Determination of Selenium Speciation in High Se-Enriched Edible Fungus Ganoderma lucidum Via Sequential Extraction
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of High Se-Enriched G. lucidum
2.2. Experimental Procedures
2.3. Determination of Total Se and Se Speciation
2.4. Statistical Analysis
3. Results and Discussion
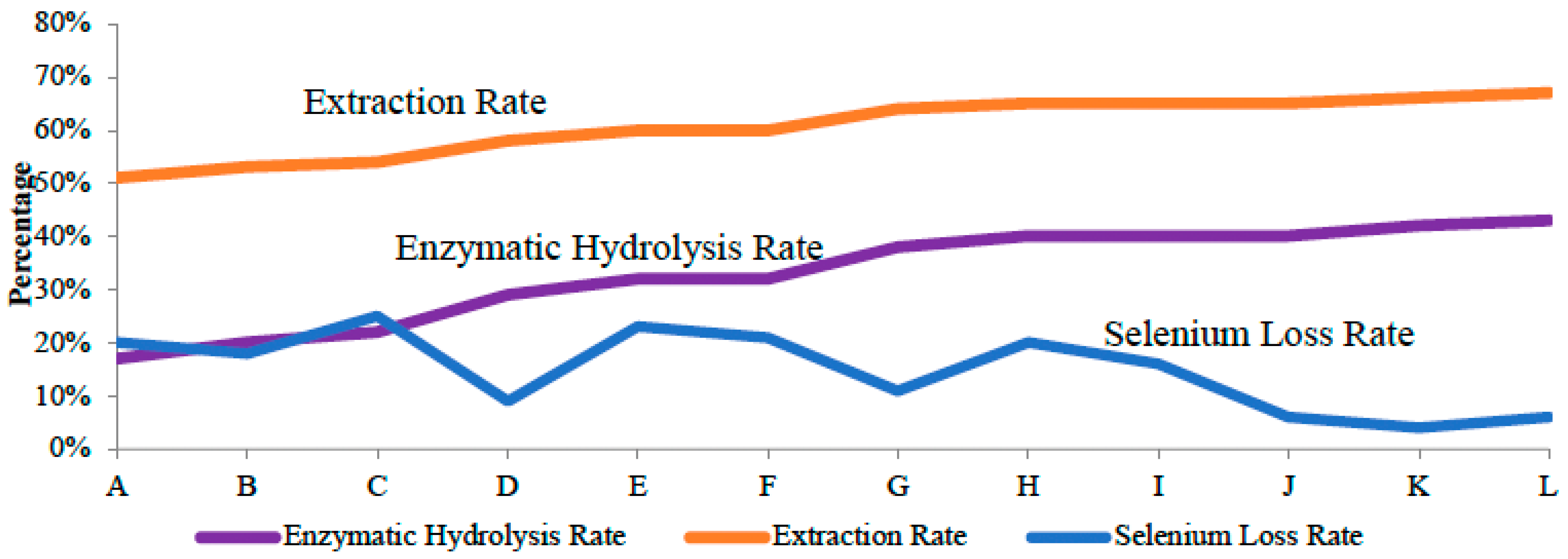
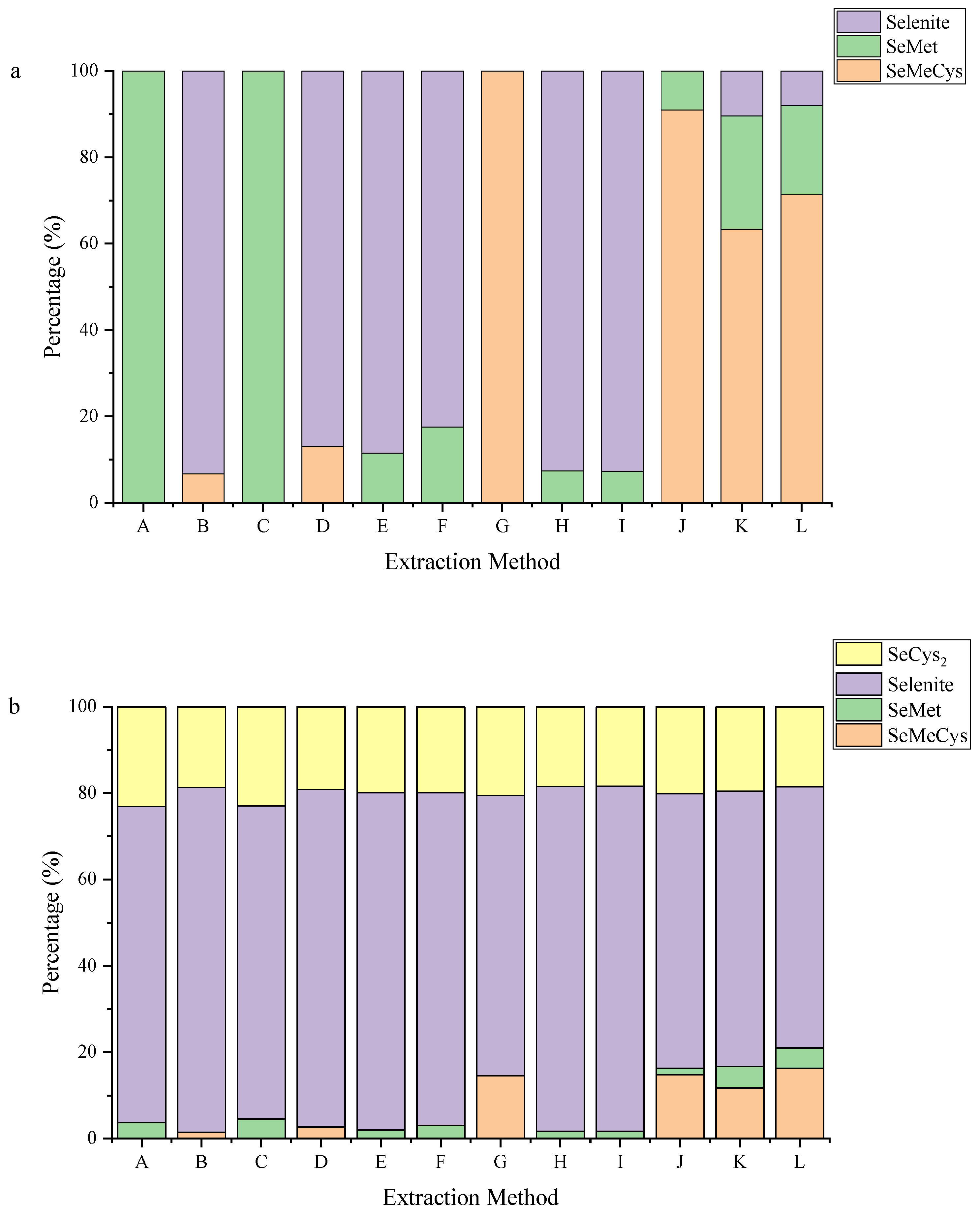
3.1. One-Step Extraction without Enzyme Has Low Extraction Rate
3.2. Sequential Extraction with Enzymes Increased Se Extraction Efficiency
3.3. Co-Application of Pepsin and Trypsin to Better Extract Se-Containing Amnio Acids
3.4. Se Species in Microorganisms
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wachtel-Galor, S.; Yuen, J.; Buswell, J.A.; Benzie, I.F. Ganoderma lucidum (Lingzhi or Reishi), Herbal Medicine: Biomolecular and Clinical Aspects, 2nd ed.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2011; pp. 1–35. [Google Scholar]
- Lin, P.-S.; Chen, S.-D.; Chen, H.-H. Antioxidant and Hypoglycemic Effect of Chromium Enriched Ganoderma lucidum Fermented Rice Flour. J. Food Sci. 2013, 2, 56–62. [Google Scholar]
- Yu, Y.; Fu, P.; Huang, Q.; Zhang, J.; Li, H. Accumulation, subcellular distribution, and oxidative stress of cadmium in Brassica chinensis supplied with selenite and selenate at different growth stages. Chemosphere 2019, 216, 331–340. [Google Scholar] [CrossRef]
- Wang, M.; Ali, F.; Qi, M.; Peng, Q.; Wang, M.; Bañuelos, G.S.; Miao, S.; Li, Z.; Dinh, Q.T.; Liang, D. Insights into uptake, accumulation, and subcellular distribution of selenium among eight wheat (Triticum aestivum L.) cultivars supplied with selenite and selenate. Ecotoxicol. Environ. Saf. 2021, 207, 111544. [Google Scholar] [CrossRef]
- Rayman, M.P. Selenium and human health. Lancet 2012, 379, 1256–1268. [Google Scholar] [CrossRef]
- Zhao, L.; Zhao, G.; Zhao, Z.; Chen, P.; Tong, J.; Hu, X. Selenium distribution in a Se-enriched mushroom species of the genus Ganoderma. J. Agric. Food Chem. 2004, 52, 3954–3959. [Google Scholar] [CrossRef] [PubMed]
- Zhao, P.; Wang, Y.; Zhang, Y.; Guo, T.; Zhang, Z.; Zhang, W.; Zhang, X.; Ashraf, M.A. The extraction of different proteins in selenium enriched peanuts and their antioxidant properties. Saudi J. Biol. Sci. 2016, 23, 353–357. [Google Scholar] [CrossRef]
- Slekovec, M.; Goessler, W. Accumulation of selenium in natural plants and selenium supplemented vegetable and selenium speciation by HPLC-ICPMS. Chem. Speciat. Bioavailab. 2005, 17, 63–73. [Google Scholar] [CrossRef]
- Kieliszek, M. Selenium–fascinating microelement, properties and sources in food. Molecules 2019, 24, 1298. [Google Scholar] [CrossRef]
