Abstract
Horticultural crops suffer from bacterial, fungal, and oomycete pathogens. Effectors are one of the main weapons deployed by those pathogens, especially in the early stages of infection. Pathogens secrete effectors with diverse functions to avoid recognition by plants, inhibit or manipulate plant immunity, and induce programmed cell death. Most identified effectors are proteinaceous, such as the well-studied type-III secretion system effectors (T3SEs) in bacteria, RXLR and CRN (crinkling and necrosis) motif effectors in oomycetes, and LysM (lysin motifs) domain effectors in fungi. In addition, some non-proteinaceous effectors such as toxins and sRNA also play crucial roles in infection. To cope with effectors, plants have evolved specific mechanisms to recognize them and activate effector-triggered immunity (ETI). This review summarizes the functions and mechanisms of action of typical proteinaceous and non-proteinaceous effectors secreted by important horticultural crop pathogens. The defense responses of plant hosts are also briefly introduced. Moreover, potential application of effector biology in disease management and the breeding of resistant varieties is discussed.
1. Introduction
Horticultural crops typically include fruits, vegetables, and ornamental plants. They are not only an excellent source of daily nutrition for people, but also have important economic value. Common fruits and vegetables can provide a variety of nutrients that the human body needs, including fiber, vitamins, organic acids, and mineral elements [1]. Like other plants, horticultural crops are infected by various bacterial, oomycete, and fungal pathogens that cause enormous economic losses and can have great impacts on human society. The Irish potato famine in the 19th century caused by the oomycete Phytophthora infestans is considered to be a historical tragedy [2]. According to nutrient-acquisition strategies, horticultural crop pathogens can be categorized as biotroph, hemibiotroph, or necrotroph [3]. Biotrophic pathogens infect living plant cells and feed on them. In contrast, necrotrophic pathogens acquire nutrients from dead cells after killing plant tissues. Hemibiotrophic pathogens exhibit two forms of nutrition acquisition, the early biotrophic stage and the late necrotrophic stage. Some typical pathogens of horticultural crops include hemibiotrophic bacteria Pseudomonas syringae and Ralstonia solanacearum, necrotrophic bacteria Xanthomonas spp., biotrophic oomycetes Phytophthora spp., biotrophic fungus Cladosporium fulvum, hemibiotrophic fungus Fusarium oxysporum, and necrotrophic fungi Botrytis cinerea, Alternaria alternata, and Penicillium spp. [4,5].
Effectors are unified agents used by bacterial, oomycete and fungal pathogens to facilitate infection. They are broadly defined as biological molecules secreted by pathogens, including proteins, small RNA, and toxic metabolites [6]. Effectors play dual roles during the complicated plant–pathogen interaction and trigger a series of molecular events. In the early stages of infection, plants recognize pathogen-associated molecular patterns (PAMPs) and activate PAMP-triggered immunity (PTI). At the same time, pathogens can secrete effectors to suppress PTI, and plants recognize effectors thereby inducing effector-triggered immunity (ETI) which may lead to hypersensitive response (HR) in the host [7]. The gene-for-gene model suggests that there is a one-to-one correspondence between an avirulence gene (Avr) of the pathogen and a resistance gene (R) of the plant [8]. In this model, effectors are often referred to as Avr proteins that can be recognized by R proteins (immunity receptors), triggering defense responses leading to host resistance [9]. Avr effectors identified from C. fulvum, the causal agent of tomato leaf mold, are a classic example of the gene-for-gene model [10].
Functions of effectors are diverse, for example, preventing recognition of pathogens by plants, suppressing host immune responses, and manipulating host-cell physiology to help colonization. Most effectors are secreted biomacromolecules and have an effect on the apoplastic area or in the cells of host. Different localization is closely associated with specific functions [11]. To date, numerous effectors have been identified from horticultural crop pathogens (Table 1), most of which are proteinaceous effectors. Modes of action of some type-III-secreted effectors (T3SEs) in bacteria, RXLR and CRN (crinkling and necrosis) motif effectors in oomycetes, and LysM (lysin motifs) domain effectors in fungi have been well characterized in various horticultural host plants, especially in tomato and potato [11]. In addition, these findings on effectors and recognition receptors are valuable for the breeding of resistant cultivars [12,13,14,15].
Though a number of reviews have successfully documented the advances in effector biology [3,16,17,18,19], most of them have been mainly focused on pathogens of model plants (such as Arabidopsis or tobacco) or cereal crops (such as rice or wheat). The crucial roles of effectors in interactions between horticultural crops and pathogens have rarely been summarized. This review focuses on proteinaceous and non-proteinaceous effectors in horticultural crop pathogens, with emphasis on the functions and action mechanisms of proteinaceous effectors from bacterial, oomycete, and fungal pathogens, respectively. Moreover, resistance responses in horticultural crops from recognition to fightback are also discussed.

Table 1.
Examples of identified and characterized effectors in horticultural crop pathogens.
Table 1.
Examples of identified and characterized effectors in horticultural crop pathogens.
| Pathogen | Effector Name | Host | Functions | Refs. |
|---|---|---|---|---|
| Proteinaceous effectors | ||||
| Pseudomonas syringae pv. syringae | PsyB728a | Tomato | T3SEs; acetyltransferase activity; acetylates other effectors and plant immunity components; suppresses defense response. | [20] |
| P. syringae | AvrPtoB | Tomato | T3SE; E3 ubiquitin ligase activity; mediates ubiquitination and degradation of Fen; suppresses both PTI and ETI. | [21] |
| Ralstonia solanacearum | Rip36 | Eggplant | T3SE; Zn-dependent protease activity; induces HR. | [22] |
| R. solanacearum | RipAB | Potato | T3SE; regulates Ca2+-dependent gene expression; promotes disease development. | [23] |
| R. solanacearum | RipI | Tomato | T3SE; interacts with bHLH9 to induce host defense response; deletion of ripI leads to increased virulence. | [24] |
| R. solanacearum | RipAY | Eggplant | T3SE; interacts with host catalases; suppresses HR. | [25] |
| R. solanacearum | RipAX2 | Eggplant | T3SE; zinc-binding motif; triggers resistance in eggplant AG91-25. | [26] |
| Xanthomonas campestris pv. vesicatoria (Xcv) | AvrBs3 | Pepper | T3SE-TALE; targets Bs3 and induces expression of upa genes; triggers resistance response. | [27] |
| X. campestris pv. vesicatoria (Xcv) | AvrBs4 | Pepper | T3SE-TALE; targets Bs4 and triggers Bs4-dependent HR; suppresses defense responses. | [28] |
| Xanthomonas gardneri | AvrHah1 | Tomato | T3SE-TALE; activates transcription of bHLH, pectate lyase and pectinesterase genes; enhances water-soaking in leaves. | [29] |
| Xanthomonas citri subsp. Citri (Xcc) | pthA4, pthAw, pthA, pthB, and pthC | Orange | T3SEs-TALEs; promotes the expression of CsLOB1; increases host disease susceptibility. | [30,31] |
| Phytophthora brassicae | RXLR24 | Potato | RXLR; targets RABA GTPases; inhibits RABA GTPase mediated vesicular secretion of PR-1, PDF1.2. | [32] |
| Phytophthora capsica | CRN12_997 | Tomato | CRN; targets TCP transcription factor SlTCP14-2; inhibits host-immunity-associated activity. | [33] |
| Phytophthora infestans | INF1 | Potato; tomato | Elicitin; activates basal defense pathways; enhances resistance response. | [34] |
| P. infestans | INF2a, INF5, INF6 | Potato | Elicitins; induces cell death in transgenic potato plants expressing ELR (elicitin response). | [35] |
| P. infestans | PiNPP1.1 | Tomato | NLP; induces cell death. | [36] |
| P. infestans | EPI1 and EPI10 | Tomato | Protease inhibitors; kazal-like domains; inhibits the pathogenesis-related protein P69B; EPI10 inhibits activity of subtilisin A; enhances host susceptibility. | [37] |
| P. infestans | EPIC1 and EPIC2B | Tomato | Protease inhibitors; cystatin-like proteins; target papain-like cysteine protease C14; inhibits protease Rcr3pim; suppresses defense. | [38,39] |
| P. infestans | PiSFI3 | Potato | RXLR; targets UBK; suppresses early immunity response. | [40] |
| P. infestans | Pi03192 | Potato | RXLR; targets NAC transcription factors NTP1 and NTP2. | [41] |
| P. infestans | PITG_15718.2 | Potato | RXLR; regulates the host transcriptome; suppresses immunity and reduces vegetative growth. | [42] |
| P. infestans | Pi22798 | Potato | RXLR; promotes negative regulator StKNOX3 homodimerization; enhances host susceptibility. | [43] |
| P. infestans, Phytophthora parasitica and Phytophthora palmivora | AVRamr3 | Potato | RXLR; targets Rpi-amr3; recognition of AVRamr3 enhancing resistance against Phytophthora spp. | [44] |
| Phytophthora sojae | AVR3a | Potato | RXLR; suppresses infestin 1 (INF1)-triggered cell death. (ICD); stabilizes E3 ligase CMPG1; manipulates plant immunity. | [45] |
| Plasmopara viticola | RXLR50253 | Grapevine | RXLR; suppresses ICD; targets VpBPA1; promotes pathogen colonization. | [46] |
| Botrytis cinerea/Sclerotinia sclerotiorum | BcSSP2/3 and SsSSP3 | Camelliae | Induces rapid necrosis. | [47] |
