Genotypic and Sanitary Characterization of Minority Grapevine Varieties Prospected in Andalusia, Spain
Abstract
1. Introduction
2. Materials and Methods
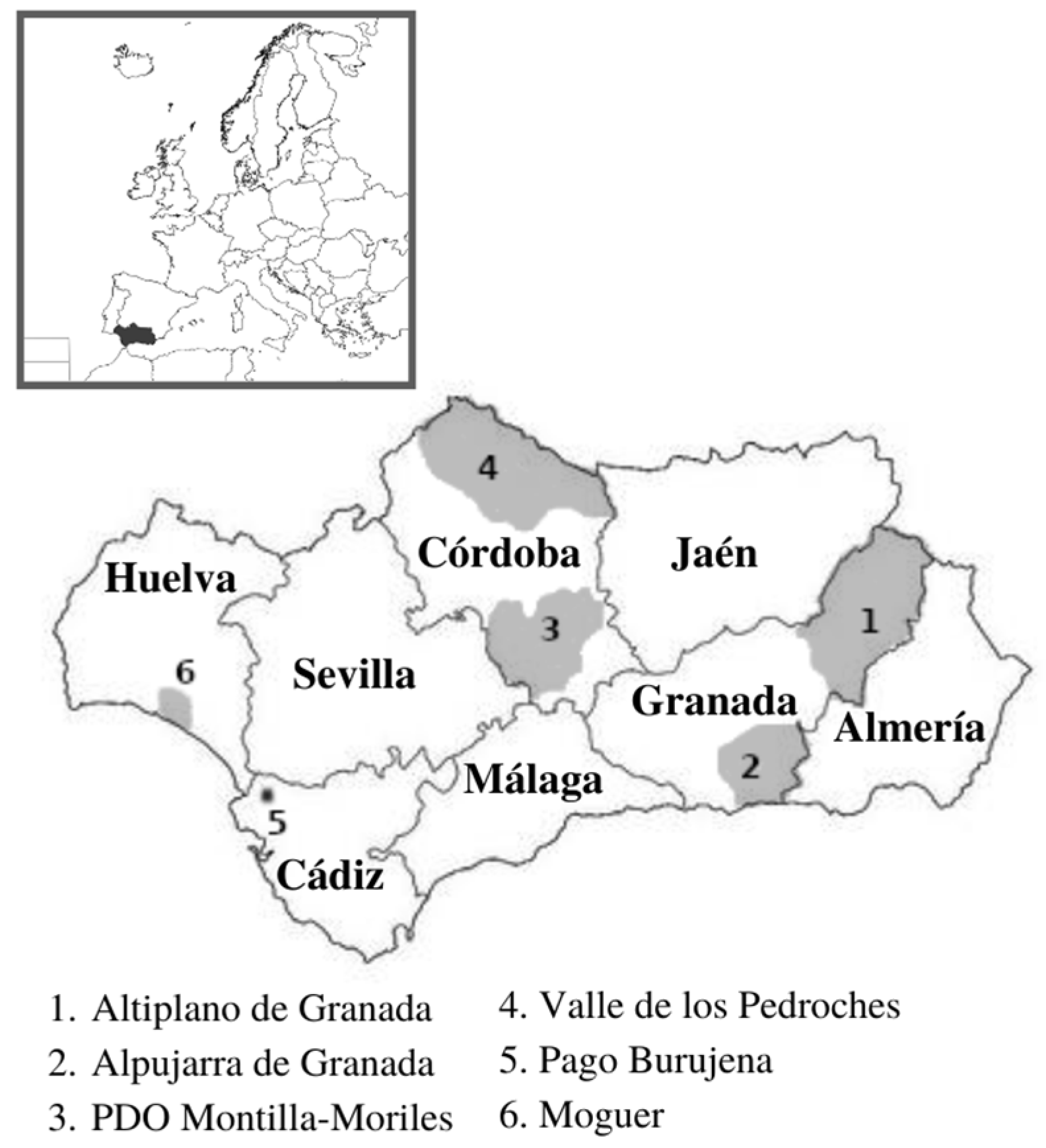
2.1. Areas and Specimens Prospected
- “Altiplano de Granada”, a region in the northeast of the Granada province that includes the Huéscar and Baza municipalities, is characterized by its high average altitude (between 700 and 1200 m). Grapevine cultivation occupies an area of about 350 ha and it is widely distributed in more than 1000 small vineyards with an average surface of 0.3 ha, thus showing great fragmentation and being owned by many local winemakers for domestic consumption. This area is protected by the PDO “Granada” and the PGI “Altiplano de Sierra Nevada”. Given that an exhaustive survey had been previously carried out in this area, only two plants have been studied in this work [7].
- “Alpujarra de Granada”, located in the southeastern part of the Granada province. It is characterized by its rugged relief, occupying most of the southern face of the Sierra Nevada. The 25 municipalities that make it up are located at an altitude between 1200 and 1500 m. Consisting of 2000 ha of vineyards with slate and stony soils and a slightly limestone subsoil. This region is protected by the PDO “Granada” and the PGI “Cumbres del Guadalfeo”. Nine specimens were surveyed in this area from two plots whose vineyards are more than 60 years old, and their origin could be dated back to the pre-phylloxera period.
- PDO “Montilla-Moriles”, in the central area of Andalusia belonging to the Córdoba province, has the Guadalquivir River to the north, the Subbética Mountains to the south, the Genil River to the east, and the Guadajoz River to the west. This zone includes 17 municipalities. This PDO has nearly 6000 ha of vineyards that produce fortified wines (Fino, Amontillado, Oloroso, and Palo Cortado) and sweet wines with the ‘Pedro Ximénez’ variety. There has been a commitment to the parallel production of other types of wine in the recent years, possibly accompanied by a greater varietal diversification. In this area, thirty-two specimens were surveyed from eight plots located in the municipalities of Montilla, Moriles, and Cabra, with the aim of clarifying the situation of a supposed different variety locally called “Montepila”, given the strong interest of the winemaking sector that requires its inclusion in the registry of authorized varieties in Andalusia.
- “Valle de los Pedroches”, a region of 3162 km2 also located in the Córdoba province, represents the northernmost territory of Andalusia. This area is characterized by its high diversity of landscapes (it goes from the holm oak dehesa to the peneplain until reaching Sierra Morena) and soils (with a predominance of slate in agricultural areas) and an average altitude of between 500 and 700 m. Valle de los Pedroches is protected by the PGI “Córdoba”. The vineyards of this area reached their maximum splendor in the 17th and 18th centuries, when they exceeded 2000 ha. It has been estimated that about 1800 ha were destined for the cultivation of vines shortly before the arrival of phylloxera at the end of 1800. Grapevine cultivation started disappearing shortly after this plague, although isolated or semi-isolated vine plants remained in the area, mainly for the production of self-consumption wines. In recent years, there has been a growing interest in these family orchards in order to preserve the biodiversity and traditional landscapes of the area. This indicates that there has been no influence from modern viticulture in this area, making it particularly attractive in terms of prospecting for possibly pre-phylloxera varieties since the introduction and spread of major national and/or international varieties have been avoided. Thirty-six vines were surveyed in this area, most of them isolated specimens located near the dividing boundaries between the plots.
