Microbial Communities on Samples of Commercially Available Fresh-Consumed Leafy Vegetables and Small Berries
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction, Amplification, and Sequencing
2.2. Sequences Analysis
3. Results
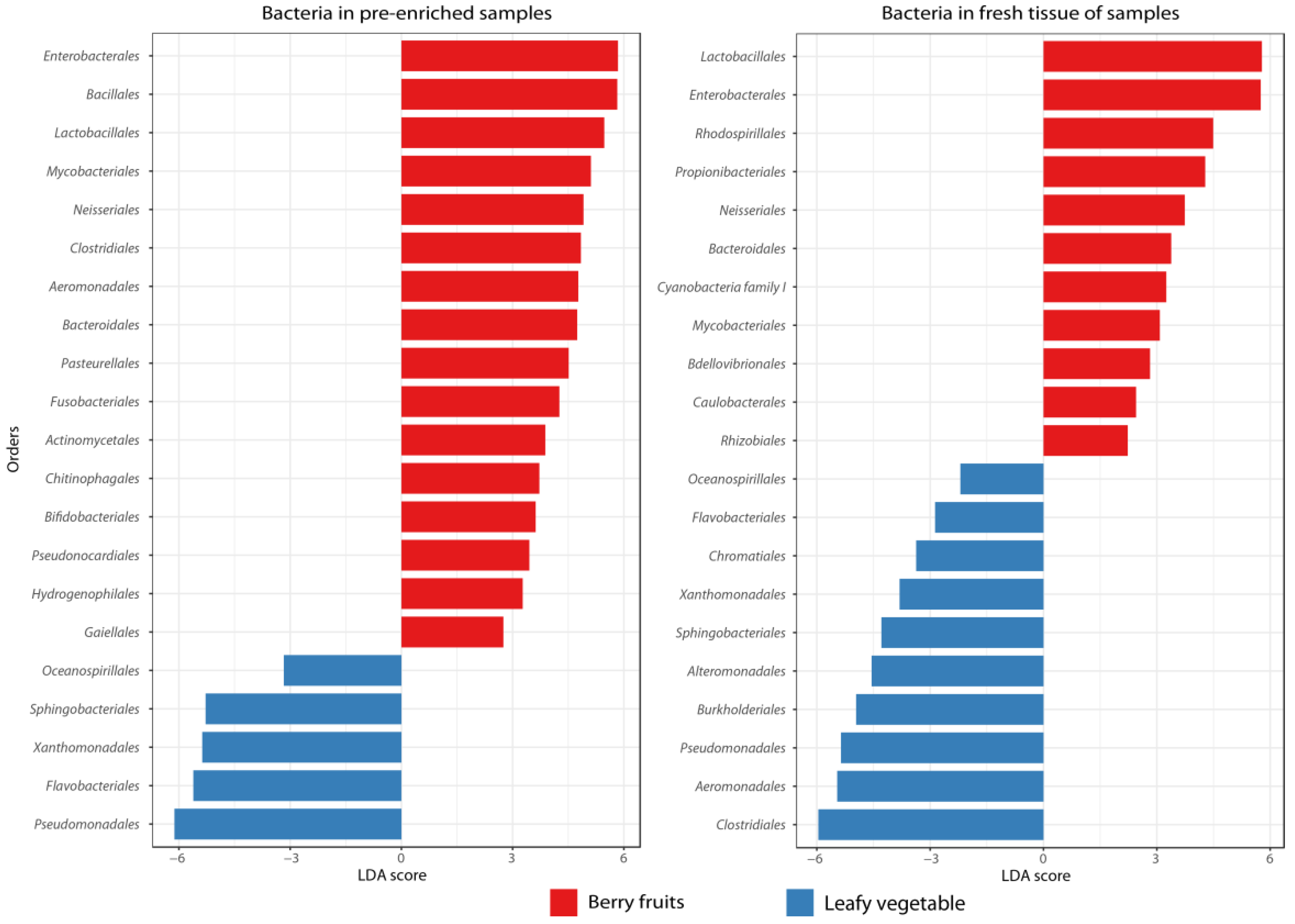
3.1. Diversity and Structure of Microbial Communities
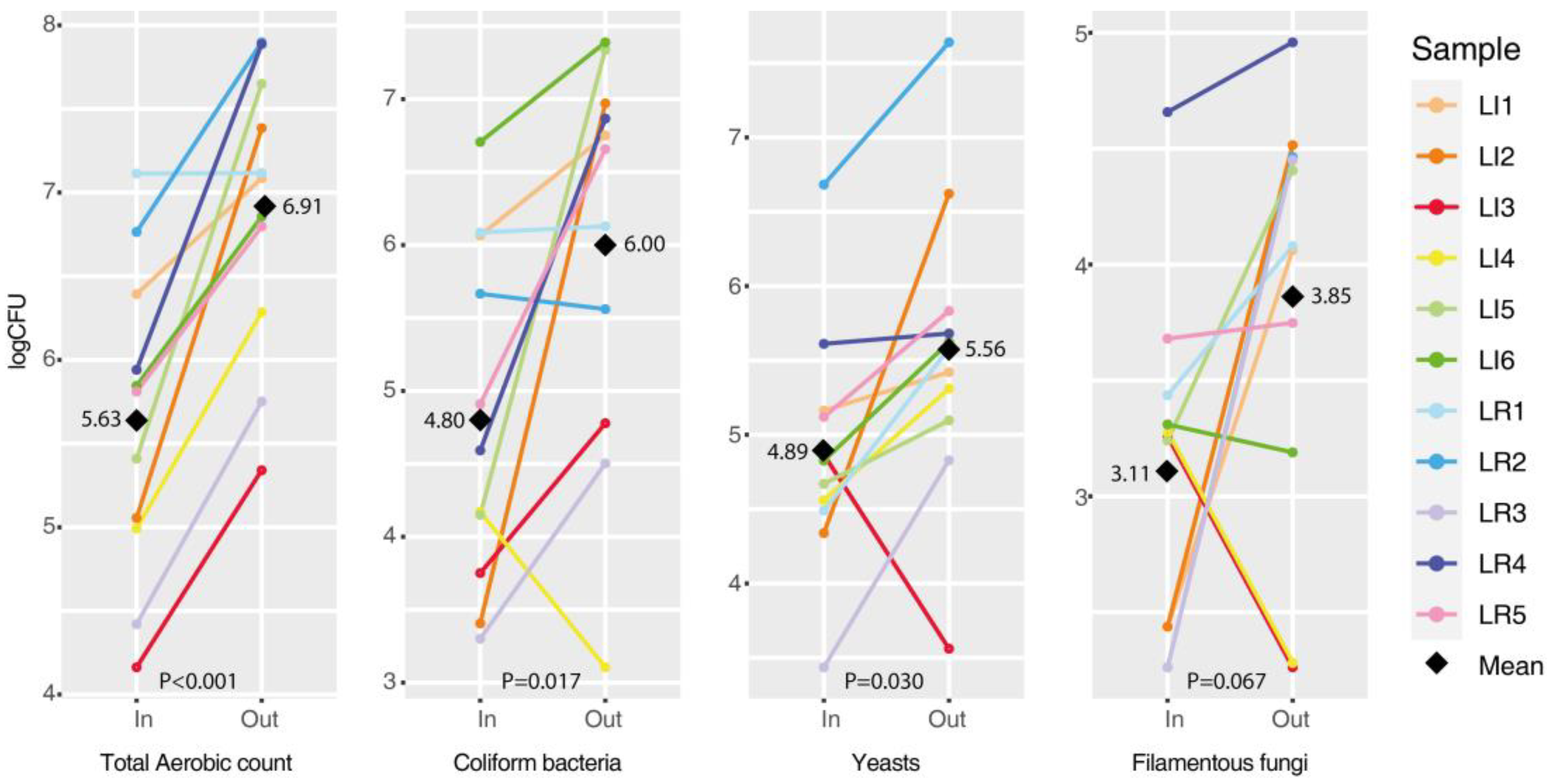
3.2. Comparison of Microbial Colonization Inside/Outside Lettuce Rosette
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Amiot-Carlin, M.-J. Fruit and vegetable consumption: What benefits, what risks? Rev. Prat. 2019, 69, 139–142. [Google Scholar] [PubMed]
- Ramya, V.; Patel, P. Health benefits of vegetables. Int. J. Chem. Stud. 2019, 7, 82–87. [Google Scholar]
- Mason-D’Croz, D.; Bogard, J.R.; Sulser, T.B.; Cenacchi, N.; Dunston, S.; Herrero, M.; Wiebe, K. Gaps between fruit and vegetable production, demand, and recommended consumption at global and national levels: An integrated modelling study. Lancet Planet. Health 2019, 3, e318–e329. [Google Scholar] [CrossRef] [PubMed]
- Balali, G.I.; Yar, D.D.; Afua Dela, V.G.; Adjei-Kusi, P. Microbial Contamination, an Increasing Threat to the Consumption of Fresh Fruits and Vegetables in Today’s World. Int. J. Microbiol. 2020, 2020, 3029295. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, E. Is the Japanese O157: H7 E. coli epidemic over? Lancet 1996, 348, 1371. [Google Scholar] [CrossRef]
- Buchholz, U.; Bernard, H.; Werber, D.; Böhmer, M.M.; Remschmidt, C.; Wilking, H.; Deleré, Y.; an der Heiden, M.; Adlhoch, C.; Dreesman, J.; et al. German Outbreak of Escherichia coli O104:H4 Associated with Sprouts. N. Engl. J. Med. 2011, 365, 1763–1770. [Google Scholar] [CrossRef]
- EU Directorate General for Health and Food Safety; Rapid Alert System for Food and Feed (RASFF): 2022. Available online: https://webgate.ec.europa.eu/rasff-window/screen/search (accessed on 2 December 2022).
- USA Centers for Disease Control and Prevention; List of Outbreaks: 2022. Available online: https://www.cdc.gov/foodsafety/outbreaks/lists/outbreaks-list.html (accessed on 2 December 2022).