| B. cinerea | BcPG1/2 | Tomato | CWDEs; endopolygalacturonases; induces necrosis; strongly affects virulence. | [48] |
| B. cinerea | BcGs1 | Tomato | CWDE; glucan 1,4-alphaglucosidase; causes necrosis; triggers host immunity. | [49] |
| B. cinerea | BcXyn11/BcXyn11A | Tomato | CWDEs; xylanase; induces necrosis; inhibits seedling growth, induces defense response. | [50] |
| B. cinerea | Crh1 | Tomato | Glycosyl hydrolase (GH16) family; transglycosylase activity; catalyzes crosslinking of chitin and glucan polymers; induces cell death. | [51] |
| Cladosporium fulvum | Ecp6 | Tomato | LysM; sequesters chitin oligosaccharides; evades host immunity. | [52] |
| C. fulvum | Avr2 | Tomato | Cysteine protease inhibitor; targets cysteine protease Rcr3; induces HR. | [53,54] |
| C. fulvum | Avr4 | Tomato | Chitin-binding domain; protects cell walls against plant chitinases; affects virulence. | [55] |
| C. fulvum | Avr9 | Tomato | Targets Cf-9; induces HR; elicits protein kinase ACIK1; affects host resistance. | [56] |
| Colletotrichum higginsianum | ChELP1 and ChELP2 | Brassicaceae | LysM; binds chitin and chitin oligomers; suppresses chitin-triggered immunity. | [57] |
| Colletotrichum orbiculare | NLP1 | Cucumber | NLP; cytotoxic activity; induces cell death; C-terminal region of NLP1 enhances host defense. | [58] |
| Fusarium oxysporum f. sp. lycopersici (Fol) | Fol-EC19 and Fol-EC14 | Tomato | Guanyl-specific ribonuclease; triggers cell death. | [59] |
| F. oxysporum f. sp. lycopersici (Fol) | Fol-EC14 | Tomato | Glucanase and trypsin activities; suppresses Bax-mediated cell death; suppresses I-2/Avr2- and I/Avr1-mediated cell death. | [59] |
| F. oxysporum f. sp. conglutinans (Foc) | Foc-SIX1 | Cabbage | Secreted-in-xylem (SIX) effector; affects virulence. | [60] |
| Moniliophthora perniciosa | MpCP1 | Cacao | Cerato-platanin-like proteins (CPP); induces necrosis | [61] |
| Penicillium expansum | Penlp | Apple | NLP; induces necrosis; affects virulence. | [62] |
| S. sclerotiorum | SsPINE1 | Pea | Targets PG-inhibiting proteins (PGIPs); inactivates plant polygalacturonase-inhibiting protein; affects virulence. | [63] |
| Non-proteinaceous effectors | ||||
| B. cinerea | Bc-siR3.1, Bc-siR3.2, and Bc-siR5 | Tomato | sRNAs; binds to Argonaute 1 (AGO1); suppresses host immunity. | [64] |
| Valsa mali | unknown | Apple | sRNA; affects virulence. | [65] |
| Penicillium italicum | unknown | Citrus fruits | milRNAs; affects virulence. | [66] |
| Alternaria alternata f. sp. Lycopersici | AAL toxins | Tomato | Host-specific toxins (HSTs); target aspartate carbamoyl transferase and sphinganine N-acltransferase. | [67,68] |
| A. alternata f. sp. fragariae | AF toxins | Strawberry | HSTs; targets microsomal phospholipase A2. | [68,69] |
| A. alternata f. sp. kikuchana | AK toxins | Japanese pear | HSTs; targets sulfhydryl-containing proteins in membrane. | [68,70] |
| A. alternata f. sp. citri tangerine | ACT toxins | Tangerines and mandarins | HSTs; targets membrane protein. | [68,71,72] |
| A. alternata f. sp. citri jambhiri | ACR toxins | Lemon | HSTs; targets mitochondria. | [68,71,72] |
| A. alternata f. sp. mali | AM toxins | Apple | HSTs; targets membrane protein and chloroplasts. | [68,73] |
2. Proteinaceous Effectors
2.1. Proteinaceous Effectors Secreted by Bacteria
As single cell organisms, pathogenic bacteria have special secretion systems to translocate their effectors. Gram-positive bacteria can deliver effectors though the conserved Sec secretion system. However, horticultural crops are mainly infected by gram-negative bacteria, which have 10 special systems (type I-X secretion systems, T1SS-T10SS) to translocate different types of effectors (T1SE-T10SE) across the inner and outer membrane and sometimes even the third membrane—the cell membrane of the host [74,75]. To date, T3SS, T4SS, and T6SS have been well studied and can deliver effectors directly to the host cell [76]. Most studies about phytopathogenic bacteria effectors have focused on T3SEs secreted by common pathogens such as P. syringae, Xanthomonas spp., and R. solanacearum [77]. P. syringae is an useful model bacterium for providing insights into plant–pathogen interactions, with a very wide range of hosts, causing bacterial speck, blight, and canker diseases in many vegetables and fruits [78]. Similarly, Xanthomonas spp. are pathogens of horticultural crops such as pepper, tomato, and citrus, causing bacterial blight or leaf streak [79]. Moreover, R. solanacearum, the causal agent of bacterial wilt, is the most important soil-borne pathogen causing devastating diseases in Solanaceaeous species [80].
T3SEs possess diverse biochemical activities and have been reported to target a range of host receptors (Figure 1). The HopZ family is an important T3SE family from P. syringae and is subdivided into HopZ1-5 subfamilies. Some HopZ family effectors exhibit acetyltransferase activity. HopZ1a is an acetyltransferase targeting MAP kinase kinase 7 (MKK7) and tubulin, and can induce host HR [81,82]. HR is an immune-related programmed cell death (PCD) that usually localizes to the site of infection and limits pathogen extension. Interestingly, HopZ3 effectors acetylate not only components of plant immune complexes, but also other effectors activating the complexes [20]. HopZ3 effector B728a can acetylate effector AvrPto1Psy and the host kinases PTO, critical components of the immune complex [20]. Moreover, B728a disrupts the AvrPto1Psy-PTO interaction and prevents initiation of defense response in tomato (Solanum lycopersicum). Some T3SE effectors exhibit E3 ubiquitin ligase activity and promote degradation of immunity-related proteins through the proteasome pathway. AvrPtoB is an effector secreted by P. syringae pv. tomato. The C-terminal of AvrPtoB has an E3 ubiquitin ligase domain [21]. When the N-terminal of the T3SE effector interacts with host kinase Fen, the C-terminal E3 ligase mediates the ubiquitination of Fen and subsequent degradation in the proteasome, and inhibits both PTI and ETI in tomato [21]. In addition, R. solanacearum secretes Ralstonia injected proteins (Rips) RipAR and RipAW, which also exhibit E3 ligase activity, to suppress PTI responses of the host [83].
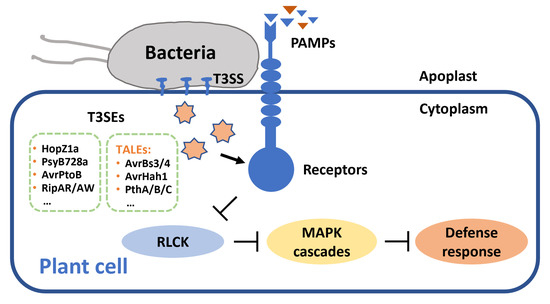
Figure 1.
Bacterial pathogens secrete T3SEs targeting receptors and suppress host defense response. T3SS: type III secretion system; T3SEs: type III secreted effectors; TALEs: transcription activator-like effectors; PAMPs: pathogen-associated molecular patterns; RLCK: receptor-like cytoplasmic kinases; MAPK: mitogen-activated protein kinase.
Xanthomonas spp. secrete T3SEs possessing transcription activators called TALEs (transcription activator-like effectors). TALEs locate themselves at the nucleus of host cells and activate the expression of target genes [79]. The conserved C-terminal region of those effectors contains a nuclear localization signal (NLS) and transcriptional activation domain (AD). TALEs usually bind to the effector binding elements (EBEs) region within the promoter region of target genes to induce expression of downstream susceptibility genes [79]. Xanthomonas campestris pv. vesicatoria (Xcv), the causal agent of pepper bacterial spot disease, secretes two TALE effectors AvrBs3 and AvrBs4 to individually target two host R genes, Bs3 and Bs4, thereby triggering a hypersensitive response in the host [28]. Moreover, AvrBs3 can induce expression of several host upa (upregulated by AvrBs3) genes, such as upa16 and upa20, thus triggering the resistance response of pepper [27,84,85]. In addition, TALE effector AvrHah1, homologous to avrBs3 in Xanthomonas gardneri, induces expression of basic helix–loop–helix (bHLH) genes bHLH3 and bHLH6, and activates transcription of pectate lyase and pectinesterase, thereby causing water soaking in tomato leaves [29]. In citrus fruit, expression of lateral organ boundaries 1 gene CsLOB1 can be induced by TALEs PthA4, PthAw, PthA, PthB, and PthC, secreted by citrus bacterial canker pathogen Xanthomonas citri subsp. citri (Xcc), thus increasing disease susceptibility in the host [31]. Likewise, AbLOB1 also is a disease susceptibility gene in Chinese box orange (Atalantia buxifolia), and mutation in the EBEs of the AbLOB1 promoter results in Xcc resistance [30].
R. solanacearum secretes many Rips, T3SEs that interfere with host immune responses during infection [86]. Rip36 is a putative Zn-dependent protease, that induces eggplant Solanum torvum HR [22]. RipAB, located in the nucleus, affects Ca2+-dependent gene expression and suppresses the immune response of potato [23]. Effectors such as RipAC, RipAY, RipAK, and RipAV contribute to bacterial fitness in eggplant or tomato [25,87,88]. RipAC and RipG are conducive to virulence in tomato or potato [88,89]. RipI and RipAB can induce cell death in tomato or potato [24,90]. RipAX2 has a zinc-binding motif and triggers resistance in eggplant AG91-25 [26].
2.2. Proteinaceous Effectors Secreted by Oomycetes
Species belonging to the genera Pythium, Phytophthora, Peronospora, and Plasmopara are important oomycetous phytopathogens [91]. Some cause serious diseases in horticultural crops including potato, tomato, pepper, and grape. After they adhere to plants, oomycetes form appressoria on the host cell surface, then hyphae penetrate the host cells to extend haustoria. Haustoria deliver virulence factors, especially effectors, that enhance pathogenicity [92]. Proteinaceous effectors in oomycetes include apoplastic and cytoplasmic effectors [93].
