Characterization of the Passion Fruit (Passiflora edulis Sim) bHLH Family in Fruit Development and Abiotic Stress and Functional Analysis of PebHLH56 in Cold Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of bHLH Members in Passion Fruit
2.2. Gene Structure, and Chromosomal Locations of PebHLHs
2.3. Cis-Acting Elements, Protein Interaction Network and Gene Collinearity of PebHLHs
2.4. Plant Materials, Transcriptome Sequencing, RNA Isolation and Reverse Transcription, Heat Map and qRT-PCR
2.5. Cloning the Promoter of PebHLH56 and Vector Construction
2.6. Cold Stress Treatment of Transgenic Lines
2.7. Detection of GUS Activity
3. Data Analysis and Results
3.1. Identification of bHLH Family of Passion Fruit
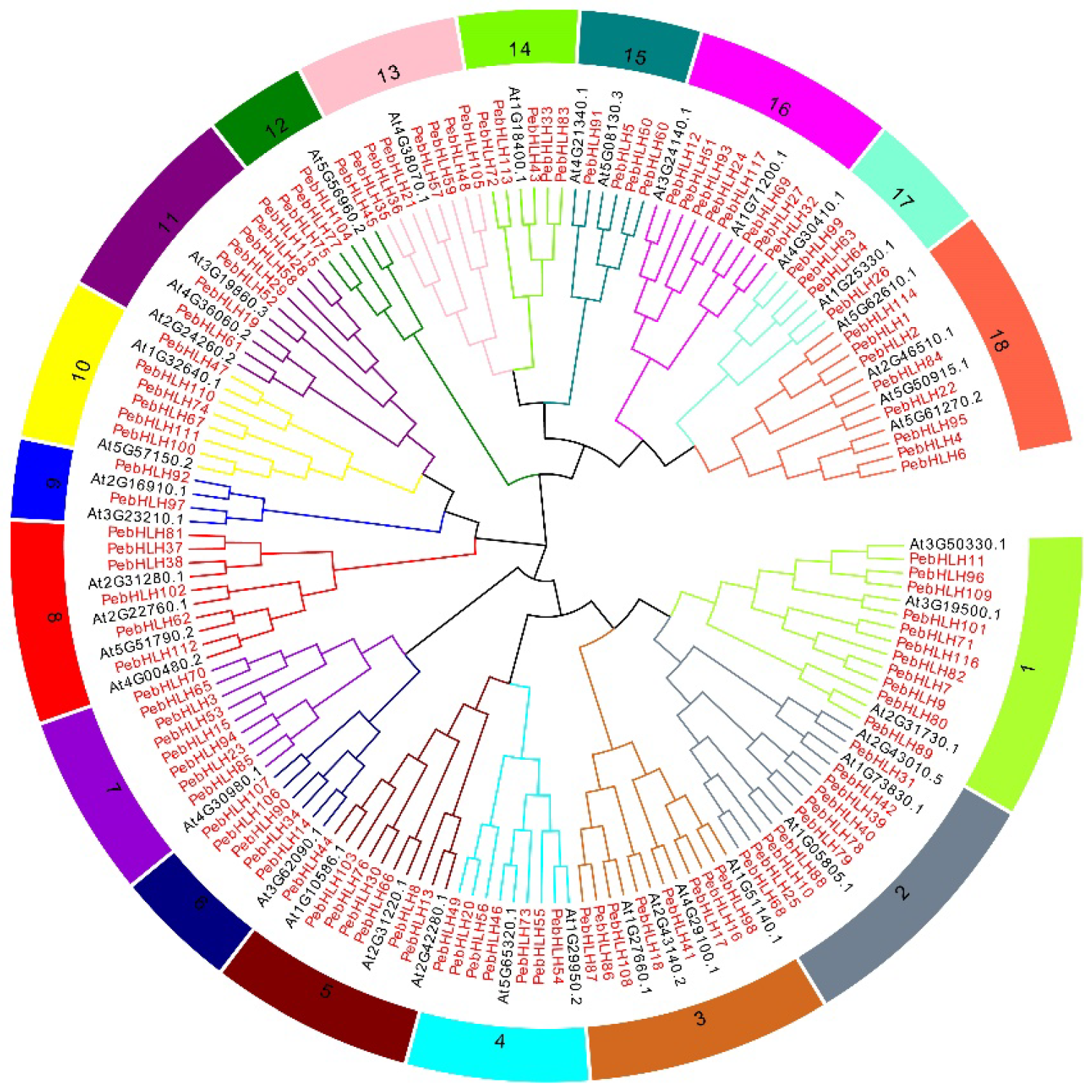
3.2. Evolutionary Analysis of the bHLH Genes
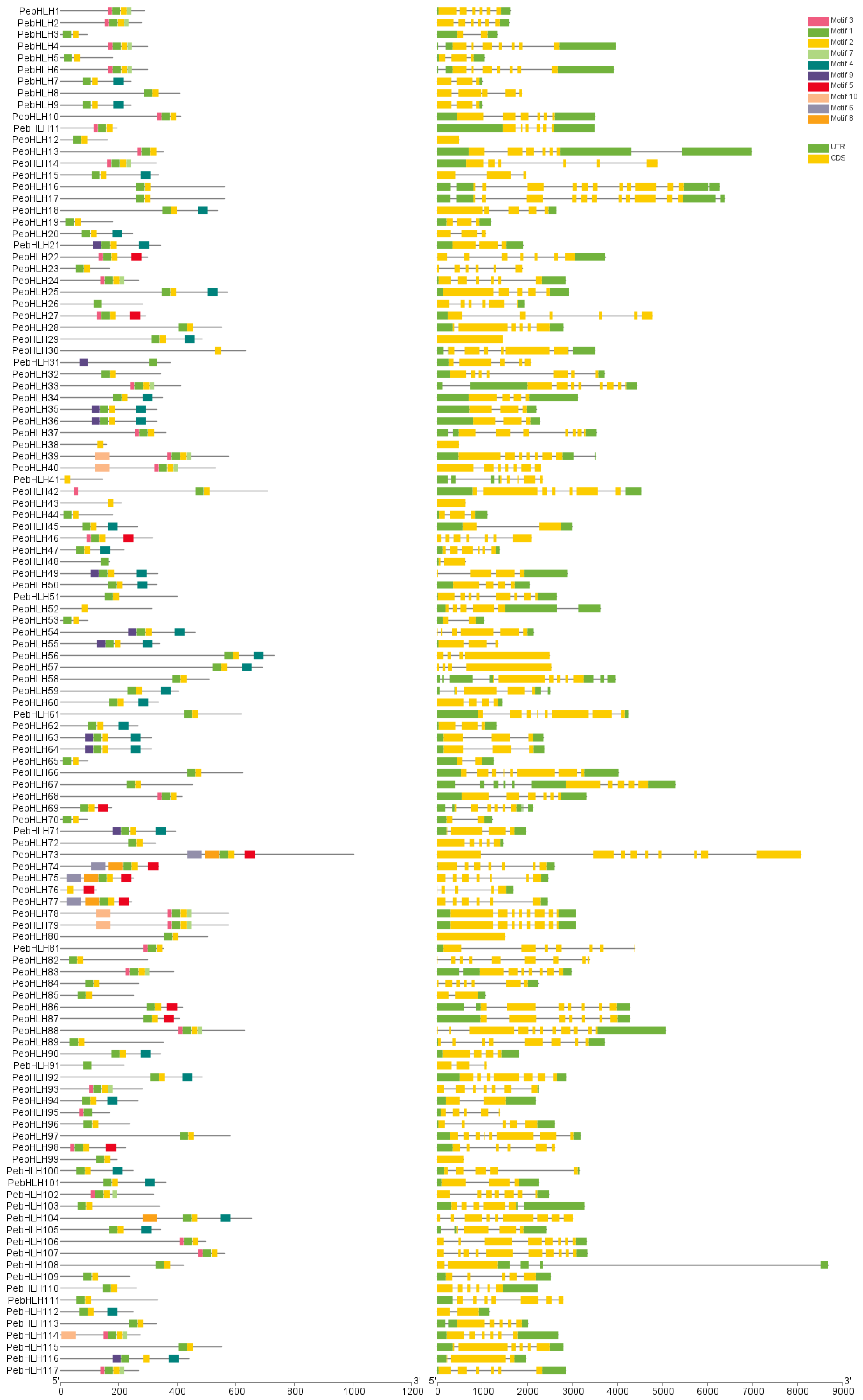
3.3. Analysis of Gene Structure and Conserved Motifs
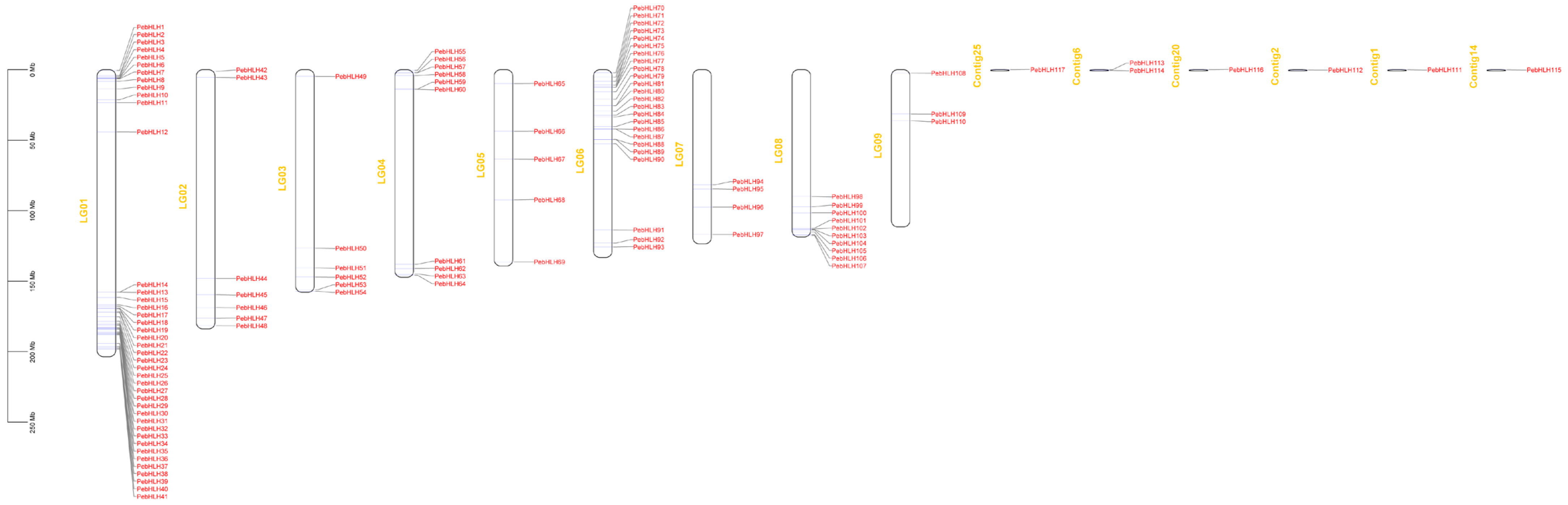
3.4. Chromosome Distribution of the PebHLHs
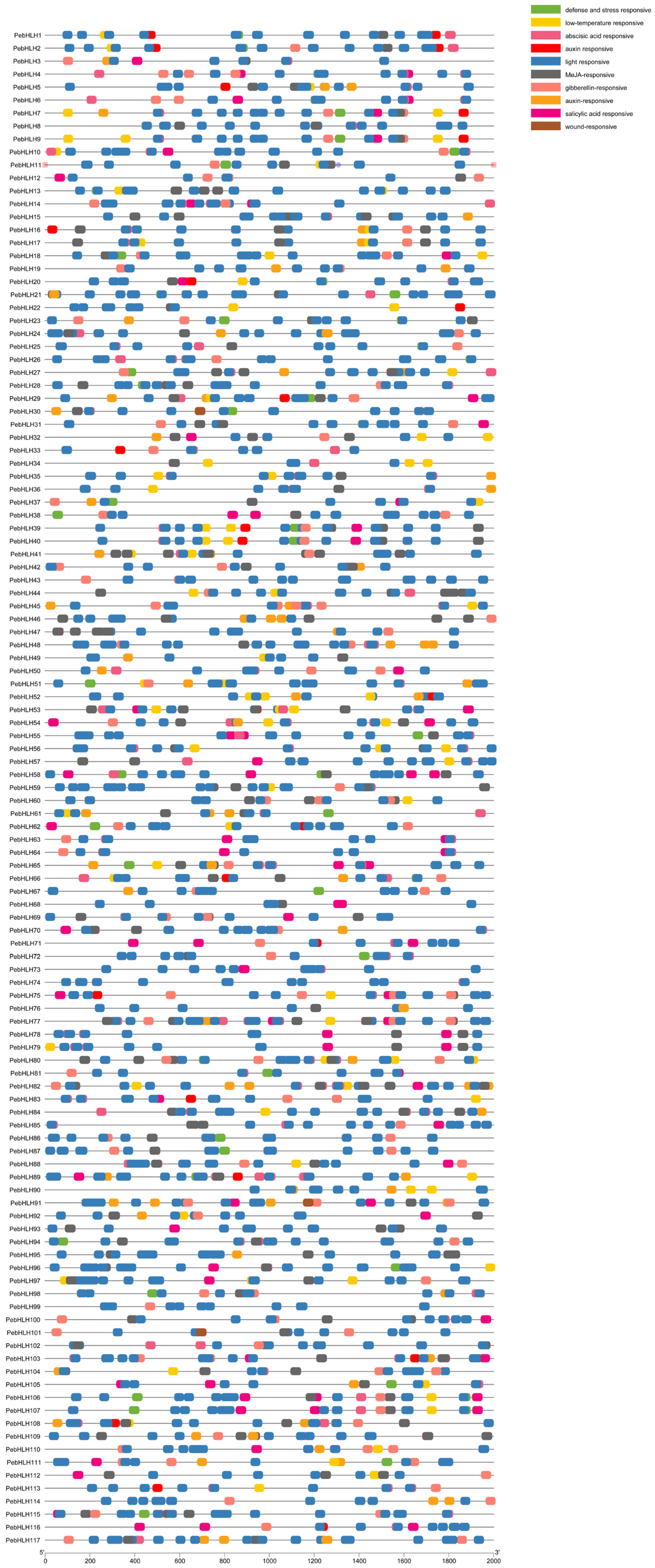
3.5. Promoter Analysis of PebHLH Genes
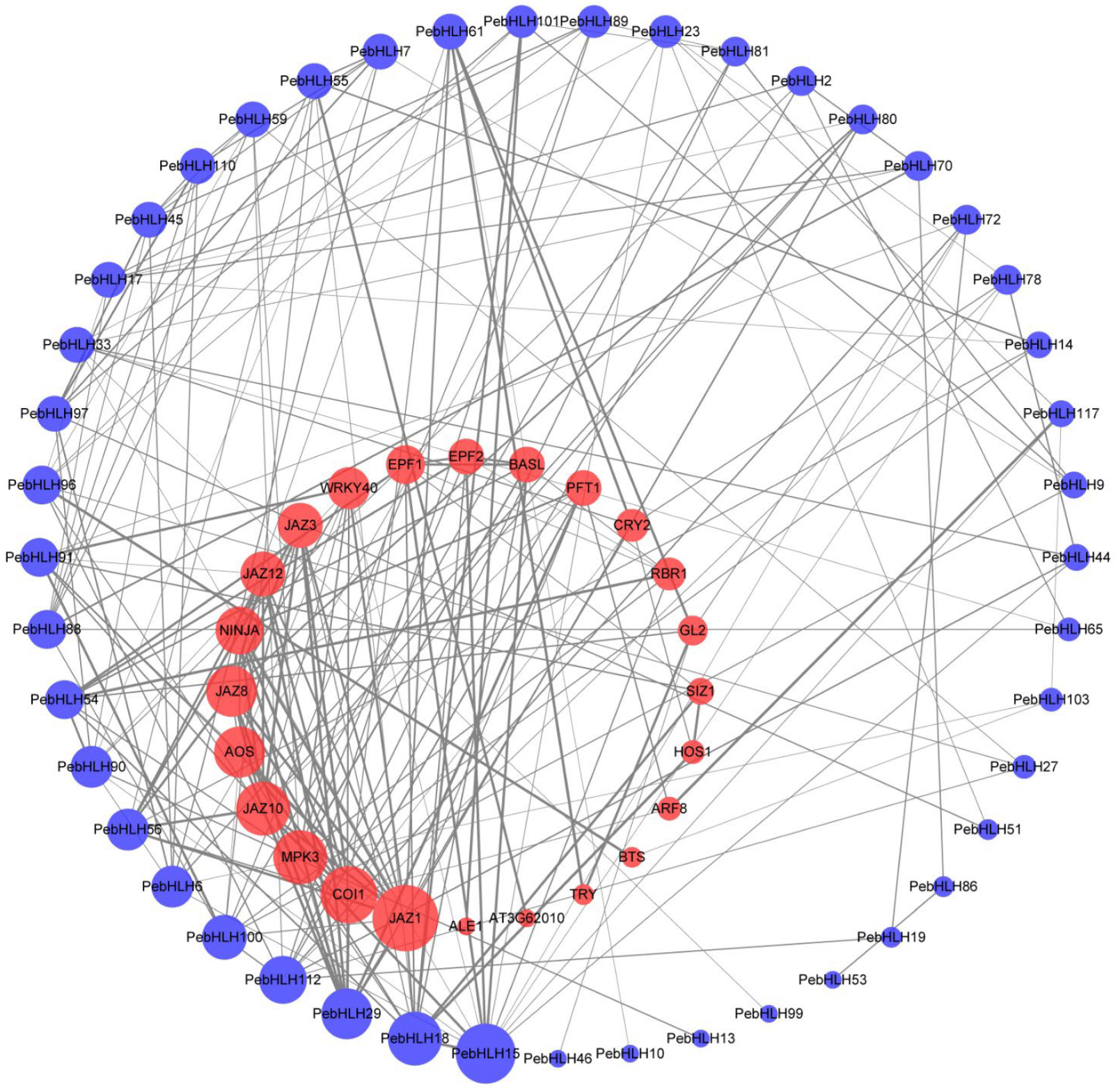
3.6. Interaction Networks Analysis of PebHLHs
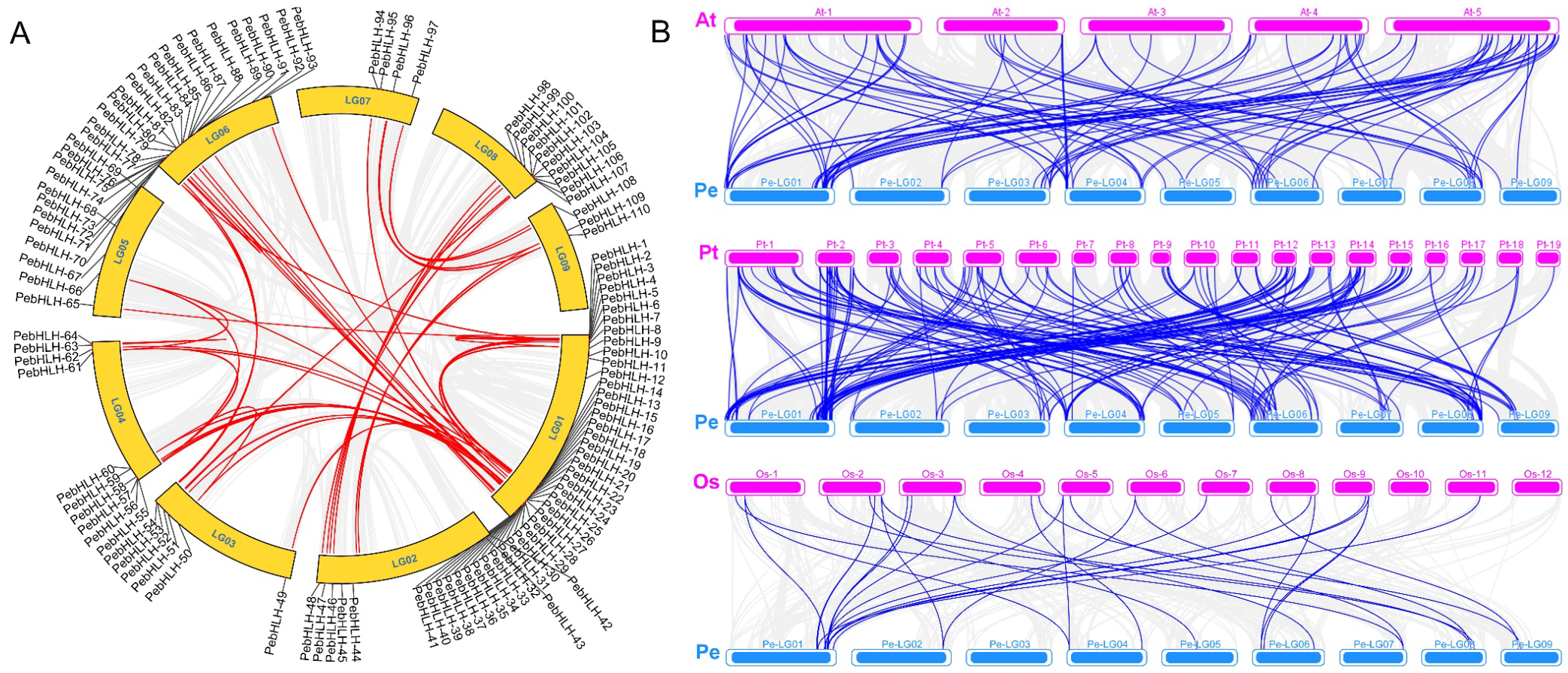
3.7. Collinearity Analysis of PebHLHs
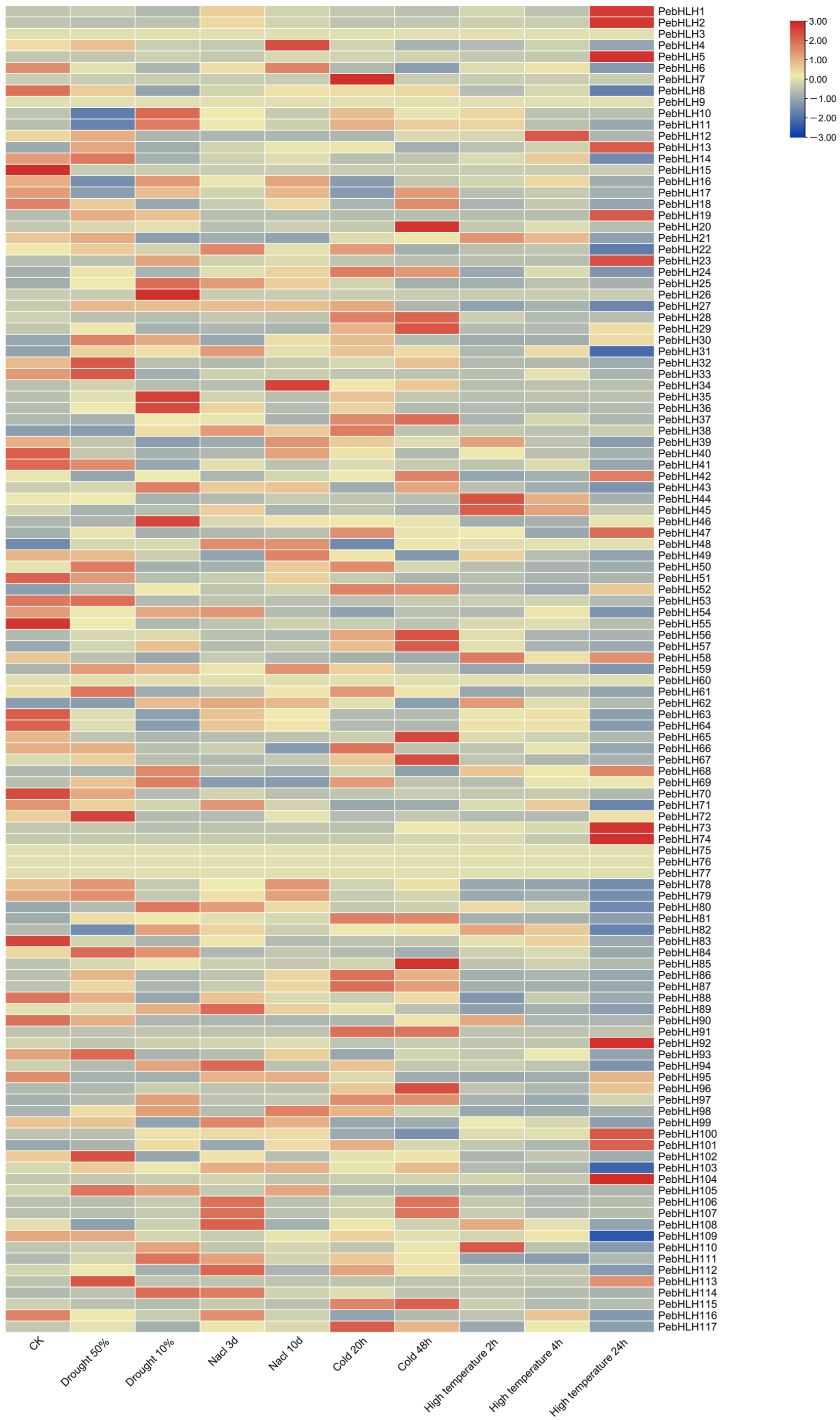
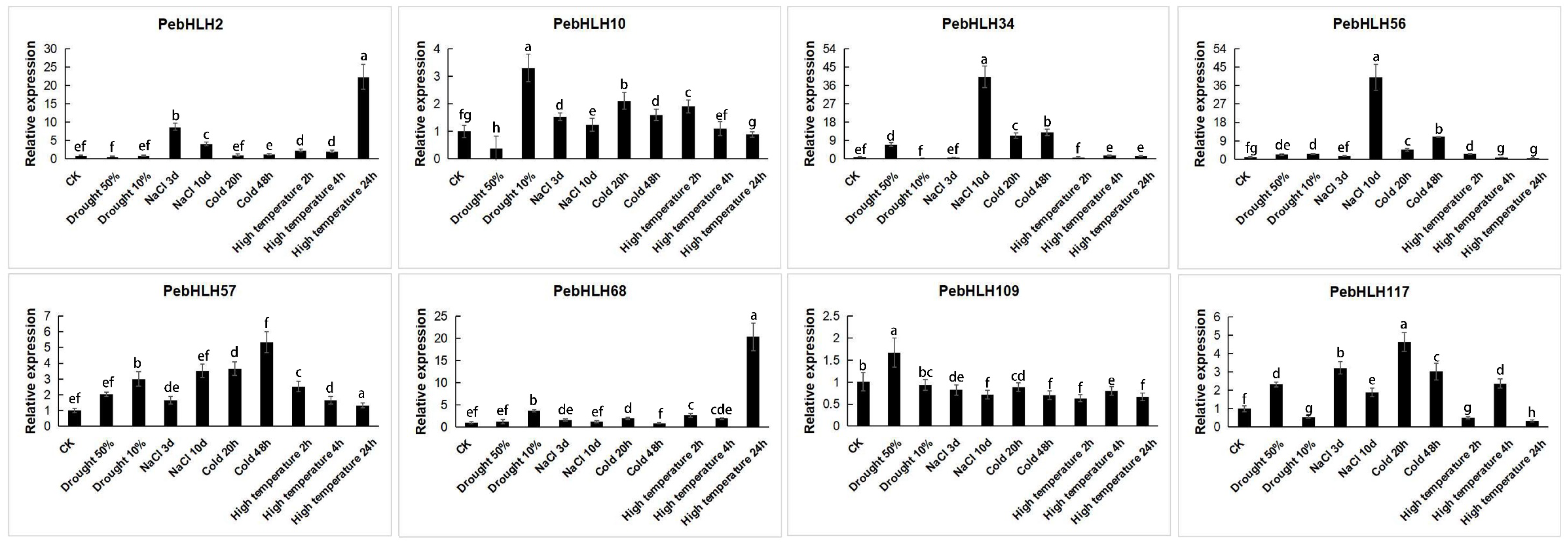
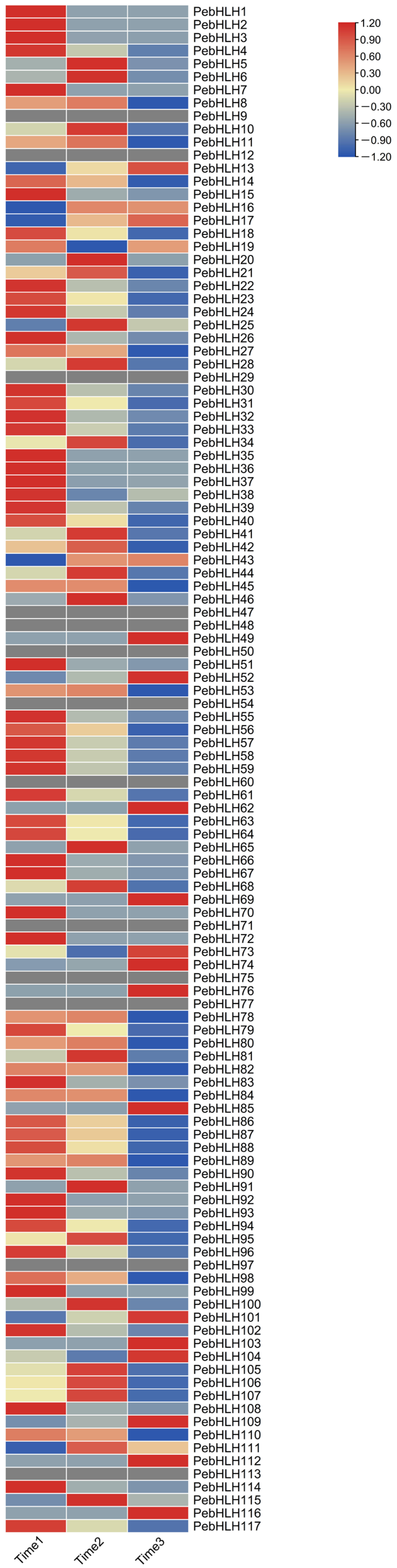
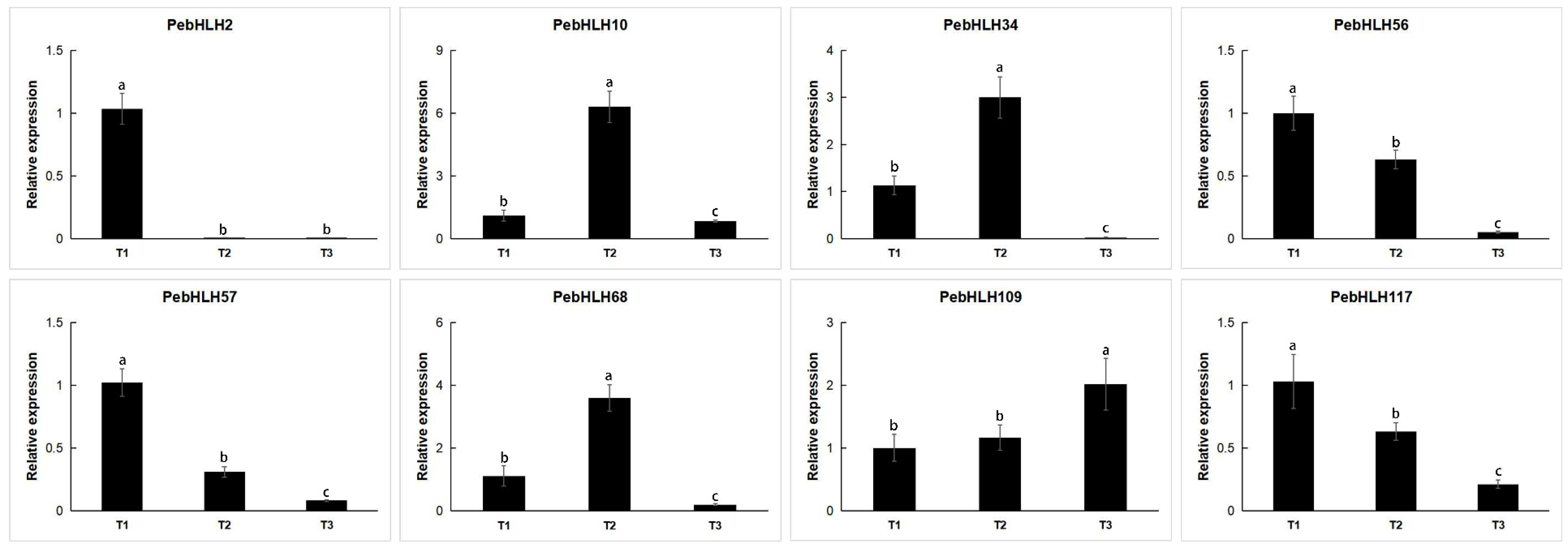
3.8. Expression Analysis of PebHLHs
3.9. Response of Transgenic Arabidopsis to Cold Stress
4. Discussion and Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Pérez-Rodríguez, P.; Riaño-Pachón, D.M.; Corrêa, L.G.; Rensing, S.A.; Kersten, B.; Mueller-Roeber, B. PlnTFDB: Updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010, 38, D822–D827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Jin, P.-J.; Tang, L.; Zhao, Y.; Gu, X.-C.; Gao, G.; Luo, J.-C. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Res. 2011, 9, D1114–D1117. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.J.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Wray, G.A.; Hahn, M.W.; Abouheif, E.; Balhoff, J.P.; Pizer, M.; Rockman, M.V.; Romano, L.A. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 2003, 20, 1377–1419. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, C.; Zourelidou, M.; Bevan, M.W. Plant transcription factor studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 127–150. [Google Scholar] [CrossRef]
- Jin, P.J.; Zhang, H.; Kong, L.; Gao, G.; Luo, J.C. PlantTFDB 3.0: A portal for the functional and evolutionary study of plant transcription factors. Nucleic Acids Res. 2014, 42, D1182–D1187. [Google Scholar] [CrossRef]