- de Oliveira Elias, S.; Noronha, T.B.; Tondo, E.C. Salmonella spp. and Escherichia coli O157:H7 prevalence and levels on lettuce: A systematic review and meta-analysis. Food Microbiol. 2019, 84, 103217. [Google Scholar] [CrossRef]
- Mohammadpour, H.; Berizi, E.; Hosseinzadeh, S.; Majlesi, M.; Zare, M. The prevalence of Campylobacter spp. in vegetables, fruits, and fresh produce: A systematic review and meta-analysis. Gut Pathog. 2018, 10, 41. [Google Scholar] [CrossRef]
- Ding, T.; Iwahori, J.i.; Kasuga, F.; Wang, J.; Forghani, F.; Park, M.-S.; Oh, D.-H. Risk assessment for Listeria monocytogenes on lettuce from farm to table in Korea. Food Control 2013, 30, 190–199. [Google Scholar] [CrossRef]
- Bozkurt, H.; Phan-Thien, K.-Y.; van Ogtrop, F.; Bell, T.; McConchie, R. Outbreaks, occurrence, and control of norovirus and hepatitis a virus contamination in berries: A review. Crit. Rev. Food Sci. Nutr. 2021, 61, 116–138. [Google Scholar] [CrossRef]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
- Schlaeppi, K.; Bulgarelli, D. The Plant Microbiome at Work. Mol. Plant-Microbe Interact. ® 2015, 28, 212–217. [Google Scholar] [CrossRef]
- van Overbeek, L.; Duhamel, M.; Aanstoot, S.; van der Plas, C.L.; Nijhuis, E.; Poleij, L.; Russ, L.; van der Zouwen, P.; Andreo-Jimenez, B. Transmission of Escherichia coli from Manure to Root Zones of Field-Grown Lettuce and Leek Plants. Microorganisms 2021, 9, 2289. [Google Scholar] [CrossRef]
- Olimi, E.; Kusstatscher, P.; Wicaksono, W.A.; Abdelfattah, A.; Cernava, T.; Berg, G. Insights into the microbiome assembly during different growth stages and storage of strawberry plants. Environ. Microbiome 2022, 17, 21. [Google Scholar] [CrossRef]
- Hou, Z.; Fink, R.C.; Radtke, C.; Sadowsky, M.J.; Diez-Gonzalez, F. Incidence of naturally internalized bacteria in lettuce leaves. Int. J. Food Microbiol. 2013, 162, 260–265. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 197. [Google Scholar] [CrossRef]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. The plant microbiome and its importance for plant and human health. Front. Microbiol. 2014, 5, 491. [Google Scholar] [CrossRef]
- Berg, G.; Eberl, L.; Hartmann, A. The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 2005, 7, 1673–1685. [Google Scholar] [CrossRef]
- Khalifa, A.Y.Z.; Alsyeeh, A.-M.; Almalki, M.A.; Saleh, F.A. Characterization of the plant growth promoting bacterium, Enterobacter cloacae MSR1, isolated from roots of non-nodulating Medicago sativa. Saudi J. Biol. Sci. 2016, 23, 79–86. [Google Scholar] [CrossRef]
- Sanzani, S.M.; Reverberi, M.; Geisen, R. Mycotoxins in harvested fruits and vegetables: Insights in producing fungi, biological role, conducive conditions, and tools to manage postharvest contamination. Postharvest Biol. Technol. 2016, 122, 95–105. [Google Scholar] [CrossRef]
- Saleh, I.; Al-Thani, R. Fungal food spoilage of supermarkets’ displayed fruits. Vet. World 2019, 12, 1877. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Awolusi, O.O.; Stenström, T.A. Organic Fertilizers: Public Health Intricacies. In Organic Fertilizers-From Basic Concepts to Applied Outcomes; Larramendy, M.L., Soloneski, S., Eds.; IntechOpen: London, UK, 2016; pp. 343–374. [Google Scholar] [CrossRef]