Apoplastic effectors secreted by oomycetes include elicitins, NLP (necrosis and ethylene-inducing peptide 1 (NEP)-like protein) family proteins, protease inhibitors, cell-wall degrading enzymes (CWDEs), and SCR (small cysteine-rich) proteins [94,95]. Elicitins are conserved, sterol-binding virulence effectors in oomycetes. Elicitins can be recognized by plant PRRs (pattern recognition receptors), trigger PTI, and protect plants by eliciting HR cell death [96]. P. infestans elicitins, such as INF1, inf2a, INF5, and INF6 can induce cell death in plants such as tobacco, but not in cultivated potatoes [34]. Wild potato Solanum microdontum cell-surface receptor-like protein ELR (elicitin response) is associated with extracellular elicitin recognition [35], and overexpression of ELR in cultivated potatoes makes the plants detect elicitins and promotes a cell-death phenotype, resulting in enhanced resistance to P. infestans [35]. NLPs also induce host cell death, but NLPs are likely to kill the host cells during the oomycetes’ necrotrophic living phase to help invasion. For example, PiNPP1.1 is upregulated during late stages of infection and induces cell death in tomato [36]. Meanwhile, oomycetes secrete protease inhibitors to protect themselves from plant defenses. Apoplastic effectors EPI1 and EPI10 of P. infestans inhibit the pathogenesis-related protein P69B in tomato. EPI10 also specifically inhibits subtilisin A to enhance host susceptibility [37]. Furthermore, P. infestans secretes EPIC1 and EPIC2B to inhibit cysteine protease Rcr3pim in tomato, and mutant Rcr3 exhibits more susceptibility to P. infestans [39]. In addition, extracellular effector PsAvh240 from Phytophthora sojae is able to prevent secretion of GmAP1 (aspartic protease) in soybean by interacting with it in the membrane and suppressing host immunity [97]. Similarly, CWDE effector PsXEG1 (endoglucanase) targets another aspartic protease GmAP5 and N-glycosylation at N174 and N190 of PsXEG1 results in binding and degradation by GmAP5 [98].
Cytoplasmic effectors need to be translocated from haustoria to the host cell. Two motifs—RXLR-dEER (X means any amino acid) and LFLAK-HVLV, referred to as CRN—are common and important cytoplasmic effectors in oomycetes [99,100] (Figure 2). N-terminal RXLR and CRN motifs play critical roles in the translocation of effectors [94]. RXLR and CRN effectors are very abundant in Phytophthora species. Almost 560 RXLR effectors and 450 CRN effectors are encoded by the P. infestans genome [94].
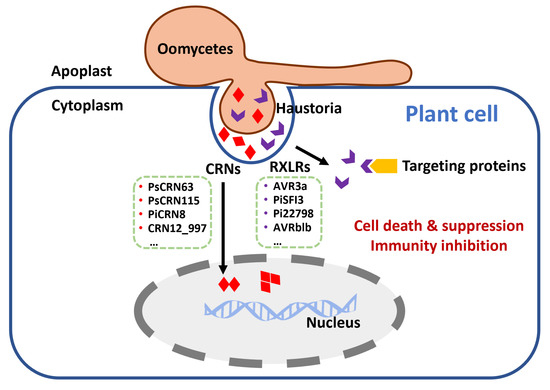
Figure 2.
Oomycetous pathogens secrete RXLR and CRN effectors to induce or suppress host cell death and inhibit host immunity. RXLR: RXLR motif (X means any amino acid); CRN: crinkling and necrosis.
RXLR and CRN effectors have been reported to induce PCD, suppress cell death, or inhibit the immune system of the host [94,101,102]. Suppressing cell death in the host promotes colonization. RXLR effector avirulence 3 (AVR3a) from P. infestans targets the R gene R3a to suppress INF1-triggered cell death (ICD) in potato. Moreover, AVR3a stabilizes ubiquitin E3 ligase CMPG1 to inhibit ICD during the biotrophic phase of infection, while the C-terminal tyrosine residue mutant of AVR3a fails to suppress ICD [45,103]. Furthermore, RXLR effectors can directly manipulate plant immunity to promote colonization by interacting with host-resistance proteins. Downy mildew pathogen Plasmopara viticola secretes RXLR50253 to inhibit grapevine ICD, while RXLR50253 can stabilize grapevine VpBPA1 and reduce the accumulation of H2O2 that promotes infection [46]. Similarly, RXLR effector Pi17316 suppresses ICD and enhances colonization in potato leaves [104], while Pi17316 activity in potato is mediated by MAP3K protein StVIK, which is a susceptibility factor and negatively regulates immunity [104]. P. infestans RXLR effectors PiSFI1-8 are essential to interfere with host PTI and influence immunity [105]. For example, PiSFI3 targets potato U-box-kinase protein (StUBK) to enhance leaf colonization of P. infestans [40]. UBK is a positive regulator of immunity in potato and Nicotiana, thus PiSFI3 can suppress early transcriptional responses of PTI by interaction with StUBK [40]. Moreover, effector Pi22798 targets negative regulator StKNOX3, a transcription factor in potato, and promotes its homodimerization [43]. StKNOX3 homodimerization is crucial for the interaction with Pi22798 and for StKNOX3 to enhance susceptibility in the host [43]. Additionally, Phytophthora brassicae RXLR effector RXLR24 has been reported to interact with potato RABA GTPases to help colonization. This interaction can repress RABA GTPases-mediated secretion of core antimicrobials, such as PR-1 and defensin (PDF1.2) [32]. RXLR effectors are also able to inhibit resistance by affecting the location of resistant proteins in the host. For instance, P. infestans RXLR effector AVRblb2 prevents papain-like cysteine protease C14 translocation to the apoplast in tomato plants. Because C14 plays a positive role in plant immunity, the inhibition of its translocation decreases the host defense [106].
Unlike RXLR effectors, all of the Phytophthora CRNs identified to date are localized in the nucleus of host cells. The CRN motif comprises 50 amino acids including the LFLAK or HVLV motif [94]. Most of the CRN effectors are enzymes, including protein kinase, nuclease, and peptidase [100]. Many CRNs themselves form dimers or multimers with other proteins. In P. sojae, PsCRN63 is able to induce necrosis and suppresses PTI by influencing MAPK cascades downstream, while PsCRN115 prevents PCD by recruiting catalase and promoting hydrogen peroxide (H2O2) accumulation [107]. PsCRN63 can homo-dimerize and dimerize with other effectors such as PsCRN115, PsCRN79, or PcCRN4. These homo-/heterodimers are indispensable for pathogen virulence [108]. PiCRN8 secreted by P. infestans is a functional RD kinase that triggers cell death in the host [109]. Its C-terminal RD kinase domain is essential for the formation of dimers or multimers. Its dominant-negative mutant shows reduced P. infestans infection [109]. Furthermore, CRNs can also interact with host proteins to facilitate infection. CRN12_997 from P. capsica interacts with TCP transcription factor SlTCP14-2 in tomato, which is an immunity regulator associated with nuclear chromatin [33]. The interaction impairs the DNA binding activity of the immune regulator. Therefore, CRN12_997 enhances the susceptibility of tomato to P. capsici.
2.3. Proteinaceous Effectors Secreted by Fungi
Pathogenic fungi are notorious enemies of horticultural crops, especially certain necrotrophic fungi including B. cinerea, A. alternata, and Penicillium spp. During the colonization stage, fungi break through plant cell walls and defense systems by secreting various virulence factors including effectors via the hyphae or appressorium [110]. Compared with bacteria and oomycetes, fungi have fewer effectors with conserved domains and clearly elucidated functions [19].
Recent studies of fungal effector domains have mainly referred to glycosyl hydrolases, lipases, cerato-platanin (CP) protein (CPP) family effectors, NLPs, and LysM effectors [17,19]. Fungi secrete CWDEs such as glycosyl hydrolases to degrade the walls of host cells, and some of them can cause the death of host cells. In B. cinerea, several CWDE effectors have been discovered, some of which induce HR cell death, such as endopolygalacturonases BcPG1 and BcPG2, glucan 1,4-alphaglucosidase BcGs1, and xylanase BcXyn11/BcXyn11A [19,48,50]. Meanwhile, effectors with cerato-platanin-domain-inducing PCD have also been widely reported. Moniliophthora perniciosa, causing witches’ broom disease in Theobroma cacao (cacao), secretes MpCP1 inducing cell death in cacao leaves [61]. Similarly, B. cinerea delivers BcSpl1 with CP domain, causing PCD in tomato, tobacco, and Arabidopsis leaves [111]. Sclerotinia sclerotiorum also secretes SsCP1, contributing to virulence and inducing cell death. Moreover, SsCP1 can interact with PR1 to promote infection [112]. Interestingly, similar to oomycetes, NLP proteins conserved in multiple fungal pathogens can induce the host cell death to enable invasion, especially in necrotrophic pathogens. A sequence called nlp24 from NLP1 is essential for the pathogenicity of Colletotrichum orbiculare, and the C-terminal region of the NLP1 can trigger defense in cucumber [58]. Penicillium expansum is one of the most destructive pathogens and causes blue mold in a wide range of harvested fruits [113]. Two NLP proteins, Penlp1 and Penlp2 from P. expansum, have been reported to induce cell death, but only the Penlp1 mutant was associated with virulence in apples [62].
It has been demonstrated that LysM domain effectors can bind glycans, thus blocking chitin sensing and then inhibiting the host immune system [114,115] (Figure 3). The tomato leaf mold pathogen C. fulvum can secrete LysM effector Ecp6 to bind chitin oligosaccharides during infection and evade plant immunity [52]. Ecp6 has three LysM domains, and two of these LysM domains can dimerize, which ensures a super chitin-binding ability [114]. Later, this mechanism was found in many other pathogens. LysM effectors ChELP1 and ChELP2 from Colletotrichum higginsianum, which can cause anthracnose disease in cultivated Brassicaceae, bind chitin and chitin oligomers [57]. RNAi of ChELP1 and ChELP2 reduces fungal pathogenicity. In P. expansum, four LysM effectors express highly during infection, but they did not affect the virulence [116]. Some effectors do not have a LysM domain, but nevertheless also present chitin-binding activity. For example, effector Avr4 secreted by C. fulvum is not an LysM effector, but it has a chitin-binding domain which protects fungal chitin from plant chitinases and induces HR in tomato [55].