- “Pago Burujena”, a territory located within El Marco de Jerez (PDO “Jerez-Xérès-Sherry” and “Manzanilla-Sanlúcar de Barrameda”), is the most emblematic and important area for the production of fortified and sweet Andalusian wines, having the same typologies mentioned for the PDO “Montilla-Moriles” but using instead the traditional ‘Palomino Fino’ variety. A greater varietal diversification has been recently pursued in the Jerez DO, and one of the objectives of producers is to rescue pre-phylloxera varieties for the production of both fortified and still wines. In the Jerez viticulture and its surroundings area, “Pagos” (rural places) represent cultivation areas geographically delimited by orographic elements and characterized by uniformity in terms of soil, microclimate, variety, and even the human qualities of the viticulturists. In this context, the definition of Pago Burujena dates back to the 16th century, under the Duchy of Medina Sidonia, and consists of an area of about 22 ha characterized by high quality limestone soils and the presence of different traditional varieties of the area. [8]. Seven specimens from a plot located in the Pago were surveyed.
- “Moguer”, whose municipality is protected by the PDO “Condado de Huelva”, is a territory with a high vocation for growing vines, characterized by vineyards at an average altitude of 25 m, loamy or sandy soils with a certain content of lime (which gives them a slightly basic pH), and a Mediterranean climate with Atlantic influences. Around 550 ha of vineyards are currently cultivated, with a high degree of fragmentation of the plots (average surfaces between 2 and 3 ha), many of them older than 40 years. For these reasons, and because of its long winemaking tradition, it is an ideal area to survey for minority varieties linked to possible wine diversification. In the province of Huelva, this production is mostly based on the cultivation of ‘Zalema’. Twelve vines over 50 years old belonging to this municipality and collected from a single plot were surveyed.
2.2. Plant Materials
2.3. Genotypic Analysis
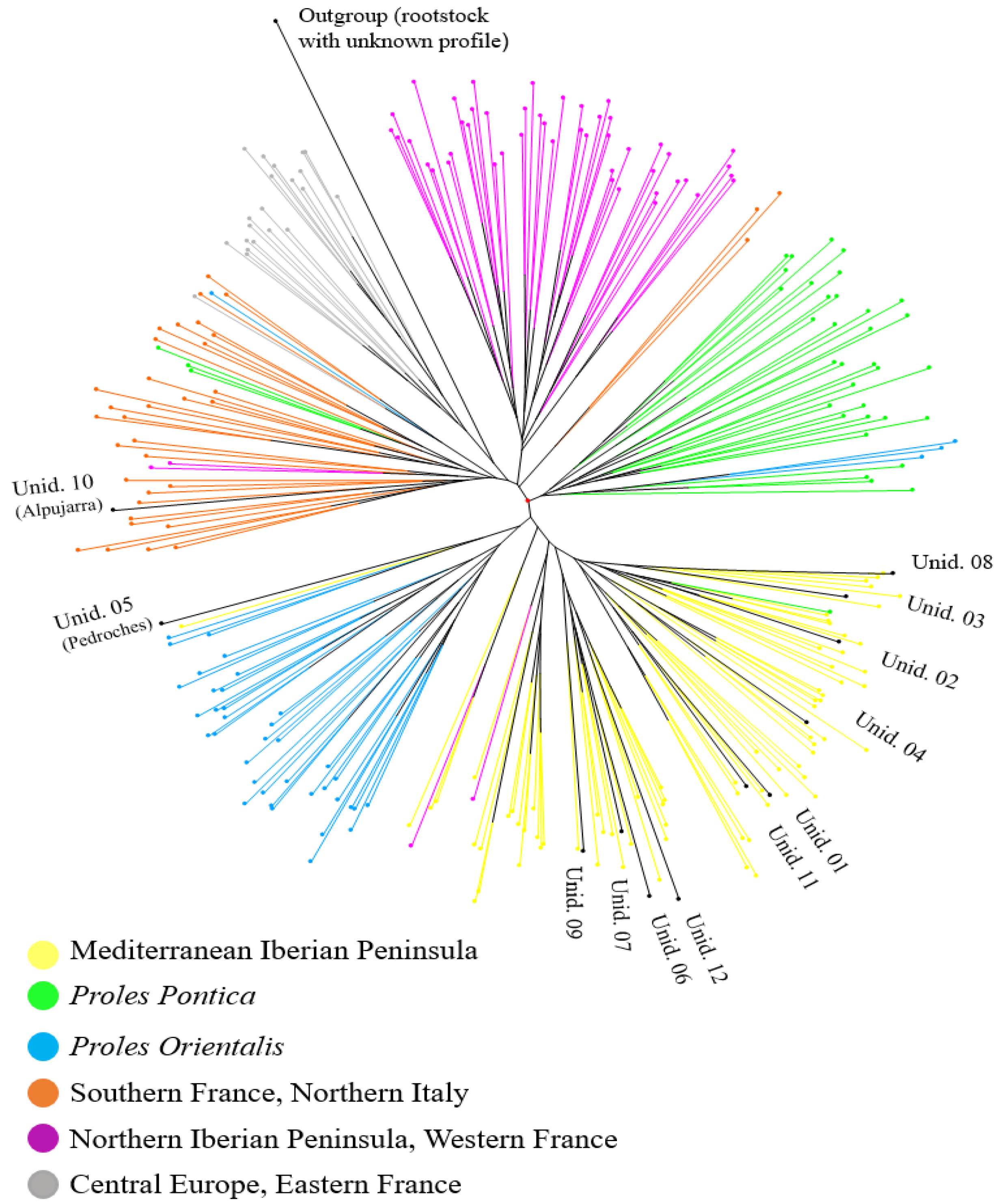
2.4. Cluster Analysis
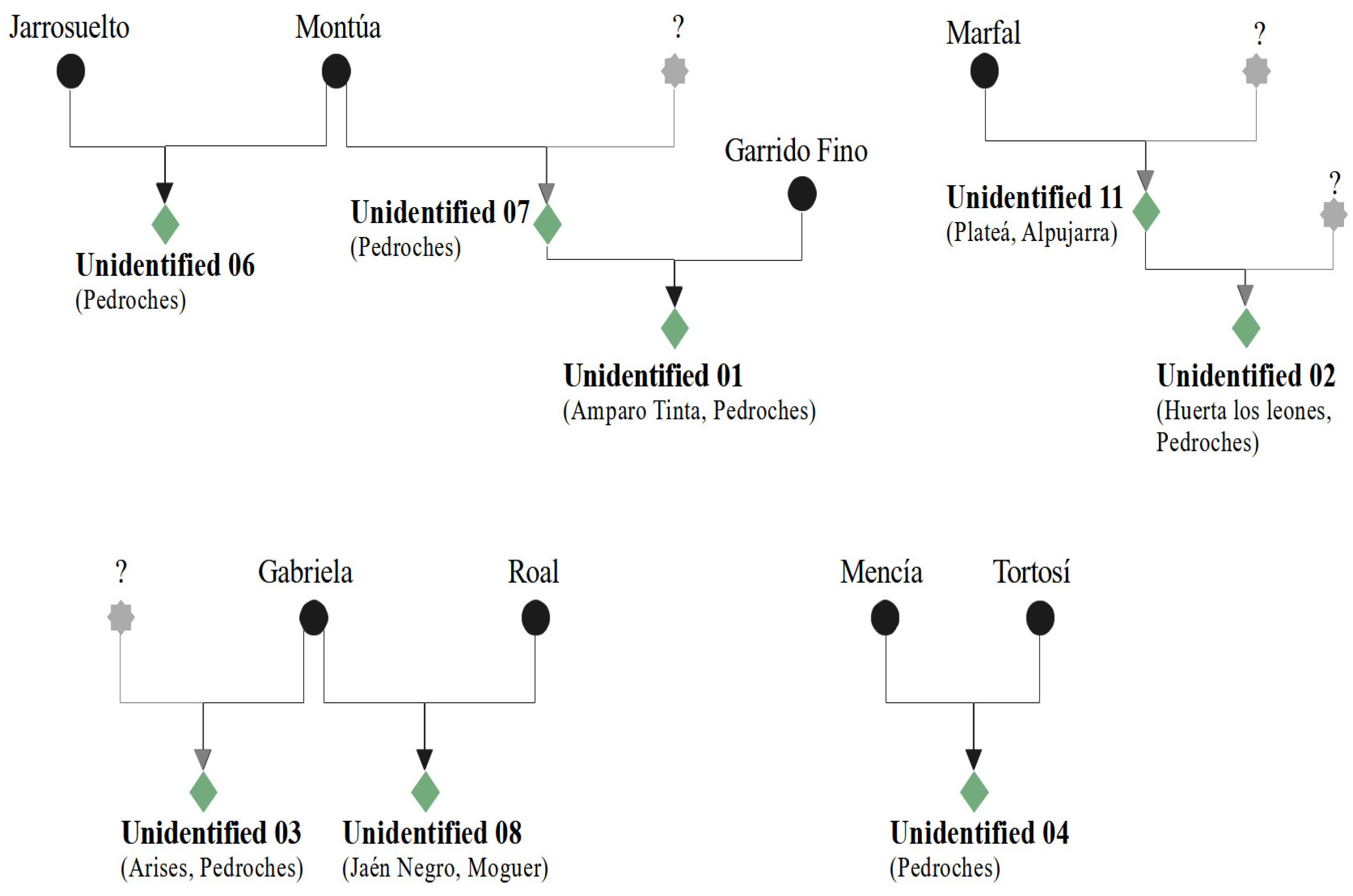
2.5. Pedigree Analysis
2.6. DAS-ELISA Test
2.7. Quantitative RT-PCR
3. Results
4. Discussion
- “Altiplano de Granada”. One specimen was shown to correspond to ‘Rojal Tinta’, a suitable variety for producing rosé wines that is not currently authorized in Andalusia but authorized in Castilla-La Mancha, an adjoining Community. Interestingly, two other prospected vines, each in both locations of La Alpujarra, corresponded to ‘Rojal Tinta’, suggesting the intentional cultivation of this variety in Eastern Andalusia. The microsatellite profile of ‘Rojal Tinta’ matched the profile described for the ‘Rojal’ variety in Castilla-La Mancha by Fernández-Gonzalez et al. [30]. The other specimen, white grape, showed an unidentified genotypic profile; thus, additional studies would be necessary to further characterize this variety. It is remarkable the low rate or absence of virus infections in the “Altiplano de Granada” area [7]. Possible explanations rely on the one hand, on the absence of massive introductions of propagation material from other origins due to the marginal viticulture in this area and, on the other hand, on the high altitude that prevents the presence of insect vectors of grapevine leafroll-associated viruses, mainly Planococcus ficus and P. citri. The results of the ELISA and RT-RT-qPCR tests of the two varieties surveyed in this study have confirmed the absence of virus infections. The presence of healthy phytosanitary material enables the possibility of certifying a local ‘Rojal Tinta’ clone in Andalusia.