- Xu, Y.-H.; Liao, Y.-C.; Lu, F.-F.; Zhang, Z.; Sun, P.-W.; Gao, Z.-H.; Hu, K.-P.; Cun, S.; Jin, Y.; Wei, J.-H. Transcription Factor AsMYC2 Controls the Jasmonate-responsive Expression of ASS1 Regulating Sesquiterpene Biosynthesis in Aquilaria sinensis (Lour.) Gilg. Plant Cell Physiol. 2017, 58, 1924–1933. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Yin, J.; Chang, X.; Kasuga, T.; Bui, M.; Reid, M.S.; Jiang, C.Z. A basic helix-loop-helix transcription factor, PhFBH4, regulates flower senescence by modulating ethylene biosynthesis pathway in petunia. Hortic. Res. 2015, 2, 15059. [Google Scholar] [CrossRef]
- Murre, C.; McCaw, P.S.; Baltimore, D. A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and mycproteins. Cell 1989, 56, 777–783. [Google Scholar] [CrossRef]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional dependence, cliques, and predictive motifs in the bHLH protein domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Ellenberger, T.; Fass, D.; Arnaud, M.; Harrison, S.C. Crystal structure of transcription factor E47: E-box recognition by a basic region helix-loop-helix dimer. Comp. Study 1994, 8, 970–980. [Google Scholar] [CrossRef] [PubMed]
- Nesi, N.; Debeaujon, I.; Jond, C.; Pelletier, G.; Caboche, M.; Lepiniec, L. The TT8 gene encodes a basic helix-loop-helix domain protein required for expression of DFR and BAN genes in Arabidopsis siliques. Plant Cell 2000, 12, 1863–1878. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.Q.; Ahmad, N.; Tian, Y.Y.; Liu, J.Y.; Wang, L.Y.; Wang, G.; Liu, X.M.; Dong, Y.Y.F.; Wang, W.; Liu, W.C.; et al. Genome-Wide Identification, Expression Analysis, and Subcellular Localization of Carthamus tinctorius bHLH Transcription Factors. Int. J. Mol. Sci. 2019, 20, 3044. [Google Scholar] [CrossRef] [PubMed]
- Li, H.T.; Gao, W.L.; Xue, C.L.; Zhang, Y.; Liu, Z.G.; Zhang, Y.; Zhang, Y.; Meng, X.W.; Liu, M.J.; Zhao, J. Genome-wide analysis of the bHLH gene family in Chinese jujube (Ziziphus jujuba Mill.) and wild jujube. BMC Genom. 2019, 20, 568. [Google Scholar] [CrossRef]
- Aslam, M.; Jakada, B.H.; Fakher, B.; Greaves, J.G.; Niu, X.; Su, Z.; Cheng, Y.; Cao, S.J.; Wang, X.M.; Qin, Y. Genome-wide study of pineapple (Ananas comosus L.) bHLH transcription factors indicates that cryptochrome-interacting bHLH2 (AcCIB2) participates in flowering time regulation and abiotic stress response. BMC Genom. 2020, 21, 735. [Google Scholar] [CrossRef]
- Zhang, Z.S.; Chen, J.; Liang, C.L.; Liu, F.; Hou, X.L.; Zou, X.X. Genome-Wide Identification and Characterization of the bHLH Transcription Factor Family in Pepper (Capsicum annuum L.). Front. Genet. 2020, 11, 570156. [Google Scholar] [CrossRef]