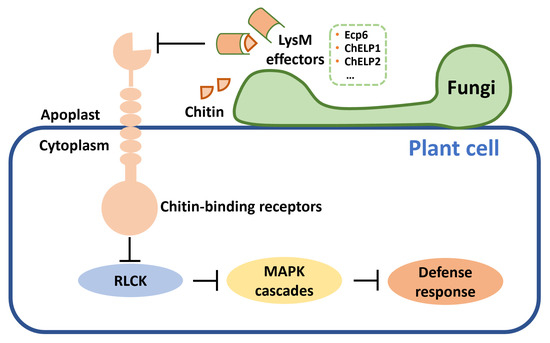
Figure 3.
Fungal pathogens secrete LysM effectors blocking chitin sensing and suppress host defense response. LysM: lysin motifs; RLCK: receptor-like cytoplasmic kinases; MAPK: mitogen-activated protein kinase.
Furthermore, some fungal effectors demonstrate other enzyme activities, such as chitinase or ribonuclease activity. A transcriptome analysis identified several effectors from the tomato vascular wilt pathogen Fusarium oxysporum f. sp. lycopersici (Fol), such as Fol-EC19 (a guanyl-specific ribonuclease) inducing cell death, as well as Fol-EC14 (a glucanase) and Fol-EC20 (a trypsin) suppressing Bax/I-2/Avr2-mediated cell death [59]. Some effectors secreted by B. cinerea and S. sclerotiorum exert chitinase activity that can break down chitin oligomers, thus preventing detection by plants [117]. In contrast, some fungal effectors show enzyme inhibitor activity. C. fulvum Avr2 is a cysteine protease inhibitor that inhibits tomato or Arabidopsis proteases such as Rcr3, Pip1, aleurain, and TDI-65 [53,54,118]. Overexpression of Avr2 in Arabidopsis thaliana increases susceptibility toward other fungi such as B. cinerea and Verticillium dahlia [53,118].
Furthermore, fungi secrete abundant uncharacterized domain effectors during infection. Some uncharacterized effectors induce cell death in the host. For example, in C. fulvum (Cf), Avr4E and Ecp1/2/4/5/7 are uncharacterized. All Avr effectors can induce HR, and Ecp (extracellular protein) effectors trigger Cf-Ecp mediated HR [55]. Similarly, small secreted proteins (SSP) BcSSP2/3 from B. cinerea and SsSSP3 from S. sclerotiorum can cause HR cell death in camelliae and N. benthamiana [47]. Apple Valsa canker fungus Valsa mali secreted 70 candidate effectors and seven of them, called VmEPs, were shown to inhibit BAX-associated PCD in N. benthamiana [119]. Some uncharacterized effectors are required for full virulence of pathogens while others are not involved in virulence. Secreted-in-xylem (SIX) effectors are secreted by Fol during infection of tomato, in which SIX1/2/3/5/6 are required for full virulence of the fungi, while SIX7/10/12 do not affect the pathogenicity [120,121]. Similarly, cabbage wilt disease pathogen F. oxysporum f. sp. conglutinans (Foc) can secrete a host specific effector Foc-SIX1 [60].
3. Non-Proteinaceous Effectors
Non-proteinaceous effectors such as sRNAs and fungal secondary metabolites (toxins) also play important biological roles. During the plant–pathogen interaction, as large biological molecular agents, sRNAs can play a crucial function through RNA interference (RNAi) [122]. From the perspective of the pathogen, sRNAs can act like effectors targeting plant gene expression [6]. Fungal sRNAs can suppress the immune system of the host by hijacking the RNAi machinery or silencing resistant genes. B. cinerea can deliver a large amount of sRNAs to many hosts such as Arabidopsis and tomato [64]. Its sRNA can bind the plant’s Argonaute 1(AGO1) to hijack host RNAi machinery and silence genes involved in host immunity [64]. Moreover, the B. cinerea mutant of two important sRNA processing enzymes, BcDCL1 and BcDCL2 (dicer-like 1, 2), displays reduced pathogenicity. Similar findings have also been reported in V. mali and Penicillium italicum [65,66]. The latter is the causal agent of blue mold in citrus.
In addition, fungi produce a variety of secondary metabolites (SMs), which also function as effectors to help colonization in hosts [6]. Generally, these SMs are phytotoxins and are classified as non-host-specific toxins (nHSTs) or host-specific toxins (HSTs). T-toxin is a well-known host-selected phytotoxin produced by Cochliobolus heterostrophus, the causal agent of corn leaf blight [123]. Among horticultural crop pathogens, Alternaria may be the most important genus, able to produce 70 mycotoxins including various HSTs [68]. For example, A. alternata has different pathotypes infecting different plants and producing various HSTs [68]. A. alternata f. sp. lycopersici produces AAL toxins that induce cell apoptosis in susceptible tomato strains without the Asc1 (Alternaria stem canker resistance gene 1) gene [67,68]. In strawberry, A. alternata f. sp. fragariae produces HST AF-toxin and causes black spot [69]. A. alternata f. sp. kikuchana (Japanese pear pathotype) produces AK-toxin [70]. Spray experiments showed that AK-toxin was only toxic to susceptible pear cultivars [124]. In lemon, A. alternata f. sp. citri secretes ACR-toxin to promote colonization [71,72]. A. alternata f. sp. mali, the apple pathotype, produces HST AM-toxin and causes apple Alternaria blotch [73].
4. Plant Defense Response against Effectors
When pathogens infect plants, plant resistance response usually begins with the recognition of the PAMPs of the pathogens and leads to PTI [125]. Flagellin and translational elongation factor Tu from bacteria and chitin from fungi are well-known PAMPs. Plant PRRs such as RLP (receptor-like proteins) with LRR (leucine-rich repeat) domain and RLK (receptor-like kinase) are usually responsible for the recognition [126]. These receptors locate in the plant cell plasma and recognize PAMPs, then the signal is transduced to transmembrane RLKS like BAK1 (BRI1-associated receptor kinase1) or the RLCKs (receptor-like cytoplasmic kinases) [127]. Afterwards, the signal continues to travel downstream through the MAP (mitogen-activated protein) cascade and finally reaches resistance genes activating the PTI [128]. This process includes the outbreak of ROS (reactive oxygen species), callose deposition, and concentration of Ca2+, which blocks further invasion [7]. To tackle these challenges, pathogens have developed various effectors to suppress this response. At the same time, plants have co-evolved specific mechanisms to recognize these effectors and initiate ETI to prevent colonization [129].
Plants use various kinds of receptors to detect effectors. Most receptors are NLRs (intracellular nucleotide-binding leucine rich repeat) and are classified into TIR (Toll/interleukin-1 receptor) domain NLRs (TNLs), CC (coiled-coil) domain NLRs (CNLs), and RPW8 (resistance to powdery mildew 8)-like coiled-coil domain NLRs (RNLs), according to their conserved domains [130]. For example, two NLRs, ZAR1 (HopZ-activated resistance1) and CAR1 (Carbonic anhydrase I) in N. benthamiana can recognize 95% of T3SEs of P. syringae [78,131]. NLR Bs4, an R protein in tomato, can suppress HR induced by effector AvrBs3 and reduce the virulence of effector AvrHah1 from tomato-pathogenic Xanthomonas strains. [132]. LRR-RLP RXEG1 of N. benthamiana detects and binds effector XEG1 from P. sojae, and triggers immune responses by changing the structure of RXEG1 and enhancing association of RXEG1 with BAK1 [133]. Rpi-amr3 from wild Solanaceaeous plant Solanum americanum can recognize conversed RXLR-WY effector AVRamr3 and activate resistance against different Phytophthora species, including P. infestans, P. parasitica, and P. palmivora [44].
Studies on the plant defense response against effectors can help explore comprehensive plant immune pathways and provide potential resistance genes for breeders to develop resistant cultivars by traditional and genetic modification-based methods. Introducing multiple R genes of Cf-2, Cf-4, Cf-4E, Cf-5, and Cf-9 into tomato has prevented outbreak of leaf mold for several decades [12]. Abundant late blight resistance genes (Rpi genes) have been identified from wild potato species [13]. The construction and utilization of Rpi gene pyramids may achieve durable and broad-spectrum late blight resistance. Resistant genes from other plants can also be deployed. Transgenic tomato containing resistant gene Bs2 from pepper demonstrated resistance to all field strains of Xanthomonas [13]. In addition, heterologous overexpression of some effectors in plants can enhance their immunity. Overexpression of oomycete effector PsCRN115 increased host resistance against two Phytophthora pathogens [134]. Interestingly, heterologous expression of non-host effector target orthologues in host plants can cause broad-spectrum immunity. Expression of cAtOrth AtPUB33, Arabidopsis target orthologues of P. infestans effector PiSFI3, in potato and tobacco significantly reduced P. infestans infection [135].
5. Conclusions and Perspectives
Horticultural crop pathogens produce proteinaceous and non-proteinaceous effectors, which play crucial roles in the interactions between pathogens and hosts. On one hand, effectors such as various enzymes, enzyme inhibitors, and phytotoxins, demonstrate diverse activities and facilitate pathogenic infection by preventing recognition of pathogens, suppressing PTI, or hijacking the host metabolism. On the other hand, plants recognize effectors and trigger ETI to prevent infection. Many different effectors have been identified among bacteria, oomycetes, and fungi, and the mechanisms of action of some effectors with conserved domain have been well described. However, the known effectors may be just the tip of the iceberg [113].
With the rapid development of techniques such as whole genome sequencing, GWAS (genome-wide association study) analysis, secretomics, and transcriptomics, a broad range of putative effectors have been identified. With a wealth of reference data, software tools such as effectR and EffectorP 3.0 are extremely useful to predict putative fungal and oomycete effectors [136,137]. In recent years, artificial intelligence (AI) has developed rapidly and demonstrated powerful capabilities in analyzing high-throughput data of life sciences [138]. AI technology will provide great help identifying new effectors in pathogens and receptors in hosts.
Effector biology will contribute to developing resistant varieties of horticultural crop varieties, and to disease control as well as prevention. With a deeper understanding of effector-mediated plant–pathogen interactions, mechanisms of pathogen infection and host immunity will be more clearly elucidated, thus new disease-management strategies can be enforced. Using the application of gene-editing technology such as CRISPR (clustered regularly interspaced short palindromic repeats), simultaneous manipulation of multiple R genes may contribute to the breeding of horticultural crop varieties with durable and broad-spectrum resistance. Moreover, new NLRs can be designed de novo using synthetic biological techniques and developed into horticultural crops.