- “La Alpujarra de Granada”. The presence of ‘Rojal Tinta’ in this area has already been highlighted. Regarding the other genotypes present in this area, there are two local varieties (‘Rome’ and ‘Mantúo de Pilas’) for which there is a strong interest among the winemakers to include them in the Register of Commercial Varieties in Spain, followed by authorization in Andalusia. ‘Rome’ has been already described in other areas of Andalusia, such as “La Axarquía” in Málaga province [31]. In addition, we identified one specimen of ‘Airén’, the most cultivated white variety in Spain; two other specimens corresponded to ‘Jacquez’, a direct producer hybrid with red berries (skin and pulp, therefore it is a dyera) and characterized by a high acidity in wines, a highly appreciated and sought-after quality in the indigenous red varieties of Andalusia, which usually lacks it; and two not previously identified genotypes, corresponding to a white grape vine and a red grape one. The red grape vine was shown to be virus-free. “La Alpujarra” is also characterized by vineyards at high altitude. However, unlike Altiplano, viticulture in La Alpujarra has historically been more intensive, and the exchange of plant materials has been frequent. This might explain why four of the vines showed simple or multiple virus infections (Appendix A, Table A2).
- PDO “Montilla-Moriles” (Córdoba). A total of 31of the 32 vines prospected from eight different locations are named “Montepila” by local viticulturists. The literature regarding the term ‘Montepila’ is unclear, with several conjectures about the origin of this name (unpublished). Microsatellite analysis allowed us to clarify that it corresponded to ‘Zalema’, the main white variety grown in the Huelva province, usually known in the Córdoba province as “Torrontés”. Moreover, 27 of the prospected specimens showed the “Zalema” genotype, three additional vines matched to ‘Cayetana Blanca’ (a common variety in several regions of Spain, known in the Córdoba province as “Baladí-Verdejo”) and another vine matched to ‘Pedro Ximénez’. Therefore, ‘Montepila’ is not an unidentified minority variety, although it is necessary to further determine by ampelography that it is not a somatic mutant. In a preliminary study by studying 11 basic grouping characters, we found no differences compared with ‘Zalema’. ‘Zalema’ is authorized in Andalusia; however, the synonymy ‘Montepila’ is not yet considered; consequently, its authorization could be undertaken through a request to the MAPA, supported by its corresponding technical report. The last specimen studied, displaying black berries, corresponded to ‘Negra Rayada’, a Spanish variety not authorized in the RVCV. This variety has also been found in the Andalusian province of Almería [32]. In one of the eight locations, two vines showing the ‘Zalema’ genotype resulted virus-free, and, therefore, they could be plausibly proposed as certified clones once the synonymy ‘Montepila’ is authorized.