- Wang, R.-Q.; Zhao, P.; Kong, N.; Lu, R.-Z.; Pei, Y.; Huang, C.-X.; Ma, H.-L.; Chen, Q. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes 2018, 22, 54. [Google Scholar] [CrossRef]
- Gao, C.; Sun, J.-L.; Wang, C.-Q.; Dong, Y.-M.; Xiao, S.-H.; Wang, X.-J.; Jiao, Z.-G. Genome-wide analysis of basic/helix-loop-helix gene family in peanut and assessment of its rolesin pod development. PLoS ONE. 2017, 27, e0181843. [Google Scholar]
- Zhu, L.; Zhao, M.-Z.; Chen, M.-Y.; Li, L.; Jiang, Y.; Liu, S.-Z.; Jiang, Y.; Wang, K.-Y.; Sun, C.-Y.; Chen, J.; et al. The bHLH gene family and its response to saline stress in Jilin ginseng, Panax ginseng C.A. Meyer. Mol. Genet. Genom. 2022, 295, 877–890. [Google Scholar] [CrossRef]
- Niu, X.; Guan, Y.-X.; Chen, S.-K.; Li, H.-F. Genome-wide analysis of basic helix-loop-helix (bHLH) transcription factors in Brachypodium distachyon. BMC Genom. 2017, 18, 619. [Google Scholar] [CrossRef] [PubMed]
- Kavas, M.; Baloğlu, M.-C.; Atabay, E.-S.; Ziplar, U.-T.; Daşgan, H.-Y.; Ünver, T. Genome-wide characterization and expression analysis of common bean bHLH transcription factors in response to excess salt concentration. Mol. Gen. Genom. 2016, 291, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Fan, H.-J.; Ling, H.-Q. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genom. 2015, 16, 9. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-X.; Duan, X.-P.; Jiang, H.-X.; Sun, Y.-J.; Tang, Y.-P.; Zhen, Y.; Guo, J.-K.; Liang, W.-Q.; Chen, L.; Yin, J.Y.; et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006, 141, 1167–1184. [Google Scholar] [CrossRef] [PubMed]
- Mao, K.; Dong, Q.-L.; Li, C.; Liu, C.-H.; Ma, F.-W. Genome Wide Identification and Characterization of Apple bHLH Transcription Factors and Expression Analysis in Response to Drought and Salt Stress. Front. Plant Sci. 2017, 11, 480. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.-T.; Lv, W.; Zhang, H.-S.; Ma, L.; Li, P.-H.; Ge, L.; Chang, L. Genome-wide analysis of the basic Helix-Loop-Helix (bHLH) transcription factor family in maize. BMC Plant Biol. 2018, 18, 235. [Google Scholar] [CrossRef]
- Song, X.-M.; Huang, Z.-N.; Duan, W.-K.; Ren, J.; Liu, T.-K.; Li, Y.; Hou, X.-L. Genome-wide analysis of the bHLH transcription factor family in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Gen. Genom. 2014, 289, 77–91. [Google Scholar] [CrossRef]
- Chen, X.-R.; Xiong, R.; Liu, H.-L.; Wu, M.; Chen, F.; Yan, H.-W.; Xiang, Y. Basic helix-loop-helix gene family: Genome wide identification, phylogeny, and expression in Moso bamboo. Plant Physiol. Biochem. 2018, 132, 104–119. [Google Scholar] [CrossRef]
- Wei, K.-F.; Chen, H.-Q. Comparative functional genomics analysis of bHLH gene family in rice, maize and wheat. BMC Plant Biol. 2018, 18, 309. [Google Scholar] [CrossRef]
