe, S.; Ovadis, M.; Tzfira, T.; Vainstein, A. Nontransgenic Genome Modification in Plant Cells. Plant Physiol. 2010, 154, 1079–1087. [Google Scholar] [CrossRef]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.D.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.L.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature. 2009, 459, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Tovkach, A.; Zeevi, V.; Tzfira, T. A toolbox and procedural notes for characterizing novel zinc finger nucleases for genome editing in plant cells. Plant J. 2009, 57, 747–757. [Google Scholar] [CrossRef]
- Zhang, F.; Maeder, M.L.; Unger-Wallace, E.; Hoshaw, J.P.; Reyon, D.; Christian, M.; Li, X.H.; Pierick, C.J.; Dobbs, D.; Peterson, T.; et al. High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. USA 2010, 107, 12028–12033. [Google Scholar] [CrossRef]
- De Pater, S.; Neuteboom, L.W.; Pinas, J.E.; Hooykaas, P.J.; van der Zaal, B.J. ZFN-induced mutagenesis and gene-targeting in Arabidopsis through Agrobacterium-mediated floral dip transformation. Plant Biotechnol. J. 2009, 7, 821–835. [Google Scholar] [CrossRef]
- Wu, J.; Kandavelou, K.; Chandrasegaran, S. Custom-designed zinc finger nucleases: What is next? Cell. Mol. Life Sci. 2007, 64, 2933–2944. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, J.S.; Lang, Z.B.; Botella, J.R.; Zhu, J.K. Genome Editing-Principles and Applications for Functional Genomics Research and Crop Improvement. Crit. Rev. Plant Sci. 2017, 36, 291–309. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhao, J.; Cai, Q.L.; Liu, G.C.; Wang, J.R.; Zhao, Z.H.; Liu, P.; Dai, L.; Yan, G.J.; Wang, W.J.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef]
- Zhang, X.H.; Tee, L.Y.; Wang, X.G.; Huang, Q.S.; Yang, S.H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 2015, 4, e264. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.M.; Yao, Y.F.; Zhang, Y.C.; Fan, G.F. CRISPR system: Discovery, development and off-target detection. Cell. Signal. 2020, 70, 109577. [Google Scholar] [CrossRef]
- Tzfira, T.; Weinthal, D.; Marton, I.; Zeevi, V.; Zuker, A.; Vainstein, A. Genome modifications in plant cells by custom-made restriction enzymes. Plant Biotechnol. J. 2012, 10, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Welker, C.M.; Balasubramanian, V.K.; Petti, C.; Rai, K.M.; DeBolt, S.; Mendu, V. Engineering Plant Biomass Lignin Content and Composition for Biofuels and Bioproducts. Energies 2015, 8, 7654–7676. [Google Scholar] [CrossRef]
- Zaidi, S.S.E.A.; Mansoor, S. Viral Vectors for Plant Genome Engineering. Front. Plant Sci. 2017, 8, 539. [Google Scholar] [CrossRef]
- Chandrasegaran, S.; Carroll, D. Origins of programmable nucleases for genome engineering. J. Mol. Biol. 2016, 428, 963–989. [Google Scholar]
- Qi, Y. High efficient genome modification by designed zinc finger nuclease. In Advances in New Technology for Targeted Modification of Plant Genomes; Springer: New York, NY, USA, 2015; pp. 39–53. [Google Scholar]
- Osborn, M.J.; DeFeo, A.P.; Blazar, B.R.; Tolar, J. Synthetic zinc finger nuclease design and rapid assembly. Hum. Gene Ther. 2011, 22, 1155–1165. [Google Scholar]
- Yasafova, D.; Gankin, D.; Sharapov, Y.; Bright, C.; Driss, F.; Campi, F. panCRISPR Toolbox—A Deep Learning Approach to Improve CRISPR/Cas Experiments. 2022. Available online: https://www.mdsi.tum.de/fileadmin/w00cet/di-lab/pdf/Helmholtz_AI_WS2021_Final_Report.pdf (accessed on 22 February 2022).
