The Role of the γ-Aminobutyric Acid (GABA) in Plant Salt Stress Tolerance
Abstract
:1. Introduction
1.1. Physiological Effects of Salt Stress on Plants
1.2. The Role of GABA in Stress Responses
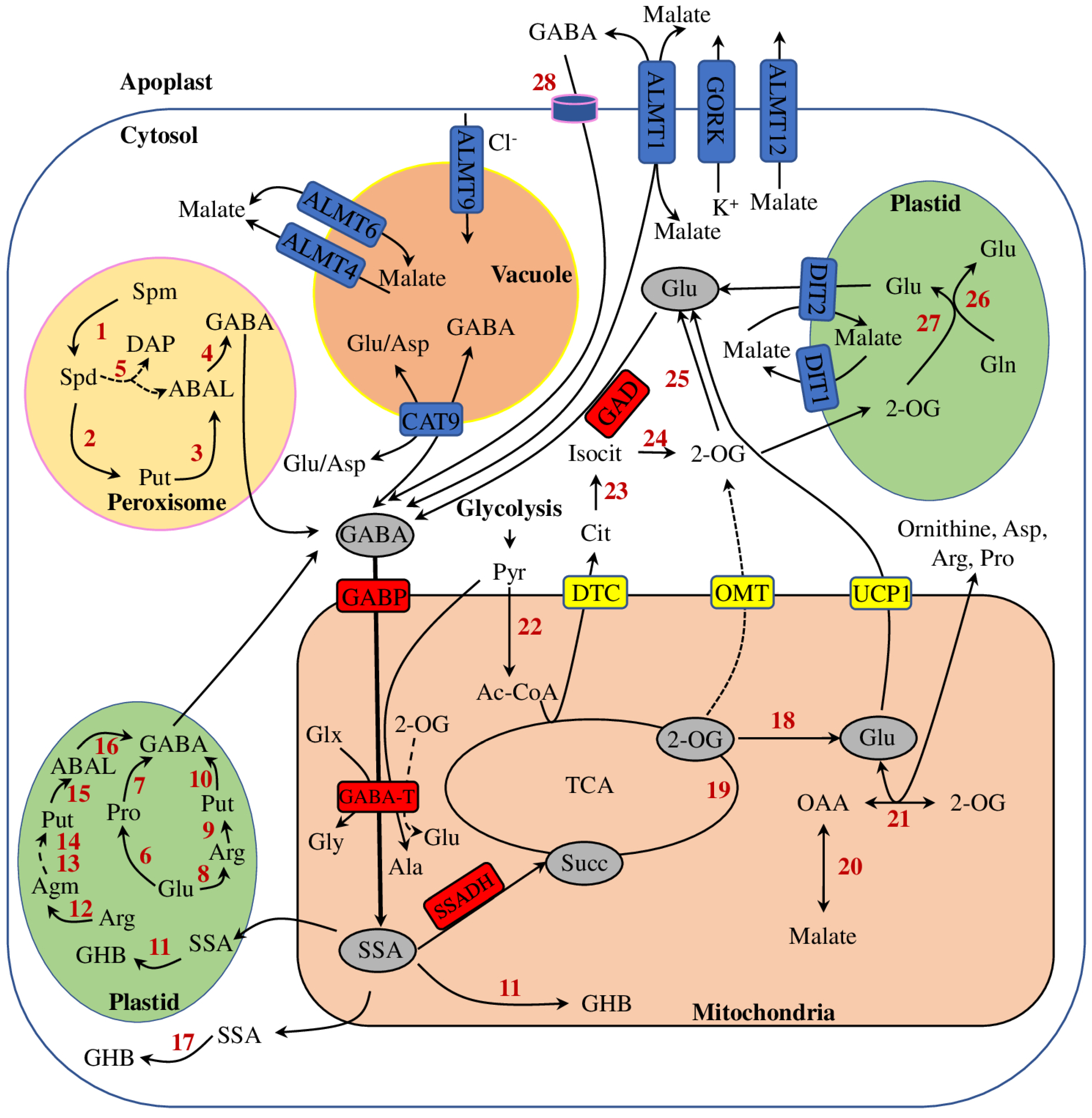
2. GABA Homeostasis in Plants
3. Physiological, Biochemical, and Molecular Aspects of GABA Metabolism in Plant Adaptation to Salt Stress
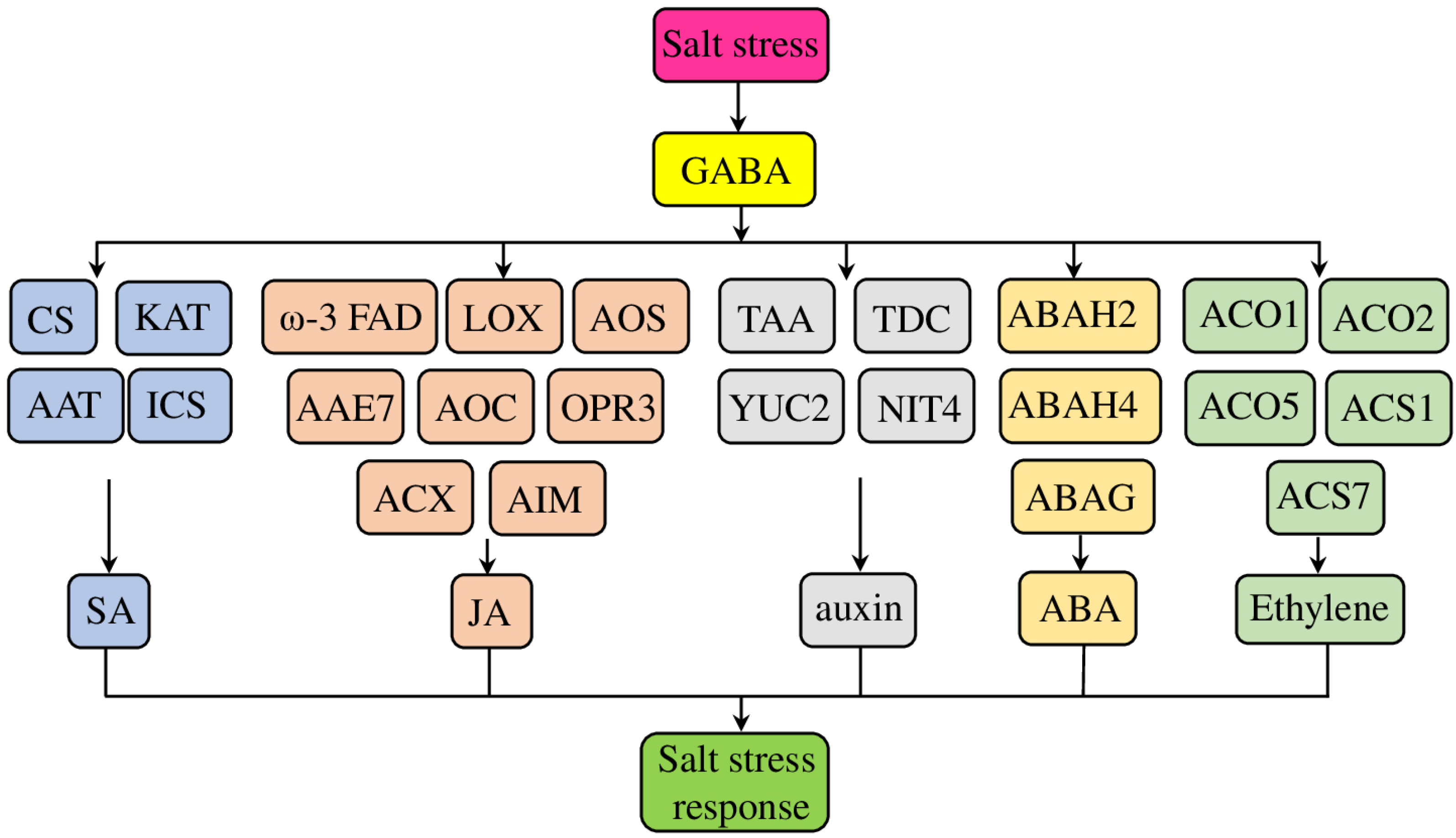
3.1. The Involvement of GABA in the Different Stress Signaling Pathways
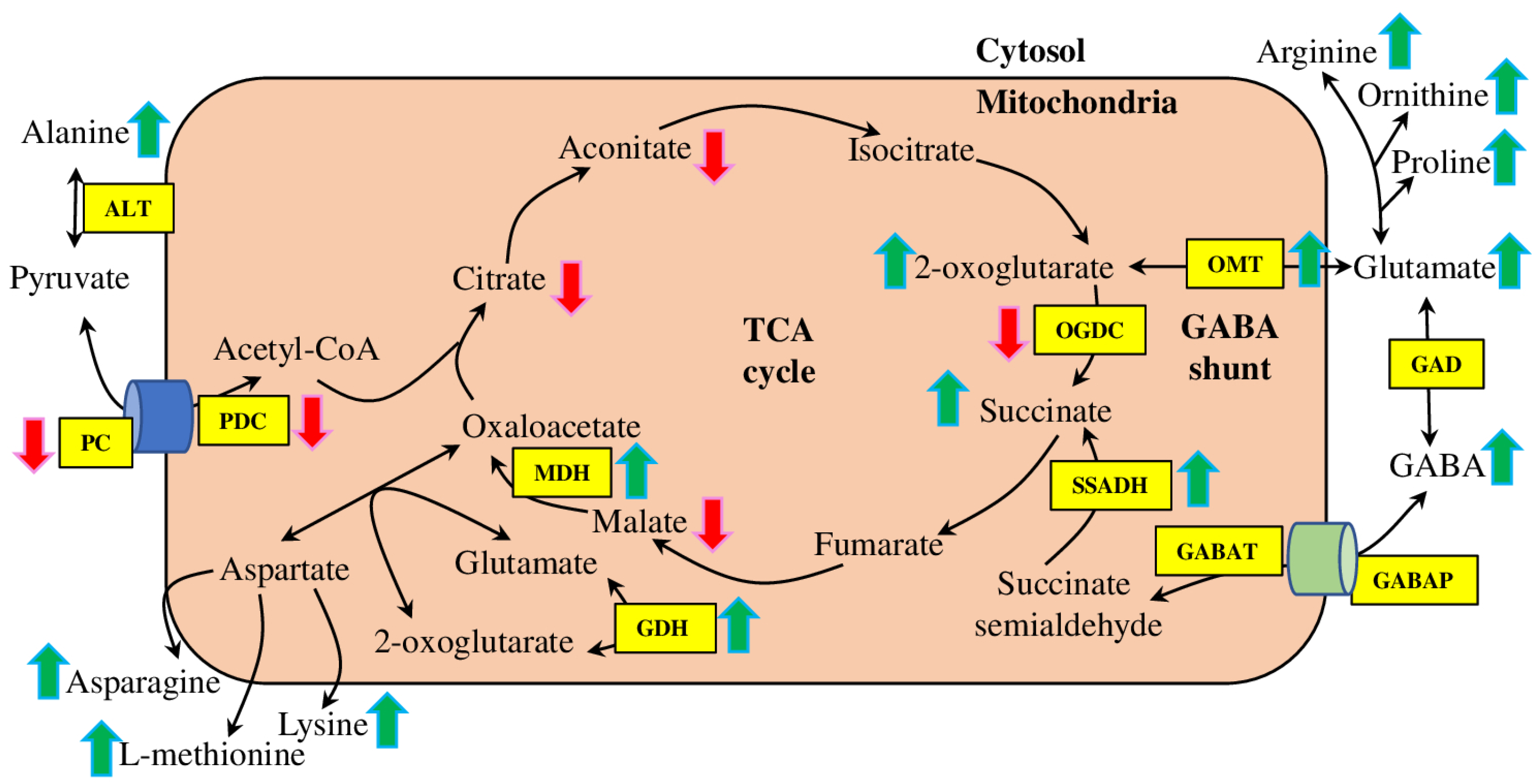
3.2. GABA Shunt and TCA cycle under Salt Stress
4. Manipulation with Genes Involved in GABA Metabolism
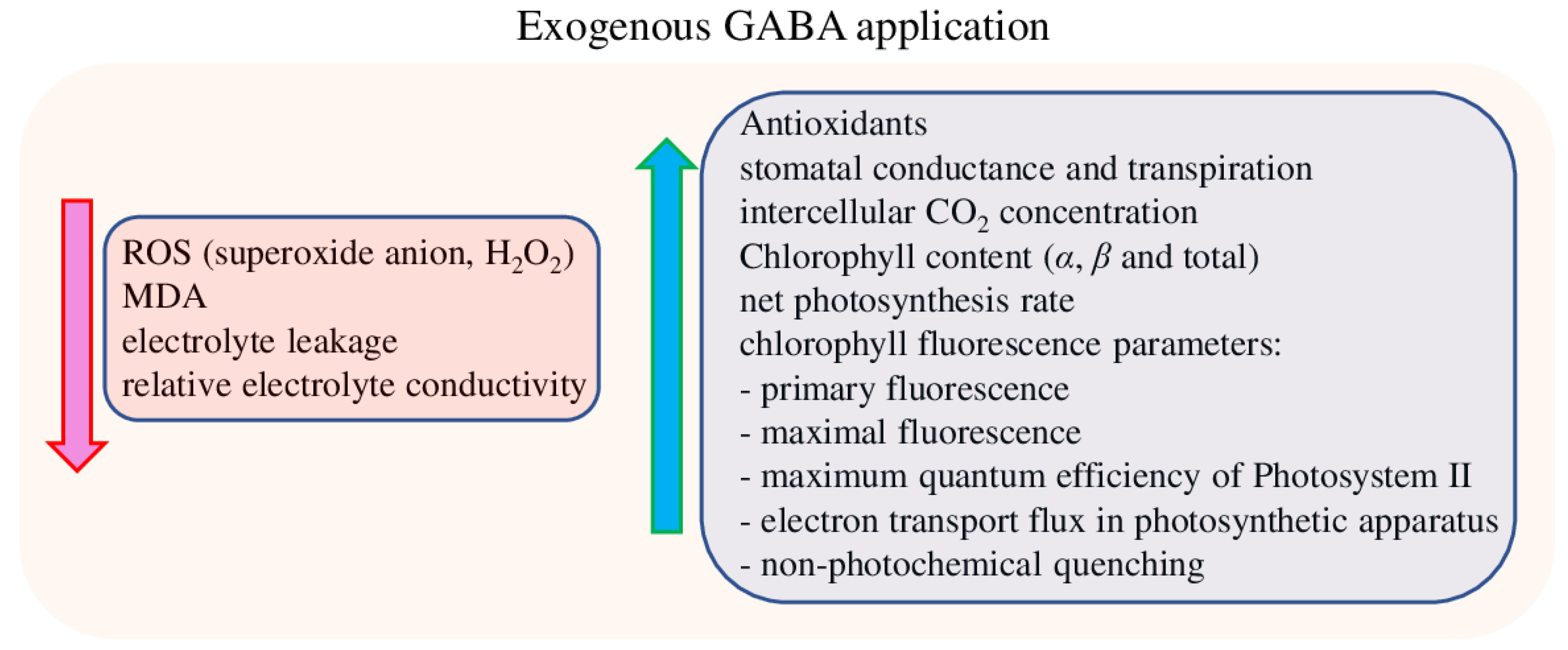
5. Manipulation of Plant GABA Levels and Salt Stress Improvement by Exogenous Application of GABA or Its Precursors
5.1. Phenolics Biosynthesis and Ca2+ Signaling
5.2. Photosynthesis and Antioxidant Defense
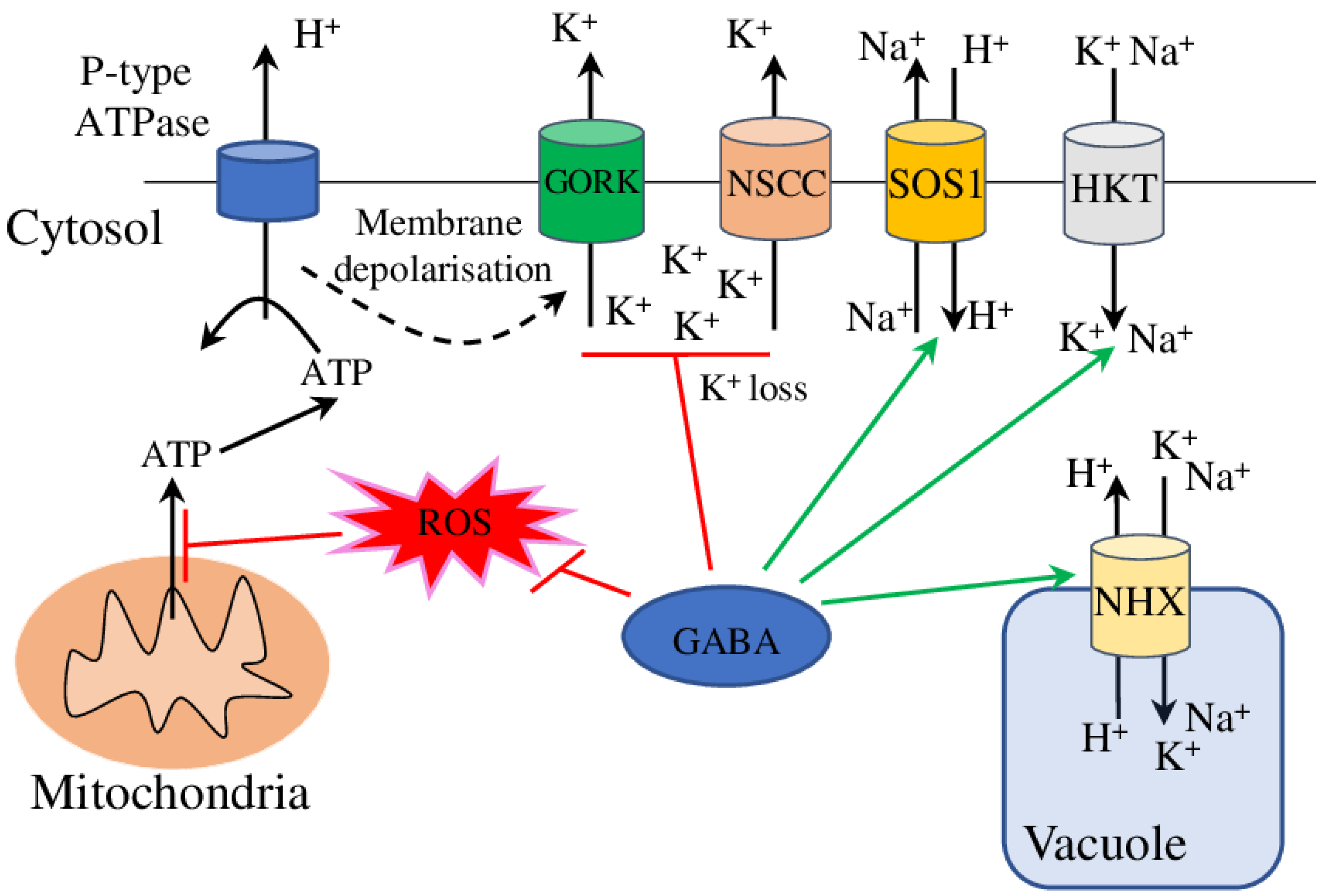
5.3. Na+/K+ Ions Homeostasis
5.4. Phytohormones
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Machado, R.; Serralheiro, R. Soil Salinity: Effect on Vegetable Crop Growth. Management Practices to Prevent and Mitigate Soil Salinization. Horticulturae 2017, 3, 30. [Google Scholar] [CrossRef]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic Strategies for Improving Crop Yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Genc, Y.; Taylor, J.; Lyons, G.; Li, Y.; Cheong, J.; Appelbee, M.; Oldach, K.; Sutton, T. Bread Wheat with High Salinity and Sodicity Tolerance. Front. Plant Sci. 2019, 10, 1280. [Google Scholar] [CrossRef] [PubMed]
- Munns, R.; Day, D.A.; Fricke, W.; Watt, M.; Arsova, B.; Barkla, B.J.; Bose, J.; Byrt, C.S.; Chen, Z.; Foster, K.J.; et al. Energy Costs of Salt Tolerance in Crop Plants. New Phytol. 2020, 225, 1072–1090. [Google Scholar] [CrossRef]
- Arzani, A.; Ashraf, M. Smart Engineering of Genetic Resources for Enhanced Salinity Tolerance in Crop Plants. Crit. Rev. Plant Sci. 2016, 35, 146–189. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. IJMS 2021, 22, 4609. [Google Scholar] [CrossRef]
- Hameed, A.; Ahmed, M.Z.; Hussain, T.; Aziz, I.; Ahmad, N.; Gul, B.; Nielsen, B.L. Effects of Salinity Stress on Chloroplast Structure and Function. Cells 2021, 10, 2023. [Google Scholar] [CrossRef]
- Chen, Y.; Li, Y.; Sun, P.; Chen, G.; Xin, J. Interactive Effects of Salt and Alkali Stresses on Growth, Physiological Responses and Nutrient (N, P) Removal Performance of Ruppia Maritima. Ecol. Eng. 2017, 104, 177–183. [Google Scholar] [CrossRef]