- “Valle de los Pedroches” (Córdoba). Eight previously unidentified genotypes were identified in this area. Two of these genotypes were detected 10 times (in eight different locations) and four times (in four different locations), respectively. Although viticulture has practically disappeared from the Valle de los Pedroches, our survey suggests that there was some intentional cultivation of these two varieties, presumably in the pre-phylloxera period. They consist of red and white varieties that are currently experimentally vinified at IFAPA. According to the classification established by Muñoz-Organero et al. [29], ‘Tinta Amparo’ and ‘Arises’ would be novel autochthonous minority varieties. Two additional novel genotypes were found twice; in both cases, they were collected in the same location, similarly to the other four unidentified genotypes that were found only once. Among the other varieties found in the area, there were two Spanish commercial varieties (‘Alarije’ and ‘Cayetana Blanca’), a table variety (‘Ahmeur bou Ahmeur’), a foreign variety (‘Schiava Grossa’), and four known minority varieties (‘Hebén’, ‘Jarrosuelto’, ‘Zurieles’, and ‘Negra Dorada’). ‘Hebén’ is a wine grape cultivar described since the 16th century [33]. It was traditionally grown in Andalusia [2], but currently it is residual in the provinces of Córdoba, Granada, Badajoz, Guadalajara, Toledo, and Cádiz [34,35]. There has been no interference through the introduction of foreign propagation material to the area in more than a century since the cultivation of these vineyards was almost abandoned. On the other hand, isolation and altitude might explain the absence of virus infections in the area. Hence, we identified healthy candidates for each variety, suitable as starting material to eventually proceed with the registration process and possible authorization in Andalusia, a strictly necessary condition for its permitted cultivation, even in plots for self-consumption.
- “Pago Burujena” (Cádiz). There has been a growing interest in recent years in the recovery of varieties grown in the past and suitable for the production of fortified and still wines by the regulatory council of the PDOs “Jerez-Xérès-Sherry” and “Manzanilla-Sanlúcar de Barrameda” [1,36]. In Pago Burujena, one of the most traditional areas in the Jerez DO, clones of ‘Mantúo Castellano’ and ‘Mantúo de Pilas’ were apparently maintained in a vineyard for which three and two vines known by these denominations were respectively surveyed. In addition, two vines known as ‘Barcelonés’ were also collected. ‘Mantúo Castellano’ was shown to be a synonym of ‘Listán del Condado’, a variety commonly cultivated in the province of Huelva, while ‘Mantúo de Pilas’ and ‘Barcelonés’ was shown to correspond to ‘Alarije’. For the former, it can be assumed that it was a naming error, while for the latter, it was a synonymy. In the case of ‘Mantúo Castellano’, it would be interesting to request the recognition of synonymy since this is the name by which it has traditionally been known in Jerez. In addition, one of the three plants was virus-free, so a certified clone could be derived from it.
- “Moguer” (Huelva). In this municipality, we identified an specimen of the most cultivated variety in Spain, ‘Airén’; two specimens of ‘Beba’, recently authorized in Andalusia, of significant interest for its cultivation in the Jerez area; a specimen of ‘Mollar Cano’; three specimens of ‘Listán del Condado’; a specimen of the table variety ‘Alphonse Lavallée’; two genetically identical specimens of an unidentified variety locally called “Jaén Negro” (it would be a homonym, since ‘Jaén Negro’ also refers to a synonym of the ‘Jaén Tinto’ variety); and two specimens of ‘Listán Prieto’. The last one is possibly the most significant result. In his book from 1513, Agricultura General Herrera [33] described the ‘Uvas Prietas’ variety cultivated in the center of the Iberian Peninsula, which could possibly correspond to ‘Listán Prieto’. This variety was also explicitly mentioned in Andalusia in the year 1807 [1]. As a result of the America and the Canary Islands conquests in the 15th century, colonists from the Iberian Peninsula (Galician, Castillan, Andalusian, Extremaduran or even Portuguese) settled down in these lands, bringing grapevines together with their customs and crops [37]. Around 1550, a Jesuit introduced the ‘Listán Prieto’ variety in Peru [38], and cultivars were introduced in Mexico between the years 1520 and 1540 [39]. In California, it is known as Mission’ [40] as an allusion to the vines carried by the friars in their evangelizing work. In the 19th century, phylloxera obliterated almost half of the peninsular vineyards [39], but this insect did not reach the Canary Islands. The use of this variety is widespread in the Canary Islands and in America [41,42]. There is no record of its cultivation on the Iberian Peninsula since the last century [40]. Therefore, the location of several specimens of this variety, some over 50 years old, in the township of Moguer has great historical relevance. ‘Listán Prieto’ usually produces wines with a high color intensity and good acidity, characteristics highly desired in Andalusian red wines, so its detection in these surveys can support the intention of authorizing it in Andalusia. We identified three virus-free vines in this zone, one belonging to ‘Beba’, another to ‘Listán Prieto’ and another to the unidentified “Jaén Negro”. They could represent the starting point for obtaining locally certified clones of these varieties.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| OIV | International Organisation of Vine and Wine |
| ELISA | Enzyme-Linked ImmunoSorbent Assay |
| PDO | Protected Designation of Origin |