- Matchett-Oates, L. Production of Novel Medicinal Cannabis Using Genome Editing Technology. Ph.D. Thesis, La Trobe University, Bundoora, Australia, 2022. [Google Scholar]
- Gao, C. Genome engineering for crop improvement and future agriculture. Cell 2021, 184, 1621–1635. [Google Scholar] [CrossRef]
- Wolt, J.D.; Wang, K.; Yang, B. The regulatory status of genome-edited crops. Plant Biotechnol. J. 2016, 14, 510–518. [Google Scholar] [CrossRef]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome edited crops touch the market: A view on the global development and regulatory environment. Front. Plant Sci. 2020, 11, 586027. [Google Scholar] [CrossRef]
- Bridgers, M. Genetically modified organisms and the precautionary principle: How the GMO dispute before the world trade organization could decide the fate of international GMO regulation. Temp. Envtl. L. Tech. J. 2003, 22, 171. [Google Scholar]
- Hundleby, P.; Harwood, W. Regulatory Constraints and Differences of Genome-Edited Crops Around the Globe. In Genome Editing: Current Technology Advances and Applications for Crop Improvement; Springer: Berlin/Heidelberg, Germany, 2022; pp. 319–341. [Google Scholar]
- Idris, S.H.; Mat Jalaluddin, N.S.; Chang, L.W. Ethical and legal implications of gene editing in plant breeding: A systematic literature review. J. Zhejiang Univ. Sci. B 2023, 1–13. [Google Scholar] [CrossRef]
- Wolt, J.D.; Wolf, C. Policy and governance perspectives for regulation of genome edited crops in the United States. Front. Plant Sci. 2018, 9, 1606. [Google Scholar] [CrossRef]
- Hartung, F.; Schiemann, J. Precise plant breeding using new genome editing techniques: Opportunities, safety and regulation in the EU. Plant J. 2014, 78, 742–752. [Google Scholar] [CrossRef]
- Eckerstorfer, M.F.; Engelhard, M.; Heissenberger, A.; Simon, S.; Teichmann, H. Plants developed by new genetic modification techniques—Comparison of existing regulatory frameworks in the EU and non-EU countries. Front. Bioeng. Biotechnol. 2019, 7, 26. [Google Scholar] [CrossRef]
- Lassoued, R.; Phillips, P.W.; Macall, D.M.; Hesseln, H.; Smyth, S.J. Expert opinions on the regulation of plant genome editing. Plant Biotechnol. J. 2021, 19, 1104–1109. [Google Scholar] [CrossRef]
- Turnbull, C.; Lillemo, M.; Hvoslef-Eide, T.A. Global regulation of genetically modified crops amid the gene edited crop boom—A review. Front. Plant Sci. 2021, 12, 630396. [Google Scholar] [CrossRef]
- Levidow, L.; Carr, S. GM Food on Trial: Testing European Democracy; Routledge: Oxfordshire, UK, 2009. [Google Scholar]
- Dederer, H.-G. Options for the Regulation of Genome Edited Plants–Framing the Issues. In Genome Editing in Agriculture; Nomos: Baden, Germany, 2019; pp. 75–122. [Google Scholar]
- Kawall, K.; Cotter, J.; Then, C. Broadening the GMO risk assessment in the EU for genome editing technologies in agriculture. Environ. Sci. Eur. 2020, 32, 106. [Google Scholar] [CrossRef]
- Entine, J.; Felipe, M.S.S.; Groenewald, J.-H.; Kershen, D.L.; Lema, M.; McHughen, A.; Nepomuceno, A.L.; Ohsawa, R.; Ordonio, R.L.; Parrott, W.A. Regulatory approaches for genome edited agricultural plants in select countries and jurisdictions around the world. Transgenic Res. 2021, 30, 551–584. [Google Scholar] [CrossRef] [PubMed]
- Ishii, T.; Araki, M. Consumer acceptance of food crops developed by genome editing. Plant Cell Rep. 2016, 35, 1507–1518. [Google Scholar] [CrossRef] [PubMed]