- Xu, M.; Zhu, S.; Wang, L.; Wei, Z.; Zhao, L.; Shi, G.; Ding, Z. Influence of selenium biofortification on the growth and bioactive metabolites of Ganoderma lucidum. Foods 2021, 10, 1860. [Google Scholar] [CrossRef]
- Kim, J.H.; Kil, D.Y. Comparison of toxic effects of dietary organic or inorganic selenium and prediction of selenium intake and tissue selenium concentrations in broiler chickens using feather selenium concentrations. Poult. Sci. 2020, 99, 6462–6473. [Google Scholar] [CrossRef] [PubMed]
- Ye, Q.; Wu, X.; Zhang, X.; Wang, S. Organic selenium derived from chelation of soybean peptide-selenium and its functional properties in vitro and in vivo. Food Funct. 2019, 10, 4761–4770. [Google Scholar] [CrossRef]
- Bureau, N.S. Available online: http://www.foodcta.com/spbz/detail75643.html# (accessed on 25 December 2022).
- Meija, J.; Montes-Bayón, M.; Le Duc, D.L.; Terry, N.; Caruso, J.A. Simultaneous monitoring of volatile selenium and sulfur species from Se accumulating plants (wild type and genetically modified) by GC/MS and GC/ICPMS using solid-phase microextraction for sample introduction. Anal. Chem. 2002, 74, 5837–5844. [Google Scholar] [CrossRef]
- Vonderheide, A.P.; Wrobel, K.; Kannamkumarath, S.S.; B’Hymer, C.; Montes-Bayón, M.; Ponce de León, C.; Caruso, J.A. Characterization of selenium species in Brazil nuts by HPLC− ICP-MS and ES-MS. J. Agric. Food Chem. 2002, 50, 5722–5728. [Google Scholar] [CrossRef]
- Ye, M.; Li, J.; Yu, R.; Cong, X.; Huang, D.; Li, Y.; Chen, S.; Zhu, S. Selenium Speciation in Selenium-Enriched Plant Foods. Food Anal. Methods. 2022, 15, 1377–1389. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, P.; Chen, J.; Shao, J.F.; Gui, R. Selenium biofortification of bamboo shoots by liquid Se fertilization in the culm pith cavity. Food Sci. Technol. 2020, 1, 27–34. [Google Scholar] [CrossRef]
- Yuan, L.; Zhu, Y.; Lin, Z.-Q.; Banuelos, G.; Li, W.; Yin, X. A novel selenocystine-accumulating plant in selenium-mine drainage area in Enshi, China. PloS ONE 2013, 8, e65615. [Google Scholar] [CrossRef]
- Roberge, M.T.; Borgerding, A.J.; Finley, J.W. Speciation of selenium compounds from high selenium broccoli is affected by the extracting solution. J. Agric. Food Chem. 2003, 51, 4191–4197. [Google Scholar] [CrossRef]
- Garcia-Rubio, R.; de Oliveira, H.C.; Rivera, J.; Trevijano-Contador, N. The fungal cell wall: Candida, Cryptococcus, and Aspergillus species. Front. Microbiol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Caridi, A. Enological functions of parietal yeast mannoproteins. Antonie Van Leeuwenhoek 2006, 89, 417–422. [Google Scholar] [CrossRef]