- Bandehagh, A.; Taylor, N.L. Can Alternative Metabolic Pathways and Shunts Overcome Salinity Induced Inhibition of Central Carbon Metabolism in Crops? Front. Plant Sci. 2020, 11, 1072. [Google Scholar] [CrossRef]
- Ramesh, S.A.; Tyerman, S.D.; Gilliham, M.; Xu, B. γ-Aminobutyric Acid (GABA) Signalling in Plants. Cell. Mol. Life Sci. 2017, 74, 1577–1603. [Google Scholar] [CrossRef]
- Sears, S.M.; Hewett, S.J. Influence of Glutamate and GABA Transport on Brain Excitatory/Inhibitory Balance. Exp. Biol. Med. 2021, 246, 1069–1083. [Google Scholar] [CrossRef]
- Ghit, A.; Assal, D.; Al-Shami, A.S.; Hussein, D.E.E. GABAA Receptors: Structure, Function, Pharmacology, and Related Disorders. J. Genet. Eng. Biotechnol. 2021, 19, 123. [Google Scholar] [CrossRef]
- Prévot, T.; Sibille, E. Altered GABA-Mediated Information Processing and Cognitive Dysfunctions in Depression and Other Brain Disorders. Mol. Psychiatry 2021, 26, 151–167. [Google Scholar] [CrossRef]
- Shelp, B.J.; Bown, A.W.; Zarei, A. 4-Aminobutyrate (GABA): A Metabolite and Signal with Practical Significance. Botany 2017, 95, 1015–1032. [Google Scholar] [CrossRef]
- Li, L.; Dou, N.; Zhang, H.; Wu, C. The Versatile GABA in Plants. Plant Signal. Behav. 2021, 16, 1862565. [Google Scholar] [CrossRef]
- Xu, B.; Sai, N.; Gilliham, M. The Emerging Role of GABA as a Transport Regulator and Physiological Signal. Plant Physiol. 2021, 187, 2005–2016. [Google Scholar] [CrossRef]
- Kaspal, M.; Kanapaddalagamage, M.H.; Ramesh, S.A. Emerging Roles of γ Aminobutyric Acid (GABA) Gated Channels in Plant Stress Tolerance. Plants 2021, 10, 2178. [Google Scholar] [CrossRef]
- Wu, Q.; Su, N.; Huang, X.; Cui, J.; Shabala, L.; Zhou, M.; Yu, M.; Shabala, S. Hypoxia-Induced Increase in GABA Content Is Essential for Restoration of Membrane Potential and Preventing ROS-Induced Disturbance to Ion Homeostasis. Plant Commun. 2021, 2, 100188. [Google Scholar] [CrossRef]
- Steward, F.C. γ-Aminobutyric Acid: A Constituent of the Potato Tuber? Science 1949, 110, 439–440. [Google Scholar]
- Roberts, E.; Frankel, S. Gamma-Aminobutyric Acid in Brain: Its Formation from Glutamic Acid. J. Biol. Chem. 1950, 187, 55–63. [Google Scholar] [CrossRef]
- Seifikalhor, M.; Aliniaeifard, S.; Hassani, B.; Niknam, V.; Lastochkina, O. Diverse Role of γ-Aminobutyric Acid in Dynamic Plant Cell Responses. Plant Cell Rep. 2019, 38, 847–867. [Google Scholar] [CrossRef] [PubMed]