| PGI | Protected Geographical Indications |
| IMIDA | Instituto Murciano de Investigación y Desarrollo Agrario y Medioambiental |
| MAPA | Ministerio de Agricultura, Pesca y Alimentación |
| CPVO | Community Plant Variety Office |
| UPOV | International Union for the Protection of New Varieties of Plants |
| GFLV | Grapevine Fanleaf Virus |
| GLRaV | Grapevine Leafroll associated Virus |
| GFkV | Grapevine Fleck Virus |
| DNA | Deoxyribonucleic Acid |
| SSR | Short Sequence Repeat |
| CREA-UTV | Consiglio per la ricerca in agricoltura e l’analisi dell’economia agraria |
| PCR | Polymerase Chain Reaction |
| VIVC | Vitis International Variety Catalog |
| IFAPA | Instituto Andaluz de Investigación y Formación Agraria, Pesquera y Alimentaria |
| RVCV | Registro de Variedades Comerciales de Vid |
| ICVG | International Council for the Study of Virus and Virus-Like Diseases of the Grapevine |
Appendix A
| Locus | k | N | HObs | HExp | PIC | NE-1P | NE-2P | NE-PP | NE-I | NE-SI | HW | F (Null) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| md7 | 14 | 541 | 0.797 | 0.803 | 0.779 | 0.551 | 0.375 | 0.189 | 0.063 | 0.364 | NS | 0.0046 |
| md32 | 11 | 540 | 0.863 | 0.820 | 0.797 | 0.527 | 0.353 | 0.173 | 0.055 | 0.354 | NS | −0.0285 |
| zag62 | 10 | 541 | 0.824 | 0.807 | 0.782 | 0.548 | 0.371 | 0.187 | 0.061 | 0.362 | * | −0.0138 |
| zag79 | 13 | 539 | 0.818 | 0.854 | 0.838 | 0.448 | 0.287 | 0.118 | 0.037 | 0.332 | NS | 0.0208 |
| eva2 | 19 | 540 | 0.854 | 0.865 | 0.851 | 0.420 | 0.264 | 0.100 | 0.031 | 0.326 | NS | 0.0060 |
| isv2 | 23 | 541 | 0.884 | 0.851 | 0.833 | 0.464 | 0.300 | 0.132 | 0.040 | 0.335 | NS | −0.0212 |
| vvs2 | 15 | 541 | 0.850 | 0.842 | 0.824 | 0.477 | 0.311 | 0.137 | 0.043 | 0.340 | ** | −0.0042 |
| md5 | 11 | 541 | 0.854 | 0.859 | 0.842 | 0.449 | 0.286 | 0.122 | 0.036 | 0.330 | NS | 0.0022 |
| md27 | 9 | 541 | 0.848 | 0.829 | 0.804 | 0.520 | 0.347 | 0.173 | 0.053 | 0.349 | NS | −0.0122 |
| md25 | 14 | 540 | 0.785 | 0.767 | 0.728 | 0.632 | 0.454 | 0.273 | 0.093 | 0.390 | NS | −0.0118 |
| md28 | 19 | 540 | 0.852 | 0.870 | 0.856 | 0.417 | 0.262 | 0.103 | 0.031 | 0.323 | NS | 0.0107 |
| isv4 | 11 | 539 | 0.844 | 0.814 | 0.788 | 0.544 | 0.368 | 0.189 | 0.060 | 0.358 | * | −0.0210 |
| isv3 | 10 | 540 | 0.830 | 0.673 | 0.613 | 0.750 | 0.590 | 0.418 | 0.167 | 0.456 | *** | −0.1167 |
| Zone | Input Name | Variety | ELISA | PCR |
|---|---|---|---|---|
| Altiplano de Granada | Rosada Hornico | Rojal Tinta | - | - |
| Blanca Hornico | Unidentified 09 | - | - | |
| Rome | Rome | - | - | |
| Mollar Cano | Rojal Tinta | - | GFkV | |
| Tinta | Rojal Tinta | - | - | |
| Tinta Cortijo La Paz | Jacquez | - | - | |
| La Alpujarra de Granada | Llaqui | Jacquez | - | - |
| Ricardera | Mantúo de Pilas | - | GfkV, GLRaV-3 | |
| Desconocida Blanca | Airén | GFLV, GLRaV-2 | GFLV, GfkV, GLRaV-2 | |
| Tinta Piedras Blancas | Unidentified 10 | - | - | |
| Plateá | Unidentified 11 | GFLV | GFLV, GFkV | |
| Peñalista | Negra Rayada | m.d. | m.d. | |
| Cp1 Marenas fila 3 cp5 | Zalema | GFkV | GFkV | |
| Cp2 Marenas fila 4 cp18 | Zalema | GFkV, GLRaV-3 | GFkV, GLRaV-3 | |
| Marenas fila 7 cp3 cp44 | Zalema | GFkV, GLRaV-3 | GFkV, GLRaV-2, GLRaV-3 | |
| Cp 4Marenas fila 8 cp16 | Zalema | GFkV, GLRaV-1 | GFkV | |
| Cp5 Marenas fila 9 cp20 | Zalema | GFkV | GFkV | |
| Cp9 La Plata fila 7 | Zalema | m.d. | m.d. | |
| La Plata fila 13 p7 | Pedro Ximénez | - | GFkV, GLRaV-2 | |
| Cp6 La Plata fila 17 | Zalema | GFkV | GFkV, GLRaV-5 | |
| PDO Montilla-Moriles | Cp3 La Plata fila 19 | Zalema | - | GLRaV-3, GLRaV-5 |
| Los Rosales 1 | Zalema | GFkV | GFkV | |
| Los Rosales 2 | Zalema | - | GFkV, GLRaV-2 | |
| Los Rosales 3 | Zalema | - | GFkV, GLRaV-3 | |
| Los Rosales 4 | Zalema | GFkV | GFkV, GLRaV-3 | |
| Colección Montepila 1 | Zalema | - | GFkV | |
| Colección Montepila 2 | Zalema | - | GFkV | |
| Montepila 1 recinto 16 | Zalema | GFkV | GFkV | |
| Montepila 2 (recinto 16) | Zalema | - | GFkV | |
| Montepila 3 recinto 16 | Zalema | GFkV, GLRaV-3 | GFkV, GLRaV-2, GLRaV-3 | |
| Montepila 4 recinto 16 | Zalema | GLRaV-3 | GFkV, GLRaV-3 | |
| Montepila los Naranjos 1 | Zalema | m.d. | m.d. | |
| Montepila (2 linde) los Naranjos | Zalema | m.d. | m.d. | |
| Montepila los Naranjos 3 | Zalema | m.d. | m.d. | |
| Montepila 4 | Zalema | m.d. | m.d. | |
| Montepila los Naranjos 5 | Cayetana Blanca | m.d. | m.d. | |
| La Primilla 1 | Zalema | - | - | |
| La Primilla 2 | Cayetana Blanca | m.d. | m.d. | |
| La Primilla 3 | Zalema | - | - | |
| La Primilla 4 | Cayetana Blanca | m.d. | m.d. | |
| Cuesta Blanca 1 | Zalema | m.d. | m.d. | |
| Cuesta Blanca 2 | Zalema | m.d. | m.d. | |
| Cuesta Blanca 3 | Zalema | m.d. | m.d. | |
| Risquez 1 | Ahmeur bou Ahmeur | - | - | |
| Risquez 2 | Ahmeur bou Ahmeur | - | - | |
| Risquez 3 | Unidentified 07 | - | - | |
| Risquez 4 | Cayetana Blanca | - | - | |
| Vieja Primera | Cayetana Blanca | - | - | |
| Vieja Primera | Cayetana Blanca | - | - | |
| Merino 1 | Cayetana Blanca | - | - | |
| Merino 2 | Cayetana Blanca | - | - | |
| Lindero C1 | Unidentified 01 | - | - | |
| Valle de los Pedroches | La Torre Amparo Tinta | Unidentified 01 | - | - |
| Camino falda de la sierra | Unidentified 01 | - | - | |
| Recio 1° Amparo Tinta | Unidentified 01 | - | - | |
| Recio 2° Amparo TInta | Unidentified 01 | - | - | |
| Recio 3° Amparo TInta | Unidentified 01 | - | - | |
| Recio 4° Amparo TInta | Unidentified 01 | - | - | |
| Recio 5° Amparo TInta | Unidentified 01 | - | - | |
| Tinta Amparo malla Rafael | Unidentified 01 | - | - | |
| Tinta Amparo huerta Rafael | Unidentified 01 | - | - | |
| Huerta Los Leones C5 | Unidentified 02 | - | - | |
| Huerta Los Leones C2 | Unidentified 02 | - | - | |
| Isleta (Arises) | Unidentified 03 | - | - | |
| Garrido (Arises) | Unidentified 03 | - | - | |