- Tinggi, U. Selenium: Its role as antioxidant in human health. Environ. Health Prev. Med. 2008, 13, 102–108. [Google Scholar] [CrossRef]
- Wickleder, M.S. Sodium selenite, Na2SeO3. Acta Crystallogr. Sect. E Struct. Rep. Online 2002, 58, i103–i104. [Google Scholar] [CrossRef]
- Rao, Y.; McCooeye, M.; Windust, A.; Bramanti, E.; D’Ulivo, A.; Mester, Z.n. Mapping of selenium metabolic pathway in yeast by Liquid Chromatography− Orbitrap Mass Spectrometry. Anal. Chem. 2010, 82, 8121–8130. [Google Scholar] [CrossRef]
- Gammelgaard, B.; Cornett, C.; Olsen, J.; Bendahl, L.; Hansen, S.H. Combination of LC-ICP-MS, LC-MS and NMR for investigation of the oxidative degradation of selenomethionine. Talanta 2003, 59, 1165–1171. [Google Scholar] [CrossRef]
- Kieliszek, M.; Błażejak, S.; Gientka, I.; Bzducha-Wróbel, A. Accumulation and metabolism of selenium by yeast cells. Appl. Microbiol. Biotechnol. 2015, 99, 5373–5382. [Google Scholar] [CrossRef]
- Mapelli, V.; Hillestrøm, P.R.; Patil, K.; Larsen, E.H.; Olsson, L. The interplay between sulphur and selenium metabolism influences the intracellular redox balance in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 20–32. [Google Scholar] [CrossRef]
- Whanger, P. Selenocompounds in plants and animals and their biological significance. J. Am. Coll. Nutr. 2002, 21, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.; Nigam, S.; McConnell, W. Biosynthesis of Se-methylselenocysteine and S-methylcysteine in Astragalus bisulcatus origin of the selenomethyl and the thiomethyl groups. Biochim. Biophys. (BBA)-General Subjects 1972, 273, 91–96. [Google Scholar] [CrossRef]
- Soda, K. Biochemical aspects of selenium amino acids and selenium peptides. Phosphorus Sulfur Silicon Relat. Elem. 1992, 67, 461–472. [Google Scholar] [CrossRef]
- Ding, W.; Ding, Y.; Xia, F.; Gu, H.; Zhang, L.; Shi, Y. Identification of 10 unsaturated fatty acid compounds in the submerged fermentation products of Ganoderma lingzhi. Mycosystema 2021, 7, 1800–1810. [Google Scholar]
- Ye, L.; Xie, F.; Zhao, L.; Fang, J.; Wu, X. Effect of selenium on main active component contents of Ganoderma lingzhi in different periods. J. Zhejiang Univ. 2017, 43, 462–468. [Google Scholar]
- Wrobel, K.; Kannamkumarath, S.S.; Wrobel, K.; Caruso, J.A. Hydrolysis of proteins with methanesulfonic acid for improved HPLC-ICP-MS determination of seleno-methionine in yeast and nuts. Anal. Bioanal. Chem. 2003, 375, 133–138. [Google Scholar] [CrossRef]