- Shelp, B.J.; Aghdam, M.S.; Flaherty, E.J. γ-Aminobutyrate (GABA) Regulated Plant Defense: Mechanisms and Opportunities. Plants 2021, 10, 1939. [Google Scholar] [CrossRef] [PubMed]
- Paschalidis, K.; Tsaniklidis, G.; Wang, B.-Q.; Delis, C.; Trantas, E.; Loulakakis, K.; Makky, M.; Sarris, P.F.; Ververidis, F.; Liu, J.-H. The Interplay among Polyamines and Nitrogen in Plant Stress Responses. Plants 2019, 8, 315. [Google Scholar] [CrossRef] [PubMed]
- Jacques, F.; Zhao, Y.; Kopečná, M.; Končitíková, R.; Kopečný, D.; Rippa, S.; Perrin, Y. Roles for ALDH10 Enzymes in γ-Butyrobetaine Synthesis, Seed Development, Germination, and Salt Tolerance in Arabidopsis. J. Exp. Bot. 2020, 71, 7088–7102. [Google Scholar] [CrossRef]
- Signorelli, S.; Dans, P.D.; Coitiño, E.L.; Borsani, O.; Monza, J. Connecting Proline and γ-Aminobutyric Acid in Stressed Plants through Non-Enzymatic Reactions. PLoS ONE 2015, 10, e0115349. [Google Scholar] [CrossRef]
- Dai, L.; Li, P.; Li, Q.; Leng, Y.; Zeng, D.; Qian, Q. Integrated Multi-Omics Perspective to Strengthen the Understanding of Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 5236. [Google Scholar] [CrossRef]
- Kumar, P.; Choudhary, M.; Halder, T.; Prakash, N.R.; Singh, V.; Vineeth, V.T.; Sheoran, S.; Ravikiran, T.K.; Longmei, N.; Rakshit, S.; et al. Salinity Stress Tolerance and Omics Approaches: Revisiting the Progress and Achievements in Major Cereal Crops. Heredity 2022, 128, 497–518. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Jung, H.S.; Jeong, M.S.; Jung, W.-H.; Choi, C.W.; Lee, S.H.; Jin, B.J.; Park, M.S.; et al. Metabolic Adjustment of Arabidopsis Root Suspension Cells During Adaptation to Salt Stress and Mitotic Stress Memory. Plant Cell Physiol. 2019, 60, 612–625. [Google Scholar] [CrossRef]
- González-Orenga, S.; Ferrer-Gallego, P.P.; Laguna, E.; López-Gresa, M.P.; Donat-Torres, M.P.; Verdeguer, M.; Vicente, O.; Boscaiu, M. Insights on Salt Tolerance of Two Endemic Limonium Species from Spain. Metabolites 2019, 9, 294. [Google Scholar] [CrossRef]
- Dell’Aversana, E.; Hessini, K.; Ferchichi, S.; Fusco, G.M.; Woodrow, P.; Ciarmiello, L.F.; Abdelly, C.; Carillo, P. Salinity Duration Differently Modulates Physiological Parameters and Metabolites Profile in Roots of Two Contrasting Barley Genotypes. Plants 2021, 10, 307. [Google Scholar] [CrossRef]
- Thouvenot, L.; Deleu, C.; Berardocco, S.; Haury, J.; Thiébaut, G. Characterization of the Salt Stress Vulnerability of Three Invasive Freshwater Plant Species Using a Metabolic Profiling Approach. J. Plant Physiol. 2015, 175, 113–121. [Google Scholar] [CrossRef]