- Huq, E.; Quail, P.-H. A phytochrome-interacting bHLH factor, functions as a negative regulator of phytochrome B signaling in Arabidopsis. EMBO J. 2022, 15, 2441–2450. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.-h.; Yang, K.-Y.; Kim, Y.-M.; Park, S.-Y.; Kim, S.-Y.; Soh, M.-S. Overexpression of PRE1 and itshomologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.-G.; Sun, C.-H.; Zhang, Q.-Y.; An, J.-P.; You, C.-X.; Hao, Y.-J. Glucose Sensor MdHXK1 Phosphorylates and Stabilizes MdbHLH3 to Promote Anthocyanin Biosynthesis in Apple. PLoS Genet. 2016, 25, e1006273. [Google Scholar]
- Bernhardt, C.; Lee, M.-M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3(GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef] [PubMed]
- Duek, P.-D.; Fankhauser, C. bHLH class transcription factors take Centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Yao, P.-F.; Li, C.-L.; Zhao, X.-R.; Li, M.-F.; Zhao, H.-X.; Guo, J.-Y.; Cai, Y.; Chen, H.; Wu, Q. Overexpression of a Tartary buckwheat gene, FtbHLH3, Enhances Drought/Oxidative Stress Tolerance in Transgenic Arabidopsis. Front. Plant Sci. 2017, 8, 625. [Google Scholar] [CrossRef]
- Yao, P.-F.; Sun, Z.-X.; Chen, Q.-L.; Zhao, X.-Y.; Li, M.-F.; Deng, R.-Y.; Huang, Y.-J.; Zhao, H.-Q.; Chen, H.; Wu, Q. Overexpression of Fagopyrum tataricum FtbHLH2 enhances tolerance to cold stress in transgenic Arabidopsis. Plant Physiol. Biochem. 2018, 125, 85–94. [Google Scholar] [CrossRef]
- Zhai, Y.-Q.; Zhang, L.-C.; Xia, C.; Fu, S.-L.; Zhao, G.-Y.; Jia, J.-Z.; Kong, X.Y. The wheat transcription factor, TabHLH39, improves tolerance to multiple abiotic stressors in transgenic plants. Biochem. Biophys. Res. Commun. 2016, 473, 1321–1327. [Google Scholar] [CrossRef]
- Jiang, Y.-Q.; Yang, B.; Deyholos, M.-K. Functional characterization of the Arabidopsis bHLH92 transcription factor in abiotic stress. Mol. Gen. Genom. 2009, 282, 503–516. [Google Scholar] [CrossRef]
- Huang, X.-S.; Wang, W.; Zhang, Q.; Liu, J.-H. A basic helix-loop-helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol. 2013, 162, 1178–1194. [Google Scholar] [CrossRef]
- Waseem, M.; Li, N.; Su, D.-D.; Chen, J.-X.; Li, Z.-G. Overexpression of a basic helix–loop–helix transcription factor gene, SlbHLH22, promotes early fowering and accelerates fruit ripening in tomato (Solanum lycopersicum L). Planta 2019, 250, 173–185. [Google Scholar] [CrossRef]