- Zhang, X.; He, H.; Xiang, J.; Hou, T. Screening and bioavailability evaluation of anti-oxidative selenium-containing peptides from soybeans based on specific structures. Food Funct. 2022, 13, 5252–5261. [Google Scholar] [CrossRef]
- Rodriguez, J.; Gupta, N.; Smith, R.D.; Pevzner, P.A. Does trypsin cut before proline? J. Proteome Res. 2008, 7, 300–305. [Google Scholar] [CrossRef]
- Ebeling, W.; Hennrich, N.; Klockow, M.; Metz, H.; Orth, H.D.; Lang, H. Proteinase K from Tritirachium album limber. Eur. J. Biochem. 1974, 47, 91–97. [Google Scholar] [CrossRef]
- Kantorová, V.; Kaňa, A.; Krausová, G.; Hyršlová, I.; Mestek, O. Effect of protease XXIII on selenium species interconversion during their extraction from biological samples. J. Food Compost. Anal. 2022, 105, 104260. [Google Scholar] [CrossRef]
- Pavlisko, A.; Rial, A.; DE VECCHI, S.; Coppes, Z. Properties of pepsin and trypsin isolated from the digestive tract of Parona signata “palometa”. J. Food Biochem. 1997, 21, 289–308. [Google Scholar] [CrossRef]
- Del Rio, A.R.; Keppler, J.K.; Boom, R.M.; Janssen, A.E. Protein acidification and hydrolysis by pepsin ensure efficient trypsin-catalyzed hydrolysis. Food Funct. 2021, 12, 4570–4581. [Google Scholar] [CrossRef]
- Gilon, N.; Astruc, A.; Astruc, M.; Potin-Gautier, M. Selenoamino acid speciation using HPLC–ETAAS following an enzymic hydrolysis of selenoprotein. Appl. Organomet. Chem. 1995, 9, 623–628. [Google Scholar] [CrossRef]
- Dernovics, M.; Stefánka, Z.; Fodor, P. Improving selenium extraction by sequential enzymatic processes for Se-speciation of selenium-enriched Agaricus bisporus. Anal. Bioanal. Chem. 2002, 372, 473–480. [Google Scholar] [CrossRef]
- Stefánka, Z.; Ipolyi, I.; Dernovics, M.; Fodor, P. Comparison of sample preparation methods based on proteolytic enzymatic processes for Se-speciation of edible mushroom (Agaricus bisporus) samples. Talanta 2001, 55, 437–447. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Sun, X.; Li, P.; Shen, X.; Fang, Y. Selenium in cereals: Insight into species of the element from total amount. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2914–2940. [Google Scholar] [CrossRef]
- Poblaciones, M.; Rodrigo, S.; Santamaria, O. Selenium: Chemistry, Analysis, Function and Effects; The Royal Society of Chemistry: Cambridge, UK, 2015; pp. 324–340. ISBN 2045-1695. [Google Scholar]