- de Oliveira, D.F.; de Sousa Lopes, L.; Gomes-Filho, E. Metabolic Changes Associated with Differential Salt Tolerance in Sorghum Genotypes. Planta 2020, 252, 34. [Google Scholar] [CrossRef]
- Ma, N.L.; Che Lah, W.A.; Kadir, N.A.; Mustaqim, M.; Rahmat, Z.; Ahmad, A.; Lam, S.D.; Ismail, M.R. Susceptibility and Tolerance of Rice Crop to Salt Threat: Physiological and Metabolic Inspections. PLoS ONE 2018, 13, e0192732. [Google Scholar] [CrossRef]
- García-Caparrós, P.; Vogelsang, L.; Persicke, M.; Wirtz, M.; Kumar, V.; Dietz, K. Differential Sensitivity of Metabolic Pathways in Sugar Beet Roots to Combined Salt, Heat, and Light Stress. Physiol. Plant. 2022, 174, e13786. [Google Scholar] [CrossRef]
- Ji, J.; Shi, Z.; Xie, T.; Zhang, X.; Chen, W.; Du, C.; Sun, J.; Yue, J.; Zhao, X.; Jiang, Z.; et al. Responses of GABA Shunt Coupled with Carbon and Nitrogen Metabolism in Poplar under NaCl and CdCl2 Stresses. Ecotoxicol. Environ. Saf. 2020, 193, 110322. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, B.; Liu, H.; Han, H.; Zhuang, H.; Wang, J.; Yang, T.; Wang, H.; Qin, Y. Comparative Proteomic Analysis for Revealing the Advantage Mechanisms of Salt-Tolerant Tomato (Solanum lycoperscium). PeerJ 2022, 10, e12955. [Google Scholar] [CrossRef]
- Yang, R.; Guo, Y.; Wang, S.; Gu, Z. Ca2+ and Aminoguanidine on γ-Aminobutyric Acid Accumulation in Germinating Soybean under Hypoxia–NaCl Stress. J. Food Drug Anal. 2015, 23, 287–293. [Google Scholar] [CrossRef]
- Yin, Y.; Cheng, C.; Fang, W. Effects of the Inhibitor of Glutamate Decarboxylase on the Development and GABA Accumulation in Germinating Fava Beans under Hypoxia-NaCl Stress. RSC Adv. 2018, 8, 20456–20461. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Zhao, Y.; Yu, X. Cross-Talk between Gama-Aminobutyric Acid and Calcium Ion Regulates Lipid Biosynthesis in Monoraphidium Sp. QLY-1 in Response to Combined Treatment of Fulvic Acid and Salinity Stress. Bioresour. Technol. 2020, 315, 123833. [Google Scholar] [CrossRef]
- Felle, H.H.; Zimmermann, M.R. Systemic Signalling in Barley through Action Potentials. Planta 2007, 226, 203. [Google Scholar] [CrossRef]
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate Triggers Long-Distance, Calcium-Based Plant Defense Signaling. Science 2018, 361, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Gao, Q.; Lhamo, D.; Zhang, H.; Luan, S. Two Glutamate- and PH-Regulated Ca 2+ Channels Are Required for Systemic Wound Signaling in Arabidopsis. Sci. Signal. 2020, 13, eaba1453. [Google Scholar] [CrossRef]