Author Contributions
Conceptualization, B.L.; writing—original draft preparation, T.L.; writing—review and editing, B.L., Y.C. and S.T.; visualization, T.L. and Y.C.; supervision, B.L.; funding acquisition, B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key R&D Program of China (grant number: 2021YFD2100501/05), the Beijing Science and Technology Program (grant number: Z201100008920007), and Youth Innovation Promotion Association, CAS (grant number: Y201919).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Perera, C.O.; Smith, B. Handbook of Farm, Dairy and Food Machinery Engineering; Academic Press: San Diego, CA, USA, 2013; Chapter 13; pp. 299–315. [Google Scholar]
- Fry, W.E.; Birch, P.R.; Judelson, H.S.; Grunwald, N.J.; Danies, G.; Everts, K.L.; Gevens, A.J.; Gugino, B.K.; Johnson, D.A.; Johnson, S.B.; et al. Five reasons to consider Phytophthora infestans a reemerging pathogen. Phytopathology 2015, 105, 966–981. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, M.C.; Valent, B. Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 2013, 11, 800–814. [Google Scholar] [CrossRef] [PubMed]
- Shao, D.; Smith, D.L.; Kabbage, M.; Roth, M.G. Effectors of plant necrotrophic fungi. Front. Plant Sci. 2021, 12, 687713. [Google Scholar] [CrossRef]
- Kraepiel, Y.; Barny, M.A. Gram-negative phytopathogenic bacteria, all hemibiotrophs after all? Mol. Plant Pathol. 2016, 17, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Collemare, J.; O’Connell, R.; Lebrun, M.H. Nonproteinaceous effectors: The terra incognita of plant-fungal interactions. New Phytol. 2019, 223, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Feng, B.; Zhou, J.M.; Tang, D. Plant immune signaling: Advancing on two frontiers. J. Integr. Plant Biol. 2020, 62, 2–24. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Koeck, M.; Hardham, A.R.; Dodds, P.N. The role of effectors of biotrophic and hemibiotrophic fungi in infection. Cell Microbiol. 2011, 13, 1849–1857. [Google Scholar] [CrossRef]
- Kanja, C.; Hammond-Kosack, K.E. Proteinaceous effector discovery and characterization in filamentous plant pathogens. Mol. Plant Pathol. 2020, 21, 1353–1376. [Google Scholar] [CrossRef]
- Iswanto, A.B.B.; Vu, M.H.; Pike, S.; Lee, J.; Kang, H.; Son, G.H.; Kim, J.Y.; Kim, S.H. Pathogen effectors: What do they do at plasmodesmata? Mol. Plant Pathol. 2022, 23, 795–804. [Google Scholar] [CrossRef]
- de Wit, P.J. Cladosporium fulvum effectors: Weapons in the arms race with tomato. Annu. Rev. Phytopathol. 2016, 54, 1–23. [Google Scholar] [CrossRef]
- Paluchowska, P.; Sliwka, J.; Yin, Z. Late blight resistance genes in potato breeding. Planta 2022, 255, 127. [Google Scholar] [CrossRef]
- Adhikari, P.; Adhikari, T.B.; Louws, F.J.; Panthee, D.R. Advances and challenges in bacterial spot resistance breeding in tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2020, 21, 1734. [Google Scholar] [CrossRef]
- Chitwood-Brown, J.; Vallad, G.E.; Lee, T.G.; Hutton, S.F. Breeding for resistance to Fusarium wilt of tomato: A review. Genes 2021, 12, 1673. [Google Scholar] [CrossRef]
- Zhang, S.; Li, C.; Si, J.; Han, Z.; Chen, D. Action mechanisms of effectors in plant-pathogen interaction. Int. J. Mol. Sci. 2022, 23, 6758. [Google Scholar] [CrossRef]
- De Wit, P.J.; Mehrabi, R.; Van den Burg, H.A.; Stergiopoulos, I. Fungal effector proteins: Past, present and future. Mol. Plant Pathol. 2009, 10, 735–747. [Google Scholar] [CrossRef]
- Jaswal, R.; Kiran, K.; Rajarammohan, S.; Dubey, H.; Singh, P.K.; Sharma, Y.; Deshmukh, R.; Sonah, H.; Gupta, N.; Sharma, T.R. Effector biology of biotrophic plant fungal pathogens: Current advances and future prospects. Microbiol. Res. 2020, 241, 126567. [Google Scholar] [CrossRef]
- Lo Presti, L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal effectors and plant susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Jelenska, J.; Lee, J.; Manning, A.J.; Wolfgeher, D.J.; Ahn, Y.; Walters-Marrah, G.; Lopez, I.E.; Garcia, L.; McClerklin, S.A.; Michelmore, R.W.; et al. Pseudomonas syringae effector HopZ3 suppresses the bacterial AvrPto1-tomato PTO immune complex via acetylation. PLoS Pathog. 2021, 17, e1010017. [Google Scholar] [CrossRef]
- Rosebrock, T.R.; Zeng, L.; Brady, J.J.; Abramovitch, R.B.; Xiao, F.; Martin, G.B. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 2007, 448, 370–374. [Google Scholar] [CrossRef]
- Nahar, K.; Matsumoto, I.; Taguchi, F.; Inagaki, Y.; Yamamoto, M.; Toyoda, K.; Shiraishi, T.; Ichinose, Y.; Mukaihara, T. Ralstonia solanacearum type III secretion system effector Rip36 induces a hypersensitive response in the nonhost wild eggplant Solanum torvum. Mol. Plant Pathol. 2014, 15, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Li, X.; Wang, B.; Cheng, D.; Li, Y.; Li, W.; Huang, M.; Tan, X.; Zhao, G.; Song, B.; et al. A systematic screen of conserved Ralstonia solanacearum effectors reveals the role of RipAB, a nuclear-localized effector that suppresses immune responses in potato. Mol. Plant Pathol. 2019, 20, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Zhuo, T.; Wang, X.; Chen, Z.; Cui, H.; Zeng, Y.; Chen, Y.; Fan, X.; Hu, X.; Zou, H. The Ralstonia solanacearum effector RipI induces a defence reaction by interacting with the bHLH93 transcription factor in Nicotiana benthamiana. Mol. Plant Pathol. 2020, 21, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Kawazoe, T.; Ohnishi, K.; Kitagawa, T.; Popa, C.; Valls, M.; Genin, S.; Nakamura, K.; Kuramitsu, Y.; Tanaka, N.; et al. RipAY, a plant pathogen effector protein, exhibits robust gamma-glutamyl cyclotransferase activity when stimulated by eukaryotic thioredoxins. J. Biol. Chem. 2016, 291, 6813–6830. [Google Scholar] [CrossRef]
- Morel, A.; Guinard, J.; Lonjon, F.; Sujeeun, L.; Barberis, P.; Genin, S.; Vailleau, F.; Daunay, M.C.; Dintinger, J.; Poussier, S.; et al. The eggplant AG91-25 recognizes the Type III-secreted effector RipAX2 to trigger resistance to bacterial wilt (Ralstonia solanacearum species complex). Mol. Plant Pathol. 2018, 19, 2459–2472. [Google Scholar] [CrossRef]
- Romer, P.; Strauss, T.; Hahn, S.; Scholze, H.; Morbitzer, R.; Grau, J.; Bonas, U.; Lahaye, T. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009, 150, 1697–1712. [Google Scholar] [CrossRef]
- Schornack, S.; Ballvora, A.; Gurlebeck, D.; Peart, J.; Baulcombe, D.; Ganal, M.; Baker, B.; Bonas, U.; Lahaye, T. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004, 37, 46–60. [Google Scholar] [CrossRef]
- Schwartz, A.R.; Morbitzer, R.; Lahaye, T.; Staskawicz, B.J. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc. Natl. Acad. Sci. USA 2017, 114, E897–E903. [Google Scholar] [CrossRef]
- Tang, X.; Wang, X.; Huang, Y.; Ma, L.; Jiang, X.; Rao, M.J.; Xu, Y.; Yin, P.; Yuan, M.; Deng, X.; et al. Natural variations of TFIIAgamma gene and LOB1 promoter contribute to citrus canker disease resistance in Atalantia buxifolia. PLoS Genet. 2021, 17, e1009316. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef]
- Tomczynska, I.; Stumpe, M.; Mauch, F. A conserved RxLR effector interacts with host RABA-type GTPases to inhibit vesicle-mediated secretion of antimicrobial proteins. Plant J. 2018, 95, 187–203. [Google Scholar] [CrossRef]
- Stam, R.; Motion, G.B.; Martinez-Heredia, V.; Boevink, P.C.; Huitema, E. A conserved oomycete CRN effector targets tomato TCP14-2 to enhance virulence. Mol. Plant Microbe Interact. 2021, 34, 309–318. [Google Scholar] [CrossRef]
- Derevnina, L.; Dagdas, Y.F.; De la Concepcion, J.C.; Bialas, A.; Kellner, R.; Petre, B.; Domazakis, E.; Du, J.; Wu, C.H.; Lin, X.; et al. Nine things to know about elicitins. New Phytol. 2016, 212, 888–895. [Google Scholar] [CrossRef]
- Du, J.; Verzaux, E.; Chaparro-Garcia, A.; Bijsterbosch, G.; Keizer, L.C.; Zhou, J.; Liebrand, T.W.; Xie, C.; Govers, F.; Robatzek, S.; et al. Elicitin recognition confers enhanced resistance to Phytophthora infestans in potato. Nat. Plants 2015, 1, 15034. [Google Scholar] [CrossRef]
- Kanneganti, T.D.; Huitema, E.; Cakir, C.; Kamoun, S. Synergistic interactions of the plant cell death pathways induced by Phytophthora infestans Nepl-like protein PiNPP1.1 and INF1 elicitin. Mol. Plant Microbe Interact. 2006, 19, 854–863. [Google Scholar] [CrossRef]