- Tan, C.; Qiao, H.-L.; Ma, M.; Wang, X.; Tian, Y.-Y.; Bai, S.; Hasi, A. Genome-Wide Identification and Characterization of Melon bHLH Transcription Factors in Regulation of Fruit Development. Plants 2021, 10, 2721. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Shen, Y.-H.; Zhou, P.; Fatima, M.; Lin, J.-S.; Yue, J.-J.; Zhang, X.-T.; Chen, L.-Y.; Ming, Y. Papaya CpbHLH1/2 regulate carotenoid biosynthesis-related genes during papaya fruit ripening. Hortic. Res. 2019, 6, 80. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.-M.; Xu, Y.; Wu, B.; Ma, F.-N.; Song, S. Comparative analysis of basic quality of passion fruits (Passiflora edulis sims) in Guangxi, Guizhou and Fujian, China. Bangladesh J. Bot. 2019, 48, 901–906. [Google Scholar]
- Costa, J.-L.; Jesus, O.-N.-D.; Oliverira, G.-A.-F.; Oliverira, E.-J.-D. Effect of selection on genetic variability in yellow passion fruit. Crop Breed. Appl. Biotechnol. 2012, 12, 253–260. [Google Scholar] [CrossRef]
- Song, S.; Zhang, D.-H.; Ma, F.-N.; Xing, W.-T.; Huang, D.-M.; Wu, B.; Chen, J.; Chen, D.; Xu, B.Q.; Xu, Y. Genome-Wide Identification and Expression Analyses of the Aquaporin Gene Family in Passion Fruit (Passiflora edulis), Revealing PeTIP3-2 to Be Involved in Drought Stress. Int. J. Mol. Sci. 2022, 23, 5720. [Google Scholar] [CrossRef]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4, molecular evolutionary genetics analysis MEGA software version 4.0. Mol. Biol. Evol. 2007, 24, 596–1599. [Google Scholar] [CrossRef]
- Chen, C.-J.; Chen, H.; Zhang, Y.; Thomas, H.-R.; Frank, M.-H.; He, Y.-H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1199–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.-V.-D.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Jefferson, R.-A.; Kavanagh, T.-A.; Bevan, M.-W. GUS fusions: Beta-glucuronidase as a sensitive and versatilegene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef]
- Xu, Y.; Jin, Z.-Q.; Xu, B.-Y.; Li, J.-Y.; Li, Y.-J.; Wang, X.-Y.; Wang, A.-B.; Hu, W.; Huang, D.-M.; Wei, Q.; et al. Identification of transcription factors interacting with a 1274bp promoter of MaPIP1;1 which confers high-level gene expression and drought stress Inducibility in transgenic Arabidopsis thaliana. BMC Plant Biol. 2020, 20, 278. [Google Scholar] [CrossRef]
- Xia, Z.-Q.; Huang, D.-M.; Zhang, S.-K.; Wang, W.-Q.; Ma, F.-N.; Wu, B.; Xu, Y.; Xu, B.-Q.; Chen, D.; Zou, M.-L.; et al. Chromosome-scale genome assembly provides insights into the evolution and flflavor synthesis of passion fruit (Passiflflora edulis Sims). Hortic. Res. 2021, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.-R.; Sun, B.-X.; He, H.-R.; Zhang, Y.-F.; Tian, H.-Y.; Wang, B.-S. Current Understanding of bHLH Transcription Factors in Plant Abiotic Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 4921. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liao, Y.-L.; Kim, S.