- Xu, B.; Long, Y.; Feng, X.; Zhu, X.; Sai, N.; Chirkova, L.; Betts, A.; Herrmann, J.; Edwards, E.J.; Okamoto, M.; et al. GABA Signalling Modulates Stomatal Opening to Enhance Plant Water Use Efficiency and Drought Resilience. Nat. Commun. 2021, 12, 1952. [Google Scholar] [CrossRef] [PubMed]
- Adem, G.D.; Chen, G.; Shabala, L.; Chen, Z.-H.; Shabala, S. GORK Channel: A Master Switch of Plant Metabolism? Trends Plant Sci. 2020, 25, 434–445. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Zhang, X.; Giraldo, J.P.; Shabala, S. It Is Not All about Sodium: Revealing Tissue Specificity and Signalling Roles of Potassium in Plant Responses to Salt Stress. Plant Soil 2018, 431, 1–17. [Google Scholar] [CrossRef]
- Isayenkov, S.V.; Maathuis, F.J.M. Plant Salinity Stress: Many Unanswered Questions Remain. Front. Plant Sci. 2019, 10, 80. [Google Scholar] [CrossRef]
- Demidchik, V.; Straltsova, D.; Medvedev, S.S.; Pozhvanov, G.A.; Sokolik, A.; Yurin, V. Stress-Induced Electrolyte Leakage: The Role of K+-Permeable Channels and Involvement in Programmed Cell Death and Metabolic Adjustment. J. Exp. Bot. 2014, 65, 1259–1270. [Google Scholar] [CrossRef]
- Che-Othman, M.H.; Jacoby, R.P.; Millar, A.H.; Taylor, N.L. Wheat Mitochondrial Respiration Shifts from the Tricarboxylic Acid Cycle to the GABA Shunt under Salt Stress. New Phytol. 2020, 225, 1166–1180. [Google Scholar] [CrossRef]
- Woodrow, P.; Ciarmiello, L.F.; Annunziata, M.G.; Pacifico, S.; Iannuzzi, F.; Mirto, A.; D’Amelia, L.; Dell’Aversana, E.; Piccolella, S.; Fuggi, A.; et al. Durum Wheat Seedling Responses to Simultaneous High Light and Salinity Involve a Fine Reconfiguration of Amino Acids and Carbohydrate Metabolism. Physiol. Plant. 2017, 159, 290–312. [Google Scholar] [CrossRef]
- Bao, H.; Chen, X.; Lv, S.; Jiang, P.; Feng, J.; Fan, P.; Nie, L.; Li, Y. Virus-Induced Gene Silencing Reveals Control of Reactive Oxygen Species Accumulation and Salt Tolerance in Tomato by γ -Aminobutyric Acid Metabolic Pathway: Effects of GABA Shunt on Tomato Salt Tolerance. Plant Cell Environ. 2015, 38, 600–613. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Z.; Fan, Y.; Liu, C.; Wang, H.; Li, Y.; Xin, Y.; Gai, Y.; Ji, X. Characterization of GABA-Transaminase Gene from Mulberry (Morus Multicaulis) and Its Role in Salt Stress Tolerance. Genes 2022, 13, 501. [Google Scholar] [CrossRef]