- Cheajesadagul, P.; Bianga, J.; Arnaudguilhem, C.; Lobinski, R.; Szpunar, J. Large-scale speciation of selenium in rice proteins using ICP-MS assisted electrospray MS/MS proteomics. Metallomics 2014, 6, 646–653. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ali, F.; Wang, M.; Dinh, Q.T.; Zhou, F.; Bañuelos, G.S.; Liang, D. Understanding boosting selenium accumulation in Wheat (Triticum aestivum L.) following foliar selenium application at different stages, forms, and doses. Environ. Sci. Pollut. Res. 2020, 27, 717–728. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.B.; Maniero, M.Á.; Londonio, A.; Smichowski, P.; Wuilloud, R.G. Effects of common cooking heat treatments on selenium content and speciation in garlic. J. Food Compos. Anal. 2018, 70, 54–62. [Google Scholar] [CrossRef]
- Wu, L.; Van Mantgem, P.; Guo, X. Effects of forage plant and field legume species on soil selenium redistribution, leaching, and bioextraction in soils contaminated by agricultural drain water sediment. Arch. Environ. Contam. Toxicol. 1996, 31, 329–338. [Google Scholar] [CrossRef]
- Shao, S.; Mi, X.; Ouerdane, L.; Lobinski, R.; García-Reyes, J.F.; Molina-Díaz, A.; Vass, A.; Dernovics, M. Quantification of Se-methylselenocysteine and its γ-glutamyl derivative from naturally Se-enriched green bean (Phaseolus vulgaris vulgaris) after HPLC-ESI-TOF-MS and orbitrap MS n-based identification. Food Anal. Methods. 2014, 7, 1147–1157. [Google Scholar] [CrossRef]
- Kápolna, E.; Fodor, P. Speciation analysis of selenium enriched green onions (Allium fistulosum) by HPLC-ICP-MS. Microchem. J. 2006, 84, 56–62. [Google Scholar] [CrossRef]
- Rao, S.; Gou, Y.; Yu, T.; Cong, X.; Gui, J.; Zhu, Z.; Zhang, W.; Liao, Y.; Ye, J.; Cheng, S. Effects of selenate on Se, flavonoid, and glucosinolate in broccoli florets by combined transcriptome and metabolome analyses. Food Res. Int. 2021, 146, 110463. [Google Scholar] [CrossRef]
- Zou, Y.; Du, F.; Zhang, H.; Hu, Q. Selenium speciation and biological characteristics of selenium-rich Bailing mushroom, Pleurotus tuoliensis. Emir. J. Food Agric. 2018, 704–708. [Google Scholar]
- Gergely, V.; Kubachka, K.M.; Mounicou, S.; Fodor, P.; Caruso, J.A. Selenium speciation in Agaricus bisporus and Lentinula edodes mushroom proteins using multi-dimensional chromatography coupled to inductively coupled plasma mass spectrometry. J. Chromatogr. A 2006, 1101, 94–102. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.; Yang, W.; Wang, M.; Miao, Y.; Cui, Z.; Li, Z.; Liang, D. Effects of selenium application on Se content and speciation in Lentinula edodes. Food Chem. 2018, 265, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Kousha, M.; Yeganeh, S.; Keramat Amirkolaie, A. Effect of sodium selenite on the bacteria growth, selenium accumulation, and selenium biotransformation in Pediococcus acidilactici. Food Sci. Biotechnol. 2017, 26, 1013–1018. [Google Scholar] [CrossRef]