- Gurtler, J.B.; Gibson, K.E. Irrigation water and contamination of fresh produce with bacterial foodborne pathogens. Curr. Opin. Food Sci. 2022, 47, 100889. [Google Scholar] [CrossRef]
- Riggio, G.M.; Jones, S.L.; Gibson, K.E. Risk of human pathogen internalization in leafy vegetables during lab-scale hydroponic cultivation. Horticulturae 2019, 5, 25. [Google Scholar] [CrossRef]
- Jay-Russell, M.T. What is the risk from wild animals in food-borne pathogen contamination of plants? CABI Rev. 2013, 8, 1–16. [Google Scholar] [CrossRef]
- Cernava, T.; Erlacher, A.; Soh, J.; Sensen, C.W.; Grube, M.; Berg, G. Enterobacteriaceae dominate the core microbiome and contribute to the resistome of arugula (Eruca sativa Mill.). Microbiome 2019, 7, 13. [Google Scholar] [CrossRef]
- Yeon-Cheol, Y.; Su-Jin, Y.; Da-Young, J. Analysis of the Microbiota on Lettuce (Lactuca sativa L.) Cultivated in South Korea to Identify Foodborne Pathogens. J. Microbiol. Biotechnol. 2018, 28, 1318–1331. [Google Scholar] [CrossRef]
- Williams, T.R.; Moyne, A.-L.; Harris, L.J.; Marco, M.L. Season, Irrigation, Leaf Age, and Escherichia coli Inoculation Influence the Bacterial Diversity in the Lettuce Phyllosphere. PLoS ONE 2013, 8, e68642. [Google Scholar] [CrossRef]
- Brandl, M.T.; Amundson, R. Leaf Age as a Risk Factor in Contamination of Lettuce with Escherichia coli O157:H7 and Salmonella enterica. Appl. Environ. Microbiol. 2008, 74, 2298–2306. [Google Scholar] [CrossRef]
- Jensen, B.; Knudsen, I.M.B.; Andersen, B.; Nielsen, K.F.; Thrane, U.; Jensen, D.F.; Larsen, J. Characterization of microbial communities and fungal metabolites on field grown strawberries from organic and conventional production. Int. J. Food Microbiol. 2013, 160, 313–322. [Google Scholar] [CrossRef]
- Baugher, J.; Jaykus, L. Natural microbiota of raspberries (Rubus idaeus) and strawberries (Fragaria × ananassa): Microbial survey, bacterial isolation and identification, and biofilm characterization. In XI International Rubus and Ribes Symposium 1133; Fernandez, G.E., Hummer, K.E., Eds.; ISHS: Leuven, Belgium, 2015; pp. 421–526. [Google Scholar] [CrossRef]
- Quansah, J.K.; Gazula, H.; Holland, R.; Scherm, H.; Li, C.; Takeda, F.; Chen, J. Microbial quality of blueberries for the fresh market. Food Control 2019, 100, 92–96. [Google Scholar] [CrossRef]
- Macori, G.; Gilardi, G.; Bellio, A.; Bianchi, D.M.; Gallina, S.; Vitale, N.; Gullino, M.L.; Decastelli, L. Microbiological Parameters in the Primary Production of Berries: A Pilot Study. Foods 2018, 7, 105. [Google Scholar] [CrossRef]
- Pavlovic, M.; Huber, I.; Skala, H.; Konrad, R.; Schmidt, H.; Sing, A.; Busch, U. Development of a multiplex real-time polymerase chain reaction for simultaneous detection of enterohemorrhagic Escherichia coli and enteropathogenic Escherichia coli strains. Foodborne Pathog. Dis. 2010, 7, 801–808. [Google Scholar] [CrossRef]