- Su, N.; Wu, Q.; Chen, J.; Shabala, L.; Mithöfer, A.; Wang, H.; Qu, M.; Yu, M.; Cui, J.; Shabala, S. GABA Operates Upstream of H+-ATPase and Improves Salinity Tolerance in Arabidopsis by Enabling Cytosolic K+ Retention and Na+ Exclusion. J. Exp. Bot. 2019, 70, 6349–6361. [Google Scholar] [CrossRef]
- Zarei, A.; Trobacher, C.P.; Shelp, B.J. Arabidopsis Aldehyde Dehydrogenase 10 Family Members Confer Salt Tolerance through Putrescine-Derived 4-Aminobutyrate (GABA) Production. Sci. Rep. 2016, 6, 35115. [Google Scholar] [CrossRef]
- Bai, X.; Xu, J.; Shao, X.; Luo, W.; Niu, Z.; Gao, C.; Wan, D. A Novel Gene Coding γ-Aminobutyric Acid Transporter May Improve the Tolerance of Populus Euphratica to Adverse Environments. Front. Plant Sci. 2019, 10, 1083. [Google Scholar] [CrossRef]
- da Silva Rodrigues-Corrêa, K.C.; Fett-Neto, A.G. Abiotic Stresses and Non-Protein Amino Acids in Plants. Crit. Rev. Plant Sci. 2019, 38, 411–430. [Google Scholar] [CrossRef]
- Li, Z.; Cheng, B.; Zeng, W.; Zhang, X.; Peng, Y. Proteomic and Metabolomic Profilings Reveal Crucial Functions of γ-Aminobutyric Acid in Regulating Ionic, Water, and Metabolic Homeostasis in Creeping Bentgrass under Salt Stress. J. Proteome Res. 2020, 19, 769–780. [Google Scholar] [CrossRef]
- Hijaz, F.; Killiny, N. Exogenous GABA Is Quickly Metabolized to Succinic Acid and Fed into the Plant TCA Cycle. Plant Signal. Behav. 2019, 14, e1573096. [Google Scholar] [CrossRef]
- Hijaz, F.; Killiny, N. The Use of Deuterium-Labeled Gamma-Aminobutyric (D6-GABA) to Study Uptake, Translocation, and Metabolism of Exogenous GABA in Plants. Plant Methods 2020, 16, 24. [Google Scholar] [CrossRef]
- Ma, Y.; Wang, P.; Chen, Z.; Gu, Z.; Yang, R. GABA Enhances Physio-Biochemical Metabolism and Antioxidant Capacity of Germinated Hulless Barley under NaCl Stress. J. Plant Physiol. 2018, 231, 192–201. [Google Scholar] [CrossRef]
- Wang, M.; Ding, Y.; Wang, Q.; Wang, P.; Han, Y.; Gu, Z.; Yang, R. NaCl Treatment on Physio-Biochemical Metabolism and Phenolics Accumulation in Barley Seedlings. Food Chem. 2020, 331, 127282. [Google Scholar] [CrossRef]
- Wang, M.; Zhu, Y.; Wang, P.; Gu, Z.; Yang, R. Effect of γ-Aminobutyric Acid on Phenolics Metabolism in Barley Seedlings under Low NaCl Treatment. Antioxidants 2021, 10, 1421. [Google Scholar] [CrossRef] [PubMed]