| Arises cuadra Rafael | Unidentified 03 | - | - | |
| Arises huerta Rafael | Unidentified 03 | - | - | |
| Camino falda de la sierra (Portillo) | Unidentified 04 | - | - | |
| Arroyo Lorito | Unidentified 05 | - | - | |
| Autóctona Miguel | Unidentified 06 | - | - | |
| Lagareyes | Unidentified 12 | - | - | |
| Lagareyes | Unidentified 12 | - | - | |
| Blanca Lagareyes | Alarije | - | - | |
| Villaharta llanos suelo | Alarije | - | - | |
| Tío Kiko Camino | Negra Dorada | - | - | |
| Hebén | Hebén | - | - | |
| Jarrosuelto | Jarrosuelto | - | - | |
| Schiava Grossa | Schiava Grossa | - | - | |
| Entreárboles | Zurieles | - | - | |
| Mantúo Castellano | Listán del Condado | GFkV, GLRaV-3 | GFkV, GLRaV-2, GLRaV-3 | |
| Pago Burujena | Mantúo Castellano | Listán del Condado | GFkV, GFLV, GLRaV-3 | GFkV, GFLV, GLRaV-2, GLRaV-3 |
| Mantúo Castellano | Listán del Condado | - | - | |
| Mantúo de Pilas | Alarije | GFkV | GFkV | |
| Mantúo de Pilas | Alarije | GFkV | GFkV | |
| Barcelonés | Alarije | GFkV | GLRaV-2, GLRaV-3, GFkV | |
| Barcelonés | Alarije | GFkV | GLRaV-2, GLRaV-3, GFkV | |
| Mollar Cano | Mollar Cano | GLRaV-3 | GLRaV-3 | |
| Listán Prieto | Beba | - | GFkV, GLRaV-3 | |
| Beba | Beba | - | - | |
| Moguer | Airén | - | - | |
| Moguer | Listán Prieto | - | - | |
| Moguer | Listán Prieto | - | GLRaV-3 | |
| Moguer | Mantúo de Sanlúcar | Listán del Condado | - | GLRaV-3 |
| Mantúo de Sanlúcar | Listán del Condado | GLRaV-3 | - | |
| Mantúo de Sanlúcar | Listán del Condado | - | GLRaV-3 | |
| Mesa Plaza Tinta | Alphonse Lavallée | - | GLRaV-3 | |
| Jaén Negro | Unidentified 08 | GFkV. GFLV | GFkV. GFLV | |
| Jaén Negro | Unidentified 08 | - | - |
References
- Clemente y Rubio, S.R. Ensayo Sobre las Variedades de la vid Común que Vegetan en Andalucía: Con un índice Etimológico y Tres Listas de Plantas en que se Ca-Racterizan Varias Especies Nuevas; Imprenta de Villalpando: Madrid, Spain, 1807; 325p. [Google Scholar]
- García de los Salmones, N. Memoria general de las sesiones del Congreso y ponencias presentadas. In Proceedings of the Congreso Nacional de Viticultura, Pamplona, Spain, 11–22 July 1912. [Google Scholar]
- Rodríguez-Torres, I.; Márquez, M.I.; Ramírez, M.P.; Cretazzo, E. Caracterización molecular e identificación de variedades minoritarias de vid de cultivo tradicional en Andalucía. In Proceedings of the Congreso Nacional de Ciencias Hortícolas, Córdoba, Spain, 17–21 October 2021. [Google Scholar]
- OIV. Distribution of the World’s Grapevine Varieties; OIV: Paris, France, 2017; p. 54. Available online: https://www.oiv.int/public/me4d66ias/5distribution-of-the-worlds-grapevine-varieties.pdf (accessed on 1 May 2023).
- Cretazzo, E.; Tomás, M.; Padilla, C.; Roselló, J.; Medrano, H.; Padilla, V.; Cifre, J. Incidence of virus infection in old vineyards of local grapevine varieties from Majorca: Implications for clonal selection strategies. Span. J. Agric. Res. 2010, 8, 409–418. [Google Scholar] [CrossRef]
- Espinosa Roldán, F.E.; Muñoz Organero, G.; Uscola Fernández, M.; de Santa María, F.C.S.; De Toda, F.M. Minority grapevine varieties as climate change adaptation strategy: Exploring heat tolerance plasticity. In Proceedings of the 43rd World Congress of Vine and Wine, Ensenada, Mexico, 31 October–4 November 2022. [Google Scholar]
- Egea Bartual, C.; León Gutiérrez, J.M.; Lasheras Ocaña, J.; Ramírez Pérez, P. Estudio de las Variedades Locales y Foráneas Cultivadas en la zona del Altiplano de Granada; SERVIFAPA: 2016; 36p. Available online: https://ifapa.junta-andalucia.es/agriculturaypesca/ifapa/ser4v7i4fapaservifapa/19f319c3-6a05-4014-85ee-f5eecb197429 (accessed on 1 May 2023).
- Cabral Fernández, J. La viña y el Vino en la Edad Moderna a Través de la Casa Ducal de Medina Sidonia; Santa Teresa Industrias Gráficas SA: Trebujena, Spain, 2021; 192p. [Google Scholar]
- Avramidou, E.V.; Masaoutis, I.; Pitsoli, T.D.; Kapazoglou, A.; Pikraki, M.; Trantas, E.A.; Markantonakis, M.; Doulis, A.G. Analysis of Wine-Producing Vitis vinifera L. Biotypes, Autochthonous to Crete (Greece), Employing Ampelographic and Microsatellite Markers. Life 2023, 13, 220. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.R.; Scott, N.S. Microsatellite repeats in grapevine reveal DNA polymorphisms when analysed as sequence-tagged sites (STSs). Theor. Appl. Genet. 1993, 86, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Dangl, G.S.; Vignani, R.; Meredith, C.P. Isolation and characterization of new polymorphic simple sequence repeat loci in grape (Vitis vinifera L.). Genome 1996, 39, 628–633. [Google Scholar] [CrossRef] [PubMed]
- Bowers, J.E.; Meredith, C.P. The parentage of a classic wine grape, Cabernet Sauvignon. Nat. Genet. 1997, 16, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Bergamini, C.; Caputo, A.R.; Gasparro, M.; Perniola, R.; Cardone, M.F.; Antonacci, D. Evidences for an alternative genealogy of ‘Sangiovese’. Mol. Biotechnol. 2013, 53, 278–288. [Google Scholar] [CrossRef]
- Maul, E.; Sudharma, K.N.; Kecke, S.; Marx, G.; Müller, C.; Audeguin, L.; Boselli, M.; Boursiquot, J.M.; Bucchetti, B.; Cabello, F.; et al. The European Vitis Database (www.eu-vitis.de): A technical innovation through an online uploading and interactive modification system. Vitis 2012, 51, 79–85. [Google Scholar]
- Cretazzo, E.; Moreno Sanz, P.; Lorenzi, S.; Benítez, M.L.; Velasco, L.; Emanuelli, F. Genetic characterization by SSR markers of a comprehensive wine grape collection conserved at Rancho de la Merced (Andalusia, Spain). Plants 2022, 11, 1088. [Google Scholar] [CrossRef]
- Negrul, A.M. Evolution of cultivated forms of grapes. CR Acad. Sci. URSS 1938, 18, 585–588. [Google Scholar]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software. 2006. Available online: http://darwin.cirad.fr/ (accessed on 1 May 2023).