- Tian, M.; Benedetti, B.; Kamoun, S. A Second Kazal-like protease inhibitor from Phytophthora infestans inhibits and interacts with the apoplastic pathogenesis-related protease P69B of tomato. Plant Physiol. 2005, 138, 1785–1793. [Google Scholar] [CrossRef]
- Kaschani, F.; Shabab, M.; Bozkurt, T.; Shindo, T.; Schornack, S.; Gu, C.; Ilyas, M.; Win, J.; Kamoun, S.; van der Hoorn, R.A. An effector-targeted protease contributes to defense against Phytophthora infestans and is under diversifying selection in natural hosts. Plant Physiol. 2010, 154, 1794–1804. [Google Scholar] [CrossRef]
- Song, J.; Win, J.; Tian, M.; Schornack, S.; Kaschani, F.; Ilyas, M.; van der Hoorn, R.A.; Kamoun, S. Apoplastic effectors secreted by two unrelated eukaryotic plant pathogens target the tomato defense protease Rcr3. Proc. Natl. Acad. Sci. USA 2009, 106, 1654–1659. [Google Scholar] [CrossRef]
- He, Q.; McLellan, H.; Hughes, R.K.; Boevink, P.C.; Armstrong, M.; Lu, Y.; Banfield, M.J.; Tian, Z.; Birch, P.R.J. Phytophthora infestans effector SFI3 targets potato UBK to suppress early immune transcriptional responses. New Phytol. 2019, 222, 438–454. [Google Scholar] [CrossRef]
- McLellan, H.; Boevink, P.C.; Armstrong, M.R.; Pritchard, L.; Gomez, S.; Morales, J.; Whisson, S.C.; Beynon, J.L.; Birch, P.R. An RxLR effector from Phytophthora infestans prevents re-localisation of two plant NAC transcription factors from the endoplasmic reticulum to the nucleus. PLoS Pathog. 2013, 9, e1003670. [Google Scholar] [CrossRef]
- Wang, J.; Gao, C.; Li, L.; Cao, W.; Dong, R.; Ding, X.; Zhu, C.; Chu, Z. Transgenic RXLR effector PITG_15718.2 suppresses immunity and reduces vegetative growth in potato. Int. J. Mol. Sci. 2019, 20, 3031. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Qi, Y.; Nie, J.; Guo, L.; Luo, M.; McLellan, H.; Boevink, P.C.; Birch, P.R.J.; Tian, Z. A Phytophthora effector promotes homodimerization of host transcription factor StKNOX3 to enhance susceptibility. J. Exp. Bot. 2022, 73, 6902–6915. [Google Scholar] [CrossRef]
- Lin, X.; Olave-Achury, A.; Heal, R.; Pais, M.; Witek, K.; Ahn, H.K.; Zhao, H.; Bhanvadia, S.; Karki, H.S.; Song, T.; et al. A potato late blight resistance gene protects against multiple Phytophthora species by recognizing a broadly conserved RXLR-WY effector. Mol. Plant 2022, 15, 1457–1469. [Google Scholar] [CrossRef] [PubMed]
- Bos, J.I.; Armstrong, M.R.; Gilroy, E.M.; Boevink, P.C.; Hein, I.; Taylor, R.M.; Zhendong, T.; Engelhardt, S.; Vetukuri, R.R.; Harrower, B.; et al. Phytophthora infestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. USA 2010, 107, 9909–9914. [Google Scholar] [CrossRef] [PubMed]
- Yin, X.; Fu, Q.; Shang, B.; Wang, Y.; Liu, R.; Chen, T.; Xiang, G.; Dou, M.; Liu, G.; Xu, Y. An RxLR effector from Plasmopara viticola suppresses plant immunity in grapevine by targeting and stabilizing VpBPA1. Plant J. 2022, 112, 104–114. [Google Scholar] [CrossRef]
- Denton-Giles, M.; McCarthy, H.; Sehrish, T.; Dijkwel, Y.; Mesarich, C.H.; Bradshaw, R.E.; Cox, M.P.; Dijkwel, P.P. Conservation and expansion of a necrosis-inducing small secreted protein family from host-variable phytopathogens of the Sclerotiniaceae. Mol. Plant Pathol. 2020, 21, 512–526. [Google Scholar] [CrossRef]
- Kars, I.; Krooshof, G.H.; Wagemakers, L.; Joosten, R.; Benen, J.A.; van Kan, J.A. Necrotizing activity of five Botrytis cinerea endopolygalacturonases produced in Pichia pastoris. Plant J. 2005, 43, 213–225. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.; Qiu, D.; Zeng, H.; Guo, L.; Yang, X. BcGs1, a glycoprotein from Botrytis cinerea, elicits defence response and improves disease resistance in host plants. Biochem. Biophys Res. Commun. 2015, 457, 627–634. [Google Scholar] [CrossRef]
- Frias, M.; Gonzalez, M.; Gonzalez, C.; Brito, N. A 25-residue peptide from Botrytis cinerea xylanase BcXyn11A elicits plant defenses. Front. Plant Sci. 2019, 10, 474. [Google Scholar] [CrossRef]
- Bi, K.; Scalschi, L.; Jaiswal, N.; Mengiste, T.; Fried, R.; Sanz, A.B.; Arroyo, J.; Zhu, W.; Masrati, G.; Sharon, A. The Botrytis cinerea Crh1 transglycosylase is a cytoplasmic effector triggering plant cell death and defense response. Nat. Commun. 2021, 12, 2166. [Google Scholar] [CrossRef]
- de Jonge, R.; van Esse, H.P.; Kombrink, A.; Shinya, T.; Desaki, Y.; Bours, R.; van der Krol, S.; Shibuya, N.; Joosten, M.H.; Thomma, B.P. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 2010, 329, 953–955. [Google Scholar] [CrossRef]
- van Esse, H.P.; Van’t Klooster, J.W.; Bolton, M.D.; Yadeta, K.A.; van Baarlen, P.; Boeren, S.; Vervoort, J.; de Wit, P.J.; Thomma, B.P. The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell 2008, 20, 1948–1963. [Google Scholar] [CrossRef]
- Rooney, H.C.; Van’t Klooster, J.W.; van der Hoorn, R.A.; Joosten, M.H.; Jones, J.D.; de Wit, P.J. Cladosporium Avr2 inhibits tomato Rcr3 protease required for Cf-2-dependent disease resistance. Science 2005, 308, 1783–1786. [Google Scholar] [CrossRef]
- van Esse, H.P.; Bolton, M.D.; Stergiopoulos, I.; de Wit, P.J.; Thomma, B.P. The chitin-binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 2007, 20, 1092–1101. [Google Scholar] [CrossRef]
- Rowland, O.; Ludwig, A.A.; Merrick, C.J.; Baillieul, F.; Tracy, F.E.; Durrant, W.E.; Fritz-Laylin, L.; Nekrasov, V.; Sjolander, K.; Yoshioka, H.; et al. Functional analysis of Avr9/Cf-9 rapidly elicited genes identifies a protein kinase, ACIK1, that is essential for full Cf-9-dependent disease resistance in tomato. Plant Cell 2005, 17, 295–310. [Google Scholar] [CrossRef]
- Takahara, H.; Hacquard, S.; Kombrink, A.; Hughes, H.B.; Halder, V.; Robin, G.P.; Hiruma, K.; Neumann, U.; Shinya, T.; Kombrink, E.; et al. Colletotrichum higginsianum extracellular LysM proteins play dual roles in appressorial function and suppression of chitin-triggered plant immunity. New Phytol. 2016, 211, 1323–1337. [Google Scholar] [CrossRef]
- Azmi, N.S.A.; Singkaravanit-Ogawa, S.; Ikeda, K.; Kitakura, S.; Inoue, Y.; Narusaka, Y.; Shirasu, K.; Kaido, M.; Mise, K.; Takano, Y. Inappropriate expression of an NLP effector in Colletotrichum orbiculare impairs infection on cucurbitaceae cultivars via plant recognition of the c-terminal region. Mol. Plant Microbe Interact. 2018, 31, 101–111. [Google Scholar] [CrossRef]
- Sun, X.; Fang, X.; Wang, D.; Jones, D.A.; Ma, L. Transcriptome analysis of Fusarium-tomato interaction based on an updated genome annotation of Fusarium oxysporum f. sp. lycopersici identifies novel effector candidates that suppress or induce cell death in Nicotiana benthamiana. J. Fungi 2022, 8, 672. [Google Scholar] [CrossRef]
- Li, E.; Wang, G.; Xiao, J.; Ling, J.; Yang, Y.; Xie, B. A SIX1 homolog in Fusarium oxysporum f. sp. conglutinans is required for full virulence on cabbage. PLoS ONE 2016, 11, e0152273. [Google Scholar] [CrossRef]
- Zaparoli, G.; Cabrera, O.G.; Medrano, F.J.; Tiburcio, R.; Lacerda, G.; Pereira, G.G. Identification of a second family of genes in Moniliophthora perniciosa, the causal agent of witches’ broom disease in cacao, encoding necrosis-inducing proteins similar to cerato-platanins. Mycol. Res. 2009, 113, 61–72. [Google Scholar] [CrossRef]
- Levin, E.; Raphael, G.; Ma, J.; Ballester, A.R.; Feygenberg, O.; Norelli, J.; Aly, R.; Gonzalez-Candelas, L.; Wisniewski, M.; Droby, S. Identification and functional analysis of NLP-encoding genes from the postharvest pathogen Penicillium expansum. Microorganisms 2019, 7, 175. [Google Scholar] [CrossRef] [PubMed]
- Wei, W.; Xu, L.; Peng, H.; Zhu, W.; Tanaka, K.; Cheng, J.; Sanguinet, K.A.; Vandemark, G.; Chen, W. A fungal extracellular effector inactivates plant polygalacturonase-inhibiting protein. Nat. Commun. 2022, 13, 2213. [Google Scholar] [CrossRef] [PubMed]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Xu, M.; Liu, Y.; Dong, R.; Gao, X.; Huang, L. Dicer-like genes are required for H2O2 and KCl stress responses, pathogenicity and small RNA generation in Valsa mali. Front. Microbiol. 2017, 8, 1166. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Zhu, H.; Jiang, Y.; Shan, Y.; Gong, L. Silencing dicer-like genes reduces virulence and sRNA generation in Penicillium italicum, the cause of citrus blue mold. Cells 2020, 9, 363. [Google Scholar] [CrossRef]