- Lee, D.-Y.; Shannon, K.; Beaudette, L.A. Detection of bacterial pathogens in municipal wastewater using an oligonucleotide microarray and real-time quantitative PCR. J. Microbiol. Methods 2006, 65, 453–467. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516–4522. [Google Scholar] [CrossRef]
- Artimová, R.; Hleba, L.; Javoreková, S.; Maková, J.; Medová, J.; Medo, J. Chloroplast Excluding Primers For Metagenomic Analysis Of Bacteria In Plant Tissues. J. Microbiol. Biotechnol. Food Sci. 2022, 12, e9650. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E. New primers to amplify the fungal ITS2 region–evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. A Guide Methods Appl. 1990, 18, 315–322. [Google Scholar]
- Lane, D.J.; Pace, B.; Olsen, G.J.; Stahl, D.A.; Sogin, M.L.; Pace, N.R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc. Natl. Acad. Sci. USA 1985, 82, 6955–6959. [Google Scholar] [CrossRef]
- Vetrovský, T.; Baldrian, P.; Morais, D.; Berger, B. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 1, 3. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Standley, D.M.; Katoh, K. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Lozupone, C.; Knight, R. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2010, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘Vegan’. Community Ecology Package, Version 2.5-6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 2 October 2022).
- Zhao, S.; Guo, Y.; Sheng, Q.; Shyr, Y. Advanced Heat Map and Clustering Analysis Using Heatmap3. BioMed Res. Int. 2014, 2014, 6. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring Correlation Networks from Genomic Survey Data. PLOS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Chong, J.; Liu, P.; Zhou, G.; Xia, J. Using MicrobiomeAnalyst for comprehensive statistical, functional, and meta-analysis of microbiome data. Nat. Protoc. 2020, 15, 799–821. [Google Scholar] [CrossRef]
- Holland, R.M.; Chen, J.; Gazula, H.; Scherm, H. Environmental and Fecal Indicator Organisms on Fruit Contact Surfaces and Fruit from Blueberry Mechanical Harvesters. Horticulturae 2022, 8, 20. [Google Scholar] [CrossRef]
- Korir, R.C.; Parveen, S.; Hashem, F.; Bowers, J. Microbiological quality of fresh produce obtained from retail stores on the Eastern Shore of Maryland, United States of America. Food Microbiol. 2016, 56, 29–34. [Google Scholar] [CrossRef]
- Kuźniar, P.; Belcar, J.; Zardzewiały, M.; Basara, O.; Gorzelany, J. Effect of Ozonation on the Mechanical, Chemical, and Microbiological Properties of Organically Grown Red Currant (Ribes rubrum L.) Fruit. Molecules 2022, 27, 8231. [Google Scholar] [CrossRef]
- Smith, J.; Fratamico, P. Escherichia coli and other Enterobacteriaceae: Food poisoning and health effects. In Encyclopedia of Food and Health; Caballer., B., Finglas, P., Toldrá, F., Eds.; AcademicPress: Waltham, MA, USA, 2016; pp. 539–544. [Google Scholar]
- Ekici, G.; Dümen, E. Escherichia coli and food safety. In The Universe of Escherichia Coli; Starčič Erjavec, M., Ed.; IntechOpen: London, UK, 2019; pp. 1–16. [Google Scholar] [CrossRef]