- Lee, M.R.; Fleming, H.R.; Cogan, T.; Hodgson, C.; Davies, D.R. Assessing the ability of silage lactic acid bacteria to incorporate and transform inorganic selenium within laboratory scale silos. Anim. Feed Sci. Technol. 2019, 253, 125–134. [Google Scholar] [CrossRef] [PubMed]
- Eszenyi, P.; Sztrik, A.; Babka, B.; Prokisch, J. Elemental, nano-sized (100–500 nm) selenium production by probiotic lactic acid bacteria. Int. j. Biosci. Biochem. Bioinforma. 2011, 1, 148. [Google Scholar] [CrossRef]
- Zommara, M.A.; Prokisch, J. Conversion of inorganic selenium to organic form (s) by Lactobacillus acidophilus. Alex. J. Food Sci. Tech. 2019, 16, 17–24. [Google Scholar] [CrossRef]
- Jin, W.; Yoon, C.; Johnston, T.V.; Ku, S.; Ji, G.E. Production of selenomethionine-enriched Bifidobacterium bifidum BGN4 via sodium selenite biocatalysis. Molecules 2018, 23, 2860. [Google Scholar] [CrossRef]
- Capelo, J.; Ximenez-Embun, P.; Madrid-Albarran, Y.; Cámara, C. Enzymatic probe sonication: Enhancement of protease-catalyzed hydrolysis of selenium bound to proteins in yeast. Anal. Chem. 2004, 76, 233–237. [Google Scholar] [CrossRef]
- Mahan, D.; Cline, T.; Richert, B. Effects of dietary levels of selenium-enriched yeast and sodium selenite as selenium sources fed to growing-finishing pigs on performance, tissue selenium, serum glutathione peroxidase activity, carcass characteristics, and loin quality. J. Anim. Sci. 1999, 77, 2172–2179. [Google Scholar] [CrossRef]
- González-Salitre, L.; Román-Gutiérrez, A.; Contreras-López, E.; Bautista-Ávila, M.; Rodríguez-Serrano, G.; González-Olivares, L. Promising use of selenized yeast to develop new enriched food: Human health implications. Food Rev. Int. 2021, 1–18. [Google Scholar] [CrossRef]
- Yang, J.; Yang, H. Recent development in Se-enriched yeast, lactic acid bacteria and bifidobacteria. Crit. Rev. Food Sci. Nutr. 2021, 63, 1–15. [Google Scholar] [CrossRef]
- Alzate, A.; Fernández-Fernández, A.; Pérez-Conde, M.; Gutiérrez, A.; Cámara, C. Comparison of biotransformation of inorganic selenium by Lactobacillus and Saccharomyces in lactic fermentation process of yogurt and kefir. J. Agric. Food Chem. 2008, 56, 8728–8736. [Google Scholar] [CrossRef] [PubMed]
- Schrauzer, G. Anticarcinogenic effects of selenium. Cell. Mol. Life Sci. 2000, 57, 1864–1873. [Google Scholar] [CrossRef] [PubMed]

| Enzyme | Medium | Origin | Function | References |
|---|---|---|---|---|
| Protease K | General | Engydontium album | cut peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups | [25] |
| Protease XIV | General | Streptomyces griseus | still under investigation | [26] |