- Brandwagt, B.F.; Mesbah, L.A.; Takken, F.L.; Laurent, P.L.; Kneppers, T.J.; Hille, J.; Nijkamp, H.J. A longevity assurance gene homolog of tomato mediates resistance to Alternaria alternata f. sp. lycopersici toxins and fumonisin B1. Proc. Natl. Acad. Sci. USA 2000, 97, 4961–4966. [Google Scholar] [CrossRef]
- Meena, M.; Samal, S. Alternaria host-specific (HSTs) toxins: An overview of chemical characterization, target sites, regulation and their toxic effects. Toxicol. Rep. 2019, 6, 745–758. [Google Scholar] [CrossRef]
- Namiki, F.; Yamamoto, M.; Nishimura, S.; Nakatsuka, S.-i.; Goto, T.; Kohmot, K.; Otani, H. Studies on host-specific AF-toxins produced by Alternaria alternata strawberry pathotype causing Alternaria black spot of strawberry (4) protective effect of AF-toxin II on AF-toxin I-induced toxic action and fungal infection. Jpn. J. Phytopathol. 1986, 52, 428–436. [Google Scholar] [CrossRef]
- Tanaka, A.; Tsuge, T. Structural and functional complexity of the genomic region controlling AK-toxin biosynthesis and pathogenicity in the Japanese pear pathotype of Alternaria alternata. Mol. Plant Microbe Interact. 2000, 13, 975–986. [Google Scholar] [CrossRef]
- Kohmoto, K. Host-selective toxins from Alternaria citri. Phytopathology 1979, 69, 667–671. [Google Scholar] [CrossRef]
- Gardner, J.M.; Kono, Y.; Tatum, J.H.; Suzuki, Y.; Takeuchi, S. Plant pathotoxins from Alternaria citri: The major toxin specific for rough lemon plants. Phytochemistry 1985, 24, 2861–2867. [Google Scholar] [CrossRef]
- Ueno, T.; Nakashima, T.; Hayashi, Y.; Fukami, H. Structures of AM-Toxin I and II, host specific phytotoxic metabolites produced by Alternaria mali. Agric. Biol. Chem. 2014, 39, 1115–1122. [Google Scholar] [CrossRef]
- Green, E.R.; Mecsas, J. Bacterial secretion systems: An overview. Microbiol. Spectr. 2016, 4, 213–239. [Google Scholar] [CrossRef]
- Braet, J.; Catteeuw, D.; Van Damme, P. Recent advancements in tracking bacterial effector protein translocation. Microorganisms 2022, 10, 260. [Google Scholar] [CrossRef]
- Galan, J.E.; Waksman, G. Protein-injection machines in bacteria. Cell 2018, 172, 1306–1318. [Google Scholar] [CrossRef]
- Schreiber, K.J.; Chau-Ly, I.J.; Lewis, J.D. What the wild things do: Mechanisms of plant host manipulation by bacterial type III-secreted effector proteins. Microorganisms 2021, 9, 1029. [Google Scholar] [CrossRef]
- Bundalovic-Torma, C.; Lonjon, F.; Desveaux, D.; Guttman, D.S. Diversity, evolution, and function of Pseudomonas syringae effectoromes. Annu. Rev. Phytopathol. 2022, 60, 211–236. [Google Scholar] [CrossRef]
- Zhang, B.; Han, X.; Yuan, W.; Zhang, H. TALEs as double-edged swords in plant-pathogen interactions: Progress, challenges, and perspectives. Plant Commun. 2022, 3, 100318. [Google Scholar] [CrossRef]
- Huang, J.; Wei, Z.; Tan, S.; Mei, X.; Yin, S.; Shen, Q.; Xu, Y. The rhizosphere soil of diseased tomato plants as a source for novel microorganisms to control bacterial wilt. Appl. Soil Ecol. 2013, 72, 79–84. [Google Scholar] [CrossRef]
- Rufian, J.S.; Rueda-Blanco, J.; Lopez-Marquez, D.; Macho, A.P.; Beuzon, C.R.; Ruiz-Albert, J. The bacterial effector HopZ1a acetylates MKK7 to suppress plant immunity. New Phytol. 2021, 231, 1138–1156. [Google Scholar] [CrossRef]
- Lee, A.H.; Hurley, B.; Felsensteiner, C.; Yea, C.; Ckurshumova, W.; Bartetzko, V.; Wang, P.W.; Quach, V.; Lewis, J.D.; Liu, Y.C.; et al. A bacterial acetyltransferase destroys plant microtubule networks and blocks secretion. PLoS Pathog. 2012, 8, e1002523. [Google Scholar] [CrossRef] [PubMed]
- Nakano, M.; Oda, K.; Mukaihara, T. Ralstonia solanacearum novel E3 ubiquitin ligase (NEL) effectors RipAW and RipAR suppress pattern-triggered immunity in plants. Microbiology 2017, 163, 992–1002. [Google Scholar] [CrossRef] [PubMed]
- Morbitzer, R.; Romer, P.; Boch, J.; Lahaye, T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc. Natl. Acad. Sci. USA 2010, 107, 21617–21622. [Google Scholar] [CrossRef] [PubMed]
- Bonas, U.; Stall, R.E.; Staskawicz, B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 1989, 218, 127–136. [Google Scholar] [CrossRef]
- Landry, D.; Gonzalez-Fuente, M.; Deslandes, L.; Peeters, N. The large, diverse, and robust arsenal of Ralstonia solanacearum type III effectors and their in planta functions. Mol. Plant Pathol. 2020, 21, 1377–1388. [Google Scholar] [CrossRef]
- Nakano, M.; Mukaihara, T. Comprehensive identification of PTI suppressors in Type III effector repertoire reveals that Ralstonia solanacearum activates jasmonate signaling at two different steps. Int. J. Mol. Sci. 2019, 20, 5992. [Google Scholar] [CrossRef]
- Macho, A.P.; Guidot, A.; Barberis, P.; Beuzon, C.R.; Genin, S. A competitive index assay identifies several Ralstonia solanacearum type III effector mutant strains with reduced fitness in host plants. Mol. Plant Microbe Interact. 2010, 23, 1197–1205. [Google Scholar] [CrossRef]
- Remigi, P.; Anisimova, M.; Guidot, A.; Genin, S.; Peeters, N. Functional diversification of the GALA type III effector family contributes to Ralstonia solanacearum adaptation on different plant hosts. New Phytol. 2011, 192, 976–987. [Google Scholar] [CrossRef]
- Wroblewski, T.; Caldwell, K.S.; Piskurewicz, U.; Cavanaugh, K.A.; Xu, H.; Kozik, A.; Ochoa, O.; McHale, L.K.; Lahre, K.; Jelenska, J.; et al. Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol. 2009, 150, 1733–1749. [Google Scholar] [CrossRef]
- de Wit, P.J.G.M. Principles of Plant-Microbe Interactions; Springer: Cham, Switzerland, 2015; pp. 79–90. [Google Scholar]
- Dodds, P.N.; Rafiqi, M.; Gan, P.H.P.; Hardham, A.R.; Jones, D.A.; Ellis, J.G. Effectors of biotrophic fungi and oomycetes: Pathogenicity factors and triggers of host resistance. New Phytol. 2009, 183, 993–1000. [Google Scholar] [CrossRef]
- Kamoun, S. A catalogue of the effector secretome of plant pathogenic oomycetes. Annu. Rev. Phytopathol. 2006, 44, 41–60. [Google Scholar] [CrossRef]
- Chepsergon, J.; Motaung, T.E.; Bellieny-Rabelo, D.; Moleleki, L.N. Organize, don’t agonize: Strategic success of Phytophthora species. Microorganisms 2020, 8, 917. [Google Scholar] [CrossRef]
- Mazumdar, P.; Singh, P.; Kethiravan, D.; Ramathani, I.; Ramakrishnan, N. Late blight in tomato: Insights into the pathogenesis of the aggressive pathogen Phytophthora infestans and future research priorities. Planta 2021, 253, 119. [Google Scholar] [CrossRef]
- Noman, A.; Aqeel, M.; Irshad, M.K.; Qari, S.H.; Hashem, M.; Alamri, S.; AbdulMajeed, A.M.; Al-Sadi, A.M. Elicitins as molecular weapons against pathogens: Consolidated biotechnological strategy for enhancing plant growth. Crit. Rev. Biotechnol. 2020, 40, 821–832. [Google Scholar] [CrossRef]
- Guo, B.; Wang, H.; Yang, B.; Jiang, W.; Jing, M.; Li, H.; Xia, Y.; Xu, Y.; Hu, Q.; Wang, F.; et al. Phytophthora sojae effector psavh240 inhibits host aspartic protease secretion to promote infection. Mol. Plant 2019, 12, 552–564. [Google Scholar] [CrossRef]
- Xia, Y.; Ma, Z.; Qiu, M.; Guo, B.; Zhang, Q.; Jiang, H.; Zhang, B.; Lin, Y.; Xuan, M.; Sun, L.; et al. N-glycosylation shields Phytophthora sojae apoplastic effector PsXEG1 from a specific host aspartic protease. Proc. Natl. Acad. Sci. USA 2020, 117, 27685–27693. [Google Scholar] [CrossRef]
- Anderson, R.G.; Deb, D.; Fedkenheuer, K.; McDowell, J.M. Recent progress in RXLR effector research. Mol. Plant Microbe Interact. 2015, 28, 1063–1072. [Google Scholar] [CrossRef]
- Amaro, T.M.; Thilliez, G.J.; Motion, G.B.; Huitema, E. A perspective on CRN proteins in the genomics age: Evolution, classification, delivery and function revisited. Front. Plant Sci. 2017, 8, 99. [Google Scholar] [CrossRef]
- Haas, B.J.; Kamoun, S.; Zody, M.C.; Jiang, R.H.; Handsaker, R.E.; Cano, L.M.; Grabherr, M.; Kodira, C.D.; Raffaele, S.; Torto-Alalibo, T.; et al. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 2009, 461, 393–398. [Google Scholar] [CrossRef]
- Boevink, P.C.; Birch, P.R.J.; Turnbull, D.; Whisson, S.C. Devastating intimacy: The cell biology of plant-Phytophthora interactions. New Phytol. 2020, 228, 445–458. [Google Scholar] [CrossRef]