- Kalinowski, S.T.; Taper, M.L.; Marshall, T.C. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 2007, 16, 1099–1106. [Google Scholar] [CrossRef]
- Lassois, L.; Denancé, C.; Ravon, E.; Guyader, A.; Guisnel, R.; Hibrand-Saint-Oyant, L.; Poncet, C.; Lasserre-Zuber, P.; Feugey, L.; Durel, C. Genetic Diversity, Population Structure, Parentage Analysis, and Construction of Core Collections in the French Apple Germplasm Based on SSR Markers. Plant Mol. Biol. Report. 2016, 34, 827–844. [Google Scholar] [CrossRef]
- Khadari, B.; Bakkali, A.E.; Essalouh, L.; Tollon, C.; Pinatel, C.; Besnard, G. Cultivated Olive Diversification at Local and Regional Scales: Evidence from the Genetic Characterization of French Genetic Resources. Front. Plant Sci. 2019, 10, 1593. [Google Scholar] [CrossRef]
- Weng, Z.; Yang, Y.; Wang, X.; Wu, L.; Hua, S.; Zhang, H.; Meng, Z. Parentage analysis in giant grouper (Epinephelus lanceolatus) using microsatellite and SNP markers from genotyping-by-sequencing data. Genes 2021, 12, 1042. [Google Scholar] [CrossRef]
- Martelli, G.P. Grapevine virology highlights 2004–2005. In Proceedings of the 15th ICVG Conference, Stellenbosch, South Africa, 3–7 April 2006. [Google Scholar]
- Padilla, V.; Hita, I.; García de la Rosa, S.B.; Padilla, C.V.; Salmerón, E.; Cretazzo, E. Virosis de la vid.Situación por comunidades autónomas. Vitic. Enol. Prof. 2007, 113, 6–12. [Google Scholar]
- Sánchez-Vizcaino, J.M.; Cambra, M. Técnicas Inmunoenzimáaticas en Patología Animal y Vegetal; Monografía INIA 29; INIA: Madrid, Spain, 1981; 57p. [Google Scholar]
- Boscia, D.; Digiaro, M.; Fresno, J.; Greif, C.; Grenan, S.; Kassemeyer, H.H.; Prota, V.A.; De Sequueira, O.A. ELISA for the detection and identification of grapevine viruses. In Sanitary Selection of the Grapevine; Walter, B., Ed.; INRA Editions: Colmar, France, 1997; Volume 86, pp. 129–155. [Google Scholar]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Real-time RT-PCR (TaqMan®) assays for the detection of Grapevine Leafroll associated viruses 1-5 and 9. J. Virol. Methods 2007, 141, 22–29. [Google Scholar] [CrossRef]
- Osman, F.; Leutenegger, C.; Golino, D.; Rowhani, A. Comparison of low-density arrays, RT-PCR and real-time TaqMan RT-PCR in detection of grapevine viruses. J. Virol. Methods 2008, 149, 292–299. [Google Scholar] [CrossRef]
- Cepin, U.; Gutiérrez-Aguirre, I.; Balažic, L.; Pompe-Novak, M.; Gruden, K.; Ravnikar, M. A one-step reverse transcription time PCR assay for the detection and quantitation of Grapevine fanleaf virus. J. Virol. Methods 2010, 170, 47–56. [Google Scholar] [CrossRef]
- Muñoz Organero, G.; De Andrés, M.T.; Vargas, A.; Aller, M.; Serrano, M.J.; Cretazzo, E.; Pérez, J.A.; Puertas, M.B.; Gogorcena, Y.; Giménez, R.; et al. La enorme diversidad varietal de vid en España, en proceso de descubrimiento. ACE Rev. Enol. 2017, 170, ISSN-e 1697-412. [Google Scholar]
- Fernández-González, M.; Mena, A.; Izquierdo, P.; Martínez, J. Genetic characterization of grapevine (Vitis vinifera L.) cultivars from Castilla La Mancha (Spain) using microsatellite markers. Vitis 2007, 46, 126–130. [Google Scholar]
- Jiménez-Cantizano, A.; Muñoz-Martín, A.; Amores-Arrocha, A.; Sancho-Galán, P.; Palacios, V. Identification of Red Grapevine Cultivars (Vitis vinifera L.) Preserved in Ancient Vineyards in Axarquia (Andalusia, Spain). Plants 2020, 9, 1572. [Google Scholar] [CrossRef]
- Tello, J.; Galán, Á.; Rodríguez-Torres, I.; Martínez Zapater, J.M.; Rubio, A.; Ibáñez, J. Prospección, identificación genética y conservación de parras antiguas de Almería. In Proceedings of the IV Jornada del Grupo de Viticultura, Sociedad Española de Ciencias Hortícolas, Pamplona, Spain, 26–28 October 2022. [Google Scholar]
- De Herrera, A.G. Agricultura General; Corrected and Edited by the Real Sociedad Económica Matritense; Imprenta Real: Madrid, Spain, 1818–1819; 2026p. [Google Scholar]
- Cabello, F.; Ortiz, J.M.; Muñoz, G.; Rodríguez-Torres, I.; Benito, A.; Rubio, C.; García, S.; Sáiz, R.; De Andrés, M.T. Variedades de vid en España; Comunidad de Madrid y Editorial Agrícola Española: Madrid, Spain, 2019; 519p. [Google Scholar]
- Zinelabidine, L.H.; Cunha, J.; Eiras-Dias, J.E.; Cabello, F.; Martínez-Zapater, J.M.; Ibáñez, J. Pedigree analysis of the Spanish grapevine cultivar ’Hebén’. Vitis 2015, 54, 81–86. [Google Scholar]
- Fernández de Bobadilla, G. Viníferas Jerezanas y de Andalucía Occidental; Instituto Nacional de Investigaciones Agronómicas: Madrid, Spain, 1956; 271p. [Google Scholar]
- Rodríguez-Torres, I. Patrimonio Vitícola de Canarias. In Proceedings of the VII Semana Científica Telesforo Bravo, Instituto de Estudios Hispánicos de Canarias, Puerto de la Cruz, Spain, 21–25 November 2011. [Google Scholar]
- Zerolo, J.; Cabello, F.; Espino, A.; Borrego, J.; Ibañez, J.; Rodriguez-Torres, I.; Muñoz-Organero, G.; Rubio, C.; Hernández, M. Variedades de Vid de Cultivo Tradicional en Canarias; Instituto Canario de Calidad Agroalimentaria, Gobierno de Canarias: Santa Cruz de Tenerife, Spain, 2006; 224p. [Google Scholar]