- Sheteiwy, M.S.; Shao, H.; Qi, W.; Hamoud, Y.A.; Shaghaleh, H.; Khan, N.U.; Yang, R.; Tang, B. GABA-Alleviated Oxidative Injury Induced by Salinity, Osmotic Stress and Their Combination by Regulating Cellular and Molecular Signals in Rice. Int. J. Mol. Sci. 2019, 20, 5709. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cheng, B.; Peng, Y.; Zhang, Y. Adaptability to Abiotic Stress Regulated by γ-Aminobutyric Acid in Relation to Alterations of Endogenous Polyamines and Organic Metabolites in Creeping Bentgrass. Plant Physiol. Biochem. 2020, 157, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Li, M.F.; Guo, S.J.; Yang, X.H.; Meng, Q.W.; Wei, X.J. Exogenous Gamma-Aminobutyric Acid Increases Salt Tolerance of Wheat by Improving Photosynthesis and Enhancing Activities of Antioxidant Enzymes. Biol. Plant. 2016, 60, 123–131. [Google Scholar] [CrossRef]
- Wang, Y.; Gu, W.; Meng, Y.; Xie, T.; Li, L.; Li, J.; Wei, S. γ-Aminobutyric Acid Imparts Partial Protection from Salt Stress Injury to Maize Seedlings by Improving Photosynthesis and Upregulating Osmoprotectants and Antioxidants. Sci. Rep. 2017, 7, 43609. [Google Scholar] [CrossRef]
- Kalhor, M.S.; Aliniaeifard, S.; Seif, M.; Asayesh, E.J.; Bernard, F.; Hassani, B.; Li, T. Title: Enhanced Salt Tolerance and Photosynthetic Performance: Implication of ɤ-Amino Butyric Acid Application in Salt-Exposed Lettuce (Lactuca sativa L.) Plants. Plant Physiol. Biochem. 2018, 130, 157–172. [Google Scholar] [CrossRef]
- Shomali, A.; Aliniaeifard, S.; Didaran, F.; Lotfi, M.; Mohammadian, M.; Seif, M.; Strobel, W.R.; Sierka, E.; Kalaji, H.M. Synergistic Effects of Melatonin and Gamma-Aminobutyric Acid on Protection of Photosynthesis System in Response to Multiple Abiotic Stressors. Cells 2021, 10, 1631. [Google Scholar] [CrossRef]
- Jin, X.; Liu, T.; Xu, J.; Gao, Z.; Hu, X. Exogenous GABA Enhances Muskmelon Tolerance to Salinity-Alkalinity Stress by Regulating Redox Balance and Chlorophyll Biosynthesis. BMC Plant Biol 2019, 19, 48. [Google Scholar] [CrossRef]
- Khanna, R.R.; Jahan, B.; Iqbal, N.; Khan, N.A.; AlAjmi, M.F.; Tabish Rehman, M.; Khan, M.I.R. GABA Reverses Salt-Inhibited Photosynthetic and Growth Responses through Its Influence on NO-Mediated Nitrogen-Sulfur Assimilation and Antioxidant System in Wheat. J. Biotechnol. 2021, 325, 73–82. [Google Scholar] [CrossRef]
- Shang, J.-X.; Li, X.; Li, C.; Zhao, L. The Role of Nitric Oxide in Plant Responses to Salt Stress. Int. J. Mol. Sci. 2022, 23, 6167. [Google Scholar] [CrossRef]
- Aljuaid, B.S.; Ashour, H. Exogenous γ-Aminobutyric Acid (GABA) Application Mitigates Salinity Stress in Maize Plants. Life 2022, 12, 1860. [Google Scholar] [CrossRef]