-U.; Chen, Z.-X.; Nie, G.-P.; Cheng, S.-Y.; Ye, J.-B.; Xu, F. Genome-wide identification and characterization of bHLH family genes from Ginkgo biloba. Sci. Rep. 2020, 10, 13723. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-G.; Yu, N.; Wang, L.-J.; Gupta, D.-K.; He, Z.-L.; Wang, K.; Zhu, Z.-Q.; Yan, X.-C.; Li, T.-Q.; Yang, X.-E. The phytoremediation potential of bioenergy crop Ricinus communis for DDTs and cadmium co-contaminated soil. Bioresour. Technol. 2011, 102, 11034–11038. [Google Scholar] [CrossRef]
- Atchley, W.-R.; Fitch, W.-M. A natural classification of the basic helix–loop–helix class of transcription factors. Proc. Natl. Acad. Sci. USA 1997, 94, 5172–5176. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Adeleke, B.-S.; Babalola, O.-O. Oilseed crop sunflower (Helianthus annuus) as a source of food: Nutritional and health benefits. Food Sci. Nutr. 2020, 8, 4666–4684. [Google Scholar] [CrossRef]
- Xu, Z.-L.; Liu, X.-Q.; He, X.-L.; Xu, L.; Huang, Y.-H.; Shao, H.-B.; Zhang, D.-Y.; Tang, B.-P.; Ma, H.-X. The soybean basic helix-loop-helix transcription factor ORG3-like enhances cadmium tolerance via increased iron and reduced cadmium uptake and transport from roots to shoots. Front. Plant Sci. 2017, 8, 1098. [Google Scholar] [CrossRef]
- Ren, X.-Y.; Vorst, O.; Fiers, M.-W.-E.-J.; Stiekema, W.-J.; Nap, J.-P. In plants, highly expressed genes are the least compact. Trends Genet. 2006, 22, 528–532. [Google Scholar] [CrossRef]
- Jeffares, D.-C.; Penkett, C.-J.; Bahler, J. Rapidly regulated genes are intron poor. Trends Genet. 2008, 24, 375–378. [Google Scholar] [CrossRef]
- Mehan, M.-R.; Freimer, N.-B.; Ophoff, R.-A. A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum. Genom. 2004, 1, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Vision, T.-J.; Brown, D.-G.; Tanksley, S.-D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Paik, I.; Kathare, P.-K.; Kim, J.-I.; Huq, E. Expanding Roles of PIFs in Signal Integration from Multiple Processes. Mol. Plant 2017, 10, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Amoutzias, G.D.; Robertson, D.L.; Oliver, S.G.; Bornberg-Bauer, E. Convergent evolution of gene networks by single-gene duplications in higher eukaryotes. EMBO Rep. 2004, 5, 274–279. [Google Scholar] [CrossRef]
- Liu, H.-T.; Yu, X.-H.; Li, K.-W.; Klejnot, J.; Yang, H.-Y.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Liu, H.-T.; Wang, Q.; Liu, Y.-W.; Zhao, X.-Y.T.; Imaizumi, D.-E.; Somers, D.E.; Tobin, E.M.; Lin, C. Arabidopsis CRY2 and ZTL mediate blue-light regulation of the transcription factor CIB1 by distinct mechanisms. Proc. Natl. Acad.Sci. USA 2013, 110, 17582–17587. [Google Scholar] [CrossRef]