- Oliveira, M.; Rodrigues, C.M.; Teixeira, P. Microbiological quality of raw berries and their products: A focus on foodborne pathogens. Heliyon 2019, 5, e02992. [Google Scholar] [CrossRef]
- Walterson, A.M.; Stavrinides, J. Pantoea: Insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 2015, 39, 968–984. [Google Scholar] [CrossRef]
- Rahman, M.; Alam, M.-U.; Luies, S.K.; Kamal, A.; Ferdous, S.; Lin, A.; Sharior, F.; Khan, R.; Rahman, Z.; Parvez, S.M.; et al. Contamination of Fresh Produce with Antibiotic-Resistant Bacteria and Associated Risks to Human Health: A Scoping Review. Int. J. Environ. Res. Public Health 2022, 19, 360. [Google Scholar] [CrossRef]
- Johnston-Monje, D.; Lopez Mejia, J. Botanical microbiomes on the cheap: Inexpensive molecular fingerprinting methods to study plant-associated communities of bacteria and fungi. Appl. Plant Sci. 2020, 8, e11334. [Google Scholar] [CrossRef]
- Sare, A.R.; Stouvenakers, G.; Eck, M.; Lampens, A.; Goormachtig, S.; Jijakli, M.H.; Massart, S. Standardization of Plant Microbiome Studies: Which Proportion of the Microbiota is Really Harvested? Microorganisms 2020, 8, 342. [Google Scholar] [CrossRef]
- Seo, D.W.; Yum, S.-j.; Lee, H.R.; Kim, S.M.; Jeong, H.G. Microbiota Analysis and Microbiological Hazard Assessment in Chinese Chive (Allium tuberosum Rottler) Depending on Retail Types. J. Microbiol. Biotechnol. 2022, 32, 195–204. [Google Scholar] [CrossRef]
- Jarvis, K.G.; White, J.R.; Grim, C.J.; Ewing, L.; Ottesen, A.R.; Beaubrun, J.J.-G.; Pettengill, J.B.; Brown, E.; Hanes, D.E. Cilantro microbiome before and after nonselective pre-enrichment for Salmonella using 16S rRNA and metagenomic sequencing. BMC Microbiol. 2015, 15, 160. [Google Scholar] [CrossRef]
- Leff, J.W.; Fierer, N. Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Q.; Chen, S.; Zhang, Z.; Song, J.; Long, Z.; Yu, Y.; Fang, H. Enterobacteriaceae predominate in the endophytic microbiome and contribute to the resistome of strawberry. Sci. Total Environ. 2020, 727, 138708. [Google Scholar] [CrossRef]
- Zhimo, V.Y.; Kumar, A.; Biasi, A.; Salim, S.; Feygenberg, O.; Toamy, M.A.; Abdelfattaah, A.; Medina, S.; Freilich, S.; Wisniewski, M.; et al. Compositional shifts in the strawberry fruit microbiome in response to near-harvest application of Metschnikowia fructicola, a yeast biocontrol agent. Postharvest Biol. Technol. 2021, 175, 111469. [Google Scholar] [CrossRef]
- Darlison, J.; Mogren, L.; Rosberg, A.K.; Grudén, M.; Minet, A.; Liné, C.; Mieli, M.; Bengtsson, T.; Håkansson, Å.; Uhlig, E.; et al. Leaf mineral content govern microbial community structure in the phyllosphere of spinach (Spinacia oleracea) and rocket (Diplotaxis tenuifolia). Sci. Total Environ. 2019, 675, 501–512. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Naveed, M.; Mustafa, A.; Abbas, A. The good, the bad, and the ugly of rhizosphere microbiome. In Probiotics and Plant Health; Springer: Berlin/Heidelberg, Germany, 2017; pp. 253–290. [Google Scholar]
- Haelewaters, D.; Urbina, H.; Brown, S.; Newerth-Henson, S.; Aime, M.C. Isolation and Molecular Characterization of the Romaine Lettuce Phylloplane Mycobiome. J. Fungi 2021, 7, 277. [Google Scholar] [CrossRef] [PubMed]