- Wu, X.; Jia, Q.; Ji, S.; Gong, B.; Li, J.; Lü, G.; Gao, H. Gamma-Aminobutyric Acid (GABA) Alleviates Salt Damage in Tomato by Modulating Na+ Uptake, the GAD Gene, Amino Acid Synthesis and Reactive Oxygen Species Metabolism. BMC Plant Biol 2020, 20, 465. [Google Scholar] [CrossRef]
- Cheng, B.; Li, Z.; Liang, L.; Cao, Y.; Zeng, W.; Zhang, X.; Ma, X.; Huang, L.; Nie, G.; Liu, W.; et al. The γ-Aminobutyric Acid (GABA) Alleviates Salt Stress Damage during Seeds Germination of White Clover Associated with Na+/K+ Transportation, Dehydrins Accumulation, and Stress-Related Genes Expression in White Clover. Int. J. Mol. Sci. 2018, 19, 2520. [Google Scholar] [CrossRef]
- Cheng, B.; Hassan, M.J.; Feng, G.; Zhao, J.; Liu, W.; Peng, Y.; Li, Z. Metabolites Reprogramming and Na+/K+ Transportation Associated With Putrescine-Regulated White Clover Seed Germination and Seedling Tolerance to Salt Toxicity. Front. Plant Sci. 2022, 13, 856007. [Google Scholar] [CrossRef]
- Li, Z.; Cheng, B.; Liu, W.; Feng, G.; Zhao, J.; Zhang, L.; Peng, Y. Global Metabolites Reprogramming Induced by Spermine Contributing to Salt Tolerance in Creeping Bentgrass. Int. J. Mol. Sci. 2022, 23, 4472. [Google Scholar] [CrossRef]
- Hijaz, F.; Nehela, Y.; Killiny, N. Application of Gamma-Aminobutyric Acid Increased the Level of Phytohormones in Citrus Sinensis. Planta 2018, 248, 909–918. [Google Scholar] [CrossRef]
- Ji, J.; Yue, J.; Xie, T.; Chen, W.; Du, C.; Chang, E.; Chen, L.; Jiang, Z.; Shi, S. Roles of γ-Aminobutyric Acid on Salinity-Responsive Genes at Transcriptomic Level in Poplar: Involving in Abscisic Acid and Ethylene-Signalling Pathways. Planta 2018, 248, 675–690. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dabravolski, S.A.; Isayenkov, S.V. The Role of the γ-Aminobutyric Acid (GABA) in Plant Salt Stress Tolerance. Horticulturae 2023, 9, 230. https://doi.org/10.3390/horticulturae9020230
Dabravolski SA, Isayenkov SV. The Role of the γ-Aminobutyric Acid (GABA) in Plant Salt Stress Tolerance. Horticulturae. 2023; 9(2):230. https://doi.org/10.3390/horticulturae9020230
Chicago/Turabian StyleDabravolski, Siarhei A., and Stanislav V. Isayenkov. 2023. "The Role of the γ-Aminobutyric Acid (GABA) in Plant Salt Stress Tolerance" Horticulturae 9, no. 2: 230. https://doi.org/10.3390/horticulturae9020230
APA StyleDabravolski, S. A., & Isayenkov, S. V. (2023). The Role of the γ-Aminobutyric Acid (GABA) in Plant Salt Stress Tolerance. Horticulturae, 9(2), 230. https://doi.org/10.3390/horticulturae9020230