| Pepsin | Acid | Gastric chief cell of stomach | cut peptide bonds between large hydrophobic amino acid residues and acts on proteins and converts them into peptones | [27] |
| Trypsin | Alkaline | PA clan superfamily in digestive system | cut peptide bonds at the C-terminal side of lysine or arginine and converts peptones into polypeptides | [27] |
| Sample Name | Total Se (μg/g) | Proteolytic Process | Se Speciation | References |
|---|---|---|---|---|
| Pediococcus acidliatici (bacteria) | 430 ± 4.0 | 1. Lysozyme 2. Protease XIV under untrasonic | SeCys2 *, SeMecys, SeMet | [41] |
| Agaricus bisporus (mushroom) | 160 | 1. Water 2. Tris-HCl, lysing enzyme 3. Phosphate, protease XIV | SeMet *, SeCys2, Selenite | [31] |
| Pleurotus tuoliensis (mushroom) | 100 | Protease K, water | SeMet *, SeCys2, Se (IV) | [37] |
| Lentinula edodes (mushroom) | 46 ± 1.2 | 1. Tris-HCl 2. Trpsin 3. Protease XIV | SeMecys *, Se (IV), SeMet, SeCys2 | [38] |
| Agaricus bisporus (mushroom) | 770.7 ± 37.3 | 1. Tris-HCl 2. Trpsin 3. Protease XIV | SeCys2 *, Se (IV), SeMet, SeMecys | [38] |
| Lyophilized Se-enriched yeast | 1300 | Tris-HCl, protease XIV | SeMet | [45] |
| Bifidobacterium bifidum BGN4 (probiotic) | 207.5 ± 1.25 | Phophate buffer, pronase E | SeMet | [46] |
| Silage lactic acid (bacteria) | 155.7 ± 9.7 | Protease XIV | nano-Se *, SeMet, SeCys2 | [41] |
| Ganoderma Lucidum | 245.7 ± 52.9 | 1. Water 2. Glycine-Pepsin 3. Tris-HCl-Trypsin | Se (IV)*, SeCys2, SeMet, SeMecys | this study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, W.; Hou, Y.; Zhang, Z.; Yin, X.; Zhao, X.; Yuan, L. Determination of Selenium Speciation in High Se-Enriched Edible Fungus Ganoderma lucidum Via Sequential Extraction. Horticulturae 2023, 9, 161. https://doi.org/10.3390/horticulturae9020161
Shi W, Hou Y, Zhang Z, Yin X, Zhao X, Yuan L. Determination of Selenium Speciation in High Se-Enriched Edible Fungus Ganoderma lucidum Via Sequential Extraction. Horticulturae. 2023; 9(2):161. https://doi.org/10.3390/horticulturae9020161
Chicago/Turabian StyleShi, Wenyao, Yuzhu Hou, Zezhou Zhang, Xuebin Yin, Xiaohu Zhao, and Linxi Yuan. 2023. "Determination of Selenium Speciation in High Se-Enriched Edible Fungus Ganoderma lucidum Via Sequential Extraction" Horticulturae 9, no. 2: 161. https://doi.org/10.3390/horticulturae9020161
APA StyleShi, W., Hou, Y., Zhang, Z., Yin, X., Zhao, X., & Yuan, L. (2023). Determination of Selenium Speciation in High Se-Enriched Edible Fungus Ganoderma lucidum Via Sequential Extraction. Horticulturae, 9(2), 161. https://doi.org/10.3390/horticulturae9020161