- Dietzel, K.; Valle, D.; Fierer, N.; U’Ren, J.M.; Barberán, A. Geographical Distribution of Fungal Plant Pathogens in Dust Across the United States. Front. Ecol. Evol. 2019, 7, 304. [Google Scholar] [CrossRef]
- Hua, L.; Yong, C.; Zhanquan, Z.; Boqiang, L.; Guozheng, Q.; Shiping, T. Pathogenic mechanisms and control strategies of Botrytis cinerea causing post-harvest decay in fruits and vegetables. Food Qual. Saf. 2018, 2, 111–119. [Google Scholar] [CrossRef]
- Zarkani, A.A.; Schierstaedt, J.; Becker, M.; Krumwiede, J.; Grimm, M.; Grosch, R.; Jechalke, S.; Schikora, A. Salmonella adapts to plants and their environment during colonization of tomatoes. FEMS Microbiol. Ecol. 2019, 95, fiz152. [Google Scholar] [CrossRef]
- Žiarovská, J.; Urbanová, L.; Moravčíková, D.; Artimová, R.; Omelka, R.; Medo, J. Varieties of Lettuce Forming Distinct Microbial Communities Inhabiting Roots and Rhizospheres with Various Responses to Osmotic Stress. Horticulturae 2022, 8, 1174. [Google Scholar] [CrossRef]
- Taulé, C.; Vaz-Jauri, P.; Battistoni, F. Insights into the early stages of plant–endophytic bacteria interaction. World J. Microbiol. Biotechnol. 2021, 37, 13. [Google Scholar] [CrossRef]
- Schlechter, R.O.; Miebach, M.; Remus-Emsermann, M.N.P. Driving factors of epiphytic bacterial communities: A review. J. Adv. Res. 2019, 19, 57–65. [Google Scholar] [CrossRef]
- Leonard, S.R.; Simko, I.; Mammel, M.K.; Richter, T.K.S.; Brandl, M.T. Seasonality, shelf life and storage atmosphere are main drivers of the microbiome and E. coli O157:H7 colonization of post-harvest lettuce cultivated in a major production area in California. Environ. Microbiome 2021, 16, 25. [Google Scholar] [CrossRef]
- Szczech, M.; Kowalska, B.; Smolińska, U.; Maciorowski, R.; Oskiera, M.; Michalska, A. Microbial quality of organic and conventional vegetables from Polish farms. Int. J. Food Microbiol. 2018, 286, 155–161. [Google Scholar] [CrossRef]
- Kurtböke, D.I.; Palk, A.; Marker, A.; Neuman, C.; Moss, L.; Streeter, K.; Katouli, M. Isolation and characterization of Enterobacteriaceae species infesting post-harvest strawberries and their biological control using bacteriophages. Appl. Microbiol. Biotechnol. 2016, 100, 8593–8606. [Google Scholar] [CrossRef]
- Jechalke, S.; Schierstaedt, J.; Becker, M.; Flemer, B.; Grosch, R.; Smalla, K.; Schikora, A. Salmonella Establishment in Agricultural Soil and Colonization of Crop Plants Depend on Soil Type and Plant Species. Front. Microbiol. 2019, 10, 967. [Google Scholar] [CrossRef]
- Manyi-Loh, C.E.; Mamphweli, S.N.; Meyer, E.L.; Makaka, G.; Simon, M.; Okoh, A.I. An Overview of the Control of Bacterial Pathogens in Cattle Manure. Int. J. Environ. Res. Public Health 2016, 13, 843. [Google Scholar] [CrossRef]
- Moss, M.O. Fungi, quality and safety issues in fresh fruits and vegetables. J. Appl. Microbiol. 2008, 104, 1239–1243. [Google Scholar] [CrossRef]
- Remus-Emsermann, M.N.P.; Schlechter, R.O. Phyllosphere microbiology: At the interface between microbial individuals and the plant host. New Phytol. 2018, 218, 1327–1333. [Google Scholar] [CrossRef]