- Bos, J.I.B.; Armstrong, M.; Whisson, S.C.; Torto, T.A.; Ochwo, M.; Birch, P.R.J.; Kamoun, S. Intraspecific comparative genomics to identify avirulence genes from Phytophthora. New Phytol. 2003, 159, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Murphy, F.; He, Q.; Armstrong, M.; Giuliani, L.M.; Boevink, P.C.; Zhang, W.; Tian, Z.; Birch, P.R.J.; Gilroy, E.M. The potato MAP3K StVIK is required for the Phytophthora infestans RXLR effector Pi17316 to promote disease. Plant Physiol. 2018, 177, 398–410. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; McLellan, H.; Fraiture, M.; Liu, X.; Boevink, P.C.; Gilroy, E.M.; Chen, Y.; Kandel, K.; Sessa, G.; Birch, P.R.; et al. Functionally redundant RXLR effectors from Phytophthora infestans act at different steps to suppress early flg22-triggered immunity. PLoS Pathog. 2014, 10, e1004057. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, T.O.; Schornack, S.; Win, J.; Shindo, T.; Ilyas, M.; Oliva, R.; Cano, L.M.; Jones, A.M.; Huitema, E.; van der Hoorn, R.A.; et al. Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 2011, 108, 20832–20837. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Li, Q.; Liu, T.; Liu, L.; Shen, D.; Zhu, Y.; Liu, P.; Zhou, J.M.; Dou, D. Two cytoplasmic effectors of Phytophthora sojae regulate plant cell death via interactions with plant catalases. Plant Physiol. 2015, 167, 164–175. [Google Scholar] [CrossRef]
- Li, Q.; Zhang, M.; Shen, D.; Liu, T.; Chen, Y.; Zhou, J.M.; Dou, D. A Phytophthora sojae effector PsCRN63 forms homo-/hetero-dimers to suppress plant immunity via an inverted association manner. Sci. Rep. 2016, 6, 26951. [Google Scholar] [CrossRef]
- van Damme, M.; Bozkurt, T.O.; Cakir, C.; Schornack, S.; Sklenar, J.; Jones, A.M.; Kamoun, S. The Irish potato famine pathogen Phytophthora infestans translocates the CRN8 kinase into host plant cells. PLoS Pathog. 2012, 8, e1002875. [Google Scholar] [CrossRef]
- Wu, Y.; Xie, L.; Jiang, Y.; Li, T. Prediction of effector proteins and their implications in pathogenicity of phytopathogenic filamentous fungi: A review. Int. J. Biol. Macromol. 2022, 206, 188–202. [Google Scholar] [CrossRef]
- Frias, M.; Gonzalez, C.; Brito, N. BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 2011, 192, 483–495. [Google Scholar] [CrossRef]
- Yang, G.; Tang, L.; Gong, Y.; Xie, J.; Fu, Y.; Jiang, D.; Li, G.; Collinge, D.B.; Chen, W.; Cheng, J. A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 2018, 217, 739–755. [Google Scholar] [CrossRef]
- Li, B.; Chen, Y.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular basis and regulation of pathogenicity and patulin biosynthesis in Penicillium expansum. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3416–3438. [Google Scholar] [CrossRef]
- Tian, H.; Fiorin, G.L.; Kombrink, A.; Mesters, J.R.; Thomma, B. Fungal dual-domain LysM effectors undergo chitin-induced intermolecular, and not intramolecular, dimerization. Plant Physiol. 2022, 190, 2033–2044. [Google Scholar] [CrossRef]
- Hu, S.P.; Li, J.J.; Dhar, N.; Li, J.P.; Chen, J.Y.; Jian, W.; Dai, X.F.; Yang, X.Y. Lysin motif (LysM) proteins: Interlinking manipulation of plant immunity and fungi. Int. J. Mol. Sci. 2021, 22, 3114. [Google Scholar] [CrossRef]
- Chen, D.; Li, G.; Liu, J.; Wisniewski, M.; Droby, S.; Levin, E.; Huang, S.; Liu, Y. Multiple transcriptomic analyses and characterization of pathogen-related core effectors and LysM family members reveal their differential roles in fungal growth and pathogenicity in Penicillium expansum. Mol. Genet. Genom. 2020, 295, 1415–1429. [Google Scholar] [CrossRef]
- Mart Nez-Cruz, J.S.; Romero, D.; Hierrezuelo, J.S.; Thon, M.; de Vicente, A.; Pérez-García, A. Effectors with chitinase activity (EWCAs), a family of conserved, secreted fungal chitinases that suppress chitin-triggered immunity. Plant Cell 2021, 33, 1319–1340. [Google Scholar] [CrossRef]
- Shabab, M.; Shindo, T.; Gu, C.; Kaschani, F.; Pansuriya, T.; Chintha, R.; Harzen, A.; Colby, T.; Kamoun, S.; van der Hoorn, R.A. Fungal effector protein AVR2 targets diversifying defense-related cys proteases of tomato. Plant Cell 2008, 20, 1169–1183. [Google Scholar] [CrossRef]
- Li, Z.; Yin, Z.; Fan, Y.; Xu, M.; Kang, Z.; Huang, L. Candidate effector proteins of the necrotrophic apple canker pathogen Valsa mali can suppress BAX-induced PCD. Front. Plant Sci. 2015, 6, 579. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Gardiner, D.M.; Kazan, K.; Manners, J.M. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant Microbe Interact. 2012, 25, 180–190. [Google Scholar] [CrossRef]
- Gawehns, F.; Ma, L.; Bruning, O.; Houterman, P.M.; Boeren, S.; Cornelissen, B.J.; Rep, M.; Takken, F.L. The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front. Plant Sci. 2015, 6, 967. [Google Scholar] [CrossRef]
- Huang, C.Y.; Wang, H.; Hu, P.; Hamby, R.; Jin, H. Small RNAs—Big players in plant-microbe interactions. Cell Host Microbe 2019, 26, 173–182. [Google Scholar] [CrossRef]
- Inderbitzin, P.; Asvarak, T.; Turgeon, B.G. Six new genes required for production of T-toxin, a polyketide determinant of high virulence of Cochliobolus heterostrophus to maize. Mol. Plant Microbe Interact. 2010, 23, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Tsuge, T.; Harimoto, Y.; Akimitsu, K.; Ohtani, K.; Kodama, M.; Akagi, Y.; Egusa, M.; Yamamoto, M.; Otani, H. Host-selective toxins produced by the plant pathogenic fungus Alternaria alternata. FEMS Microbiol. Rev. 2013, 37, 44–66. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Chen, Y.; Li, B.; Zhang, Z.; Qin, G.; Chen, T.; Tian, S. Molecular mechanisms underlying multi-level defense responses of horticultural crops to fungal pathogens. Hortic. Res. 2022, 9, uhac066. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Wan, W.L.; Frohlich, K.; Pruitt, R.N.; Nurnberger, T.; Zhang, L. Plant cell surface immune receptor complex signaling. Curr. Opin. Plant Biol. 2019, 50, 18–28. [Google Scholar] [CrossRef]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef]
- Rodriguez, P.A.; Rothballer, M.; Chowdhury, S.P.; Nussbaumer, T.; Gutjahr, C.; Falter-Braun, P. Systems biology of plant-microbiome interactions. Mol. Plant 2019, 12, 804–821. [Google Scholar] [CrossRef]
- Maruta, N.; Burdett, H.; Lim, B.Y.J.; Hu, X.; Desa, S.; Manik, M.K.; Kobe, B. Structural basis of NLR activation and innate immune signalling in plants. Immunogenetics 2022, 74, 5–26. [Google Scholar] [CrossRef]
- Harant, A.; Pai, H.; Sakai, T.; Kamoun, S.; Adachi, H. A vector system for fast-forward studies of the HOPZ-ACTIVATED RESISTANCE1 (ZAR1) resistosome in the model plant Nicotiana benthamiana. Plant Physiol. 2022, 188, 70–80. [Google Scholar] [CrossRef]
- Schenstnyi, K.; Strauss, A.; Dressel, A.; Morbitzer, R.; Wunderlich, M.; Andrade, A.G.; Phan, T.T.; Aguilera, P.L.A.; Brancato, C.; Berendzen, K.W.; et al. The tomato resistance gene Bs4 suppresses leaf watersoaking phenotypes induced by AvrHah1, a transcription activator-like effector from tomato-pathogenic xanthomonads. New Phytol. 2022, 236, 1856–1870. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, Y.; Zhang, X.; Chen, Z.; Xia, Y.; Wang, L.; Sun, Y.; Zhang, M.; Xiao, Y.; Han, Z.; et al. Plant receptor-like protein activation by a microbial glycoside hydrolase. Nature 2022, 610, 335–342. [Google Scholar] [CrossRef]
- Zhang, M.; Ahmed Rajput, N.; Shen, D.; Sun, P.; Zeng, W.; Liu, T.; Juma Mafurah, J.; Dou, D. A Phytophthora sojae cytoplasmic effector mediates disease resistance and abiotic stress tolerance in Nicotiana benthamiana. Sci. Rep. 2015, 5, 10837. [Google Scholar] [CrossRef]
- McLellan, H.; Harvey, S.E.; Steinbrenner, J.; Armstrong, M.R.; He, Q.; Clewes, R.; Pritchard, L.; Wang, W.; Wang, S.; Nussbaumer, T.; et al. Exploiting breakdown in nonhost effector-target interactions to boost host disease resistance. Proc. Natl. Acad. Sci. USA 2022, 119, e2114064119. [Google Scholar] [CrossRef]
- Tabima, J.F.; Grunwald, N.J. effectR: An expandable r package to predict candidate RxLR and CRN effectors in oomycetes using motif searches. Mol. Plant Microbe Interact. 2019, 32, 1067–1076. [Google Scholar] [CrossRef]
- Sperschneider, J.; Dodds, P.N. EffectorP 3.0: Prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant Microbe Interact. 2022, 35, 146–156. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, X.; Cao, X.; Huang, C.; Liu, E.; Qian, S.; Liu, X.; Wu, Y.; Dong, F.; Qiu, C.W.; et al. Artificial intelligence: A powerful paradigm for scientific research. Innovation 2021, 2, 100179. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).