- Bartkowski, B.; Theesfeld, I.; Pirscher, F.; Timaeus, J. Snipping around for food: Economic, ethical and policy implications of CRISPR/Cas genome editing. Geoforum 2018, 96, 172–180. [Google Scholar]
- Bäckstrand, K. Civic science for sustainability: Reframing the role of experts, policy-makers and citizens in environmental governance. Glob. Environ. Politics 2003, 3, 24–41. [Google Scholar] [CrossRef]
- Sendhil, R.; Nyika, J.; Yadav, S.; Mackolil, J.; Prashat, G.; Workie, E.; Ragupathy, R.; Ragupathy, R.; Ramasundaram, P. Consumer Perception and Preference towards Genetically Modified (GM) Foods: Bibliometric Evidence and Policy Imperatives. agriRxiv 2021, 2021, 20210260196. [Google Scholar] [CrossRef]
- National Academies of Sciences, Engineering, and Medicine. The Current Biotechnology Regulatory System. In Preparing for Future Products of Biotechnology; National Academies Press (US): Washington, DC, USA, 2017. [Google Scholar]
- Ahmad, A.; Munawar, N.; Khan, Z.; Qusmani, A.T.; Khan, S.H.; Jamil, A.; Ashraf, S.; Ghouri, M.Z.; Aslam, S.; Mubarik, M.S. An outlook on global regulatory landscape for genome-edited crops. Int. J. Mol. Sci. 2021, 22, 11753. [Google Scholar]
- Hilder, V.A.; Boulter, D. Genetic engineering of crop plants for insect resistance—A critical review. Crop Prot. 1999, 18, 177–191. [Google Scholar] [CrossRef]
- Van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Malhotra, S.K. Horticultural crops and climate change: A review. Indian J. Agr. Sci. 2017, 87, 12–22. [Google Scholar] [CrossRef]
- Reguera, M.; Peleg, Z.; Blumwald, E. Targeting, metabolic pathways for genetic engineering abiotic stress-tolerance in crops. BBA-Gene Regul. Mech. 2012, 1819, 186–194. [Google Scholar] [CrossRef] [PubMed]
- Zsogon, A.; Peres, L.E.P.; Xiao, Y.J.; Yan, J.B.; Fernie, A.R. Enhancing crop diversity for food security in the face of climate uncertainty. Plant J. 2022, 109, 402–414. [Google Scholar] [CrossRef] [PubMed]
- DellaPenna, D. Nutritional genomics: Manipulating plant micronutrients to improve human health. Science 1999, 285, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Palve, A.; Joshi, C.; Srivastava, R.K. Crop biofortification for iron (Fe), zinc (Zn) and vitamin A with transgenic approaches. Heliyon 2019, 5, e01914. [Google Scholar] [CrossRef]
- Lemmon, Z.H.; Reem, N.T.; Dalrymple, J.; Soyk, S.; Swartwood, K.E.; Rodriguez-Leal, D.; Van Eck, J.; Lippman, Z.B. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 2018, 4, 766–770. [Google Scholar] [CrossRef]
- Wang, T.; Zhang, H.Y.; Zhu, H.L. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Hortic. Res. 2019, 6, 77. [Google Scholar] [CrossRef]
- Tiwari, M.; Trivedi, P.K.; Pandey, A. Emerging tools and paradigm shift of gene editing in cereals, fruits, and horticultural crops for enhancing nutritional value and food security. Food Energy Secur. 2021, 10, e258. [Google Scholar] [CrossRef]
- French, E.; Kaplan, I.; Lyer-Pascuzzi, A.; Nakatsu, C.H.; Enders, L. Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 2021, 7, 256–267. [Google Scholar] [CrossRef]
- Zha, F.C.; Rao, J.J.; Chen, B.C. Modification of pulse proteins for improved functionality and flavor profile: A comprehensive review. Compr. Rev. Food Sci. F 2021, 20, 3036–3060. [Google Scholar] [CrossRef]
- Lowry, G.V.; Avellan, A.; Gilbertson, L.M. Opportunities and challenges for nanotechnology in the agri-tech revolution. Nat. Nanotechnol. 2019, 14, 517–522. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).