- Hidalgo, L. Tratado de Viticultura General; Mundiprensa: Madrid, Spain, 2002; 1235p. [Google Scholar]
- Milla, A.; Cabezas, J.A.; Cabello, F.; Lacombe, T.; Martínez-Zapater, J.M.; Hinrichsen, P.; Cervera, M.T. Determining the Spanish origin of representative ancient American grapevine varieties. Am. J. Enol. Vitic. 2007, 58, 242–251. [Google Scholar]
- Rodríguez-Torres, I. Variedades de vid Cultivadas en Canarias: Descriptores Morfológicos: Caracterización Morfológica, Molecular, Agronómica y Enológica; Instituto Canario de Investigaciones Agrarias: Santa Cruz de Tenerife, Spain, 2017; 200p. [Google Scholar]
- Lacoste, P. La variedad de uva País (Listán Prieto) en el Cono Sur de América: Trayectoria histórica. Idesia 2021, 39, 75–84. [Google Scholar] [CrossRef]
- Osman, F.; Rowhani, A. Real-time RT-PCR (TaqMan®) assays for the detection of viruses associated with rugose wood complex of grapevine. J. Virol. Methods 2008, 154, 69–75. [Google Scholar] [CrossRef]
- Angelini, E.; Bertazzon, N.; Borgo, M. Diversity among Grapevine leafroll-associated virus 2 isolates detected by heteroduplex mobility assay. J. Phytopathol. 2004, 152, 416–422. [Google Scholar] [CrossRef]
- Bertazzon, N.; Bazzo, I.; Repetto, O.; Borgo, M.; Angelini, E. Advances in molecular detection of GLRaV-1. In Proceedings of the 16th Meeting of the ICVG, Dijon, France, 31 August–4 September 2009. [Google Scholar]
| Zone | Input Name | Confirmed Variety | Number of Individuals |
|---|---|---|---|
| Altiplano de Granada | Rosada Hornico | Rojal Tinta | 1 |
| Rome | Rome | 1 | |
| Mollar Cano | Rojal Tinta | 1 | |
| Tinta | Rojal Tinta | 1 | |
| La Alpujarra de Granada | Tinta Cortijo La Paz | Jacquez | 1 |
| Llaqui | Jacquez | 1 | |
| Ricardera | Mantúo de Pilas | 1 | |
| Desconocida blanca | Airén | 1 | |
| Peñalista | Negra Rayada | 1 | |
| PDO Montilla-Moriles | Montepila | Zalema | 27 |
| Montepila | Cayetana Blanca | 3 | |
| Montepila | Pedro Ximénez | 1 | |
| Risquez | Ahmeur bou Ahmeur | 2 | |
| Risquez | Cayetana Blanca | 1 | |
| Merino | Cayetana Blanca | 2 | |
| Vieja Primera | Cayetana Blanca | 2 | |
| Blanca Lagareyes | Alarije | 1 | |
| Valle de los Pedroches | Villaharta Llanos Suelo | Alarije | 1 |
| Tío Kiko Camino | Negra Dorada | 1 | |
| Hebén | Hebén | 1 | |
| Jarrosuelto | Jarrosuelto | 1 | |
| Schiava Grossa | Schiava Grossa | 1 | |
| Entreárboles | Zurieles | 1 | |
| Mantúo Castellano | Listán del Condado | 3 | |
| Pago Burujena | Mantúo de Pilas | Alarije | 2 |
| Barcelonés | Alarije | 2 | |
| Mollar Cano | Mollar Cano | 1 | |
| Beba | Beba | 1 | |
| Listán Prieta | Beba | 1 | |
| Moguer | Moguer | Airén | 1 |
| Moguer | Listán Prieto | 2 | |
| Mantúo de Sanlúcar | Listán del Condado | 3 | |
| Mesa Plaza Tinta | Alphonse Lavallée | 1 |
| Zone | Input Name | Confirmed Variety | Number of Individuals |
|---|---|---|---|
| Altiplano de Granada | Blanca Hornico | Unidentified 09 | 1 |
| La Alpujarra de Granada | Tinta Piedras Blancas | Unidentified 10 | 1 |
| Plateá | Unidentified 11 | 1 | |
| Tinta Amparo | Unidentified 01 | 10 | |
| Huerta de los Leones | Unidentified 02 | 2 | |
| Arises | Unidentified 03 | 4 | |
| Valle de los Pedroches | Falda de la Sierra | Unidentified 04 | 1 |
| Arroyo Lorito | Unidentified 05 | 1 | |
| Autóctona Miguel | Unidentified 06 | 1 | |
| Risquez | Unidentified 07 | 1 | |
| Lagarreyes | Unidentified 12 | 2 | |
| Moguer | Jaén Negro | Unidentified 08 | 2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Torres, I.; Martín Carrillo, A.; Ramírez, M.d.P.; Gómez Gálvez, F.J.; Velasco Arjona, L.; Padilla, C.; Cretazzo, E. Genotypic and Sanitary Characterization of Minority Grapevine Varieties Prospected in Andalusia, Spain. Horticulturae 2023, 9, 759. https://doi.org/10.3390/horticulturae9070759
Rodríguez-Torres I, Martín Carrillo A, Ramírez MdP, Gómez Gálvez FJ, Velasco Arjona L, Padilla C, Cretazzo E. Genotypic and Sanitary Characterization of Minority Grapevine Varieties Prospected in Andalusia, Spain. Horticulturae. 2023; 9(7):759. https://doi.org/10.3390/horticulturae9070759
Chicago/Turabian StyleRodríguez-Torres, Inmaculada, Ana Martín Carrillo, María del Pilar Ramírez, Francisco Jesus Gómez Gálvez, Leonardo Velasco Arjona, Carlos Padilla, and Enrico Cretazzo. 2023. "Genotypic and Sanitary Characterization of Minority Grapevine Varieties Prospected in Andalusia, Spain" Horticulturae 9, no. 7: 759. https://doi.org/10.3390/horticulturae9070759
APA StyleRodríguez-Torres, I., Martín Carrillo, A., Ramírez, M. d. P., Gómez Gálvez, F. J., Velasco Arjona, L., Padilla, C., & Cretazzo, E. (2023). Genotypic and Sanitary Characterization of Minority Grapevine Varieties Prospected in Andalusia, Spain. Horticulturae, 9(7), 759. https://doi.org/10.3390/horticulturae9070759