- Varaud, E.; Brioudes, F.; Szecsi, J.; Leroux, J.; Brown, S.; Perrot-Rechenmann, C. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 2011, 23, 973–983. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, Y.; Wang, B.-M.; Ran, Q.-J.; Zhang, J.-R. The bHLH family member ZmPTF1 regulates drought tolerance in maize by promoting root development and abscisic acid synthesis. J. Exp. Bot. 2019, 70, 5471–5486. [Google Scholar] [CrossRef]
- Qiu, J.-R.; Huang, Z.; Xiang, X.-Y.; Xu, W.-X.; Wang, J.-T.; Chen, J.; Song, L.; Xiao, Y.; Li, X.; Ma, J.; et al. MfbHLH38, a Myrothamnus flabellifolia bHLH transcription factor, confers tolerance to drought and salinity stresses in Arabidopsis. BMC Plant Biol. 2020, 20, 542. [Google Scholar] [CrossRef]
- Yu, C.-M.; Yan, M.; Dong, H.-Z.; Luo, J.; Ke, Y.-C.; Guo, A.-F.; Chen, Y.-H.; Zhang, J.; Huang, X.-S. Maize bHLH55 functions positively in salt tolerance through modulation of AsA biosynthesis by directly regulating GDP-mannose pathway genes. Plant Sci. 2021, 302, 110676. [Google Scholar] [CrossRef]
- Liu, Y.-J.; Ji, X.-Y.; Nie, X.-G.; Qu, M.; Heng, L.-Z.; Tan, Z.-L.; Zhao, H.-M.; Huo, L.; Liu, S.-N.; Zhang, B.; et al. Arabidopsis AtbHLH112regulates the expression of genes involved in abiotic stress tolerance bybinding to their E-box and GCG-box motifs. New Phytol. 2015, 207, 692–709. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Kim, H.-S.; Yu, T.; Zhang, A.-J.; Yang, Y.-F.; Liu, M.; Yu, W.-H.; Zhao, P.; Zhang, Q.-Q.; Cao, Q.-H.; et al. Identification and function analysis of bHLH genes in response to cold stress in sweetpotato. Plant Physiol. Biochem. 2021, 169, 224–235. [Google Scholar] [CrossRef] [PubMed]
- Zuo, Z.-F.; Sun, H.-J.; Lee, H.-Y.; Kang, H.-G. Identification of bHLH genes through genome-wide association study and antisense expression of ZjbHLH076/ZjICE1 influence tolerance to low temperature and salinity in Zoysia japonica. Plant Sci. 2021, 313, 111088. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Liu, J.-H.; Jia, C.-H.; Hu, W.; Song, S.; Xu, B.-Y.; Jin, Z.-Q. Overexpression of a Banana Aquaporin Gene MaPIP1;1 Enhances Tolerance to Multiple Abiotic Stresses in Transgenic Banana and Analysis of Its Interacting Transcription Factors. Front. Plant Sci. 2021, 12, 699230. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zheng, H.; Zhang, Q.; Liu, C.; Zhang, Z. Functional profiling of EcaICE1 transcription factor gene from Eucalyptus camaldulensis involved in cold response in tobacco plants. J. Plant Biochem. Biotechnol. 2014, 23, 141–150. [Google Scholar] [CrossRef]
- Xu, W.-R.; Jiao, Y.-T.; Li, R.-M.; Zhang, N.-B.; Xiao, D.-M.; Ding, X.-L.; Wang, Z.-P. Chinese wild-growing Vitis amurensis ICE1 and ICE2 encode MYC-type Bhlh transcription activators that regulate cold tolerance in Arabidopsis. PLoS ONE 2014, 9, e102303. [Google Scholar] [CrossRef]
- Seo, J.S.; Joo, J.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; Song, S.I.; Cheong, J.J.; Jong, S.L.; Kim, J.K.; Choi, Y.D. OsbHLH148, a basic helix-loop-helix protein, interacts with OsJAZ proteins in a jasmonate signaling pathway leading to drought tolerance in rice. Plant J. 2011, 65, 907–921. [Google Scholar] [CrossRef]
- Ko, D.K.; Lee, M.O.; Hahn, J.S.; Kim, B.G.; Hong, C.B. Submergence-inducible and circadian rhythmic basic helix-loop-helix protein gene in Nicotiana tabacum. J. Plant Physiol. 2009, 166, 1090–1100. [Google Scholar] [CrossRef]
- Liu, W.; Tai, H.-H.; Li, S.-S.; Gao, W.; Zhao, M.; Xie, C.-X.; Li, W.-X. bHLH122 is important for drought and osmotic stress resistance in Arabidopsis and in the repression of ABA catabolism. New Phytol. 2014, 201, 1192–1204. [Google Scholar] [CrossRef]
- Mao, T.-Y.; Liu, Y.-Y.; Zhu, H.-H.; Zhang, J.; Yang, J.X.; Fu, Q.; Wang, N.; Wang, Z. Genome-wide a nalyses of the bHLH gene family reveals structural and functional characteristics in the aquatic plant Nelumbo nucifera. PeerJ 2019, 7, e7153. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Zhou, W.; Ma, F.; Huang, D.; Xing, W.; Wu, B.; Sun, P.; Chen, D.; Xu, B.; Song, S. Characterization of the Passion Fruit (Passiflora edulis Sim) bHLH Family in Fruit Development and Abiotic Stress and Functional Analysis of PebHLH56 in Cold Stress. Horticulturae 2023, 9, 272. https://doi.org/10.3390/horticulturae9020272
Xu Y, Zhou W, Ma F, Huang D, Xing W, Wu B, Sun P, Chen D, Xu B, Song S. Characterization of the Passion Fruit (Passiflora edulis Sim) bHLH Family in Fruit Development and Abiotic Stress and Functional Analysis of PebHLH56 in Cold Stress. Horticulturae. 2023; 9(2):272. https://doi.org/10.3390/horticulturae9020272
Chicago/Turabian StyleXu, Yi, Weidong Zhou, Funing Ma, Dongmei Huang, Wenting Xing, Bin Wu, Peiguang Sun, Di Chen, Binqiang Xu, and Shun Song. 2023. "Characterization of the Passion Fruit (Passiflora edulis Sim) bHLH Family in Fruit Development and Abiotic Stress and Functional Analysis of PebHLH56 in Cold Stress" Horticulturae 9, no. 2: 272. https://doi.org/10.3390/horticulturae9020272
APA StyleXu, Y., Zhou, W., Ma, F., Huang, D., Xing, W., Wu, B., Sun, P., Chen, D., Xu, B., & Song, S. (2023). Characterization of the Passion Fruit (Passiflora edulis Sim) bHLH Family in Fruit Development and Abiotic Stress and Functional Analysis of PebHLH56 in Cold Stress. Horticulturae, 9(2), 272. https://doi.org/10.3390/horticulturae9020272