- Jacques, M.-A. The Effect of Leaf Age and Position on the Dynamics of Microbial Populations on Aerial Plant Surfaces. In Aerial Plant Surface Microbiology; Morris, C.E., Nicot, P.C., Nguyen-The, C., Eds.; Springer: Boston, MA, USA, 1996; pp. 233–248. [Google Scholar] [CrossRef]
- Aycicek, H.; Oguz, U.; Karci, K. Determination of total aerobic and indicator bacteria on some raw eaten vegetables from wholesalers in Ankara, Turkey. Int. J. Hyg. Environ. Health 2006, 209, 197–201. [Google Scholar] [CrossRef]
- Smolinski, H.S.; Wang, S.; Ren, L.; Chen, Y.; Kowalcyk, B.; Thomas, E.; Doren, J.V.; Ryser, E.T. Transfer and Redistribution of Salmonella Typhimurium LT2 and Escherichia coli O157:H7 during Pilot-Scale Processing of Baby Spinach, Cilantro, and Romaine Lettuce. J. Food Prot. 2018, 81, 953–962. [Google Scholar] [CrossRef]
- Gu, G.; Ottesen, A.; Bolten, S.; Luo, Y.; Rideout, S.; Nou, X. Microbiome convergence following sanitizer treatment and identification of sanitizer resistant species from spinach and lettuce rinse water. Int. J. Food Microbiol. 2020, 318, 108458. [Google Scholar] [CrossRef]
- Chun, H.H.; Kang, J.H.; Song, K.B. Effects of aqueous chlorine dioxide treatment and cold storage on microbial growth and quality of blueberries. J. Korean Soc. Appl. Biol. Chem. 2013, 56, 309–315. [Google Scholar] [CrossRef]
- Brandl, M.T. Plant Lesions Promote the Rapid Multiplication of Escherichia coli O157:H7 on Postharvest Lettuce. Appl. Environ. Microbiol. 2008, 74, 5285–5289. [Google Scholar] [CrossRef] [PubMed]
- Julien-Javaux, F.; Gérard, C.; Campagnoli, M.; Zuber, S. Strategies for the safety management of fresh produce from farm to fork. Curr. Opin. Food Sci. 2019, 27, 145–152. [Google Scholar] [CrossRef]
- Lorenzi, A.S.; Bonatelli, M.L.; Chia, M.A.; Peressim, L.; Quecine, M.C. Opposite Sides of Pantoea agglomerans and Its Associated Commercial Outlook. Microorganisms 2022, 10, 2072. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Erlacher, A.; Smalla, K.; Krause, R. Vegetable microbiomes: Is there a connection among opportunistic infections, human health and our ‘gut feeling’? Microb. Biotechnol. 2014, 7, 487–495. [Google Scholar] [CrossRef]
- Denis, N.; Zhang, H.; Leroux, A.; Trudel, R.; Bietlot, H. Prevalence and trends of bacterial contamination in fresh fruits and vegetables sold at retail in Canada. Food Control 2016, 67, 225–234. [Google Scholar] [CrossRef]

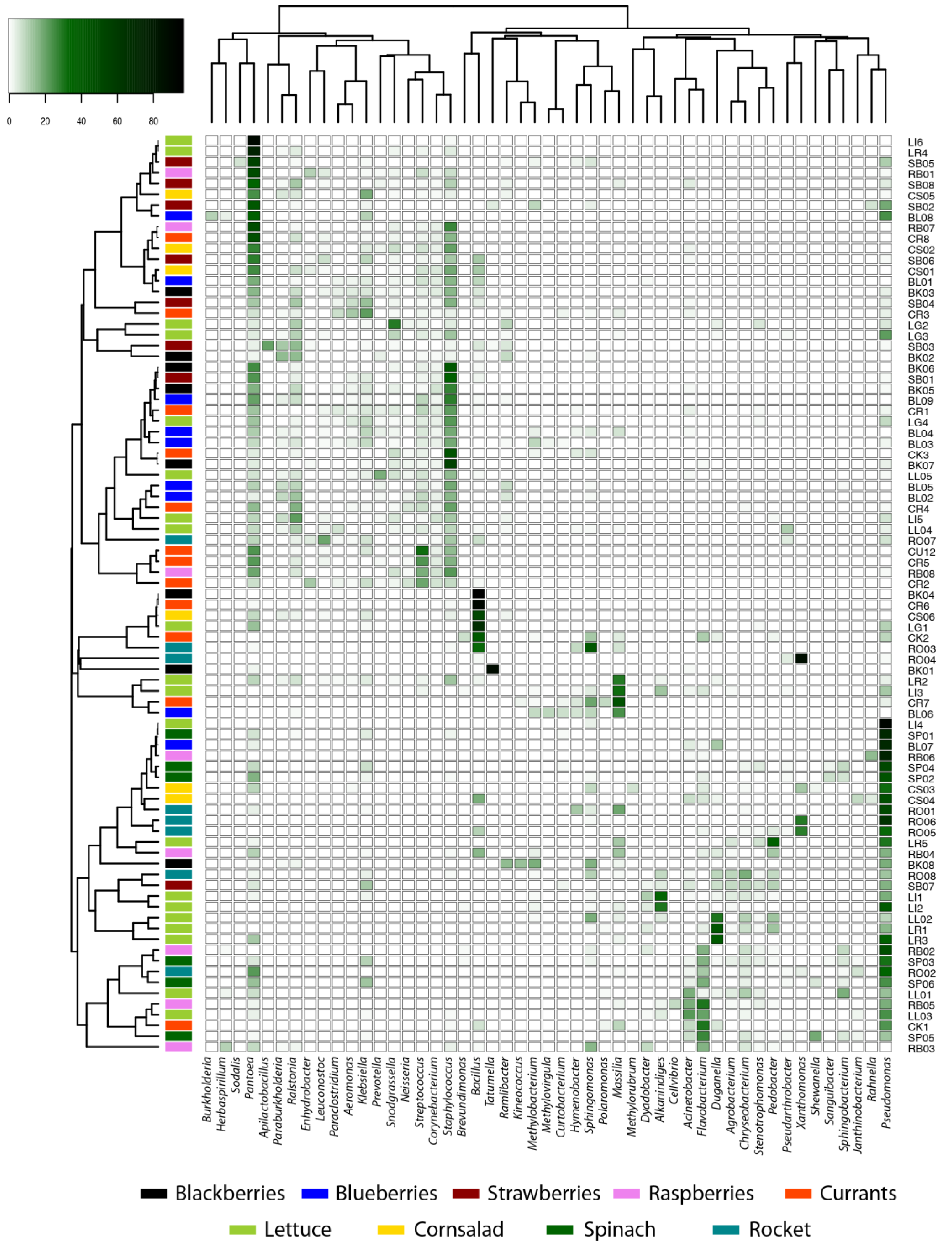
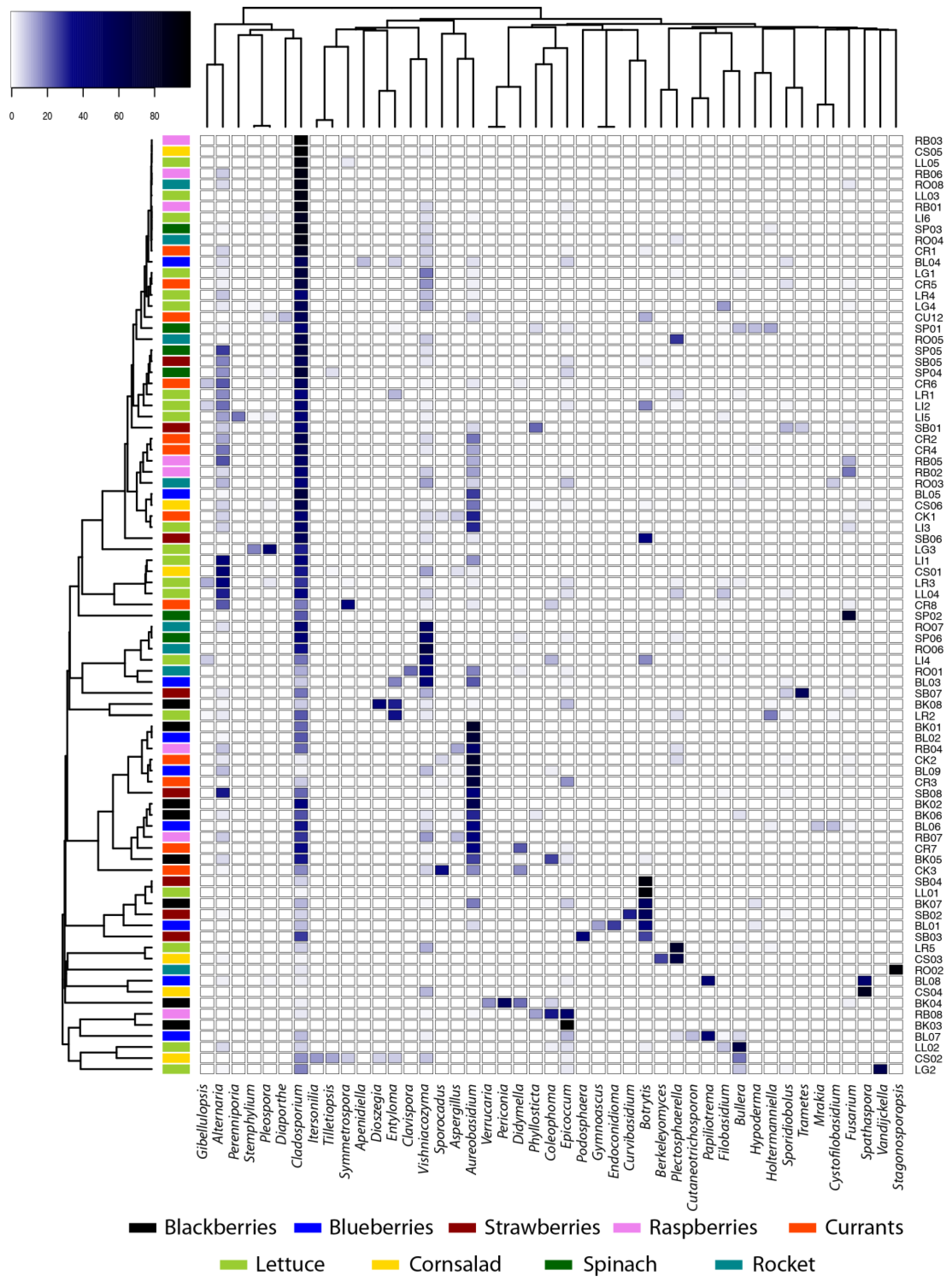
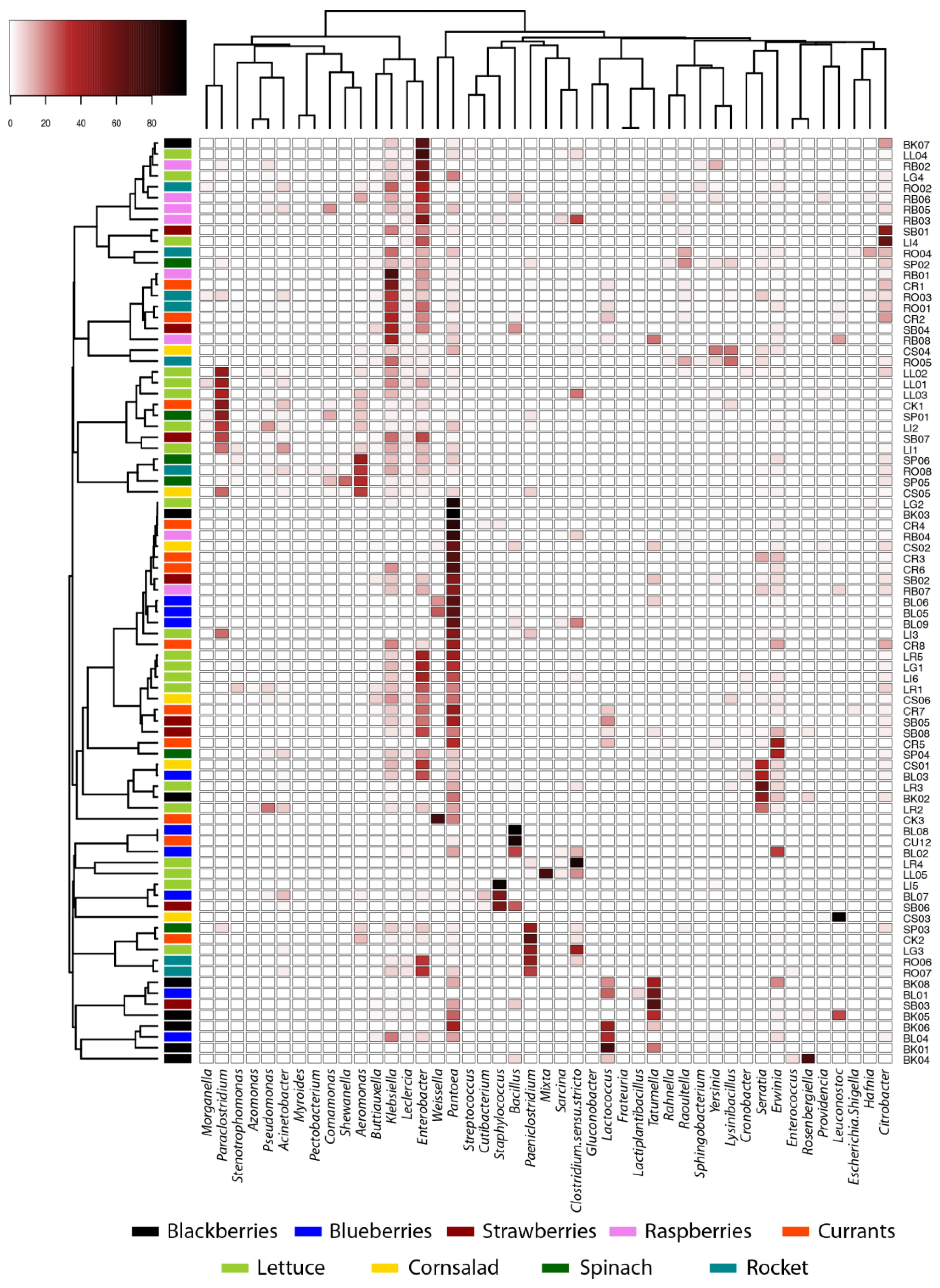


| Species | Blackberry | Blueberry | Raspberry | Strawberry | Currants | Gem | Rockett | Spinach | Valerianella | Total |
|---|---|---|---|---|---|---|---|---|---|---|
| Citrobacter braakii | – | 1 | – | – | – | – | – | – | – | 1 |
| Citrobacter freundii | – | – | 1 | – | – | 7 | – | 3 | 1 | 12 |
| Citrobacter gillenii | – | – | – | – | – | – | 1 | – | – | 1 |
| Enterobacter asburiae | – | – | – | – | 1 | 3 | 2 | – | – | 6 |
| Enterobacter bugandensis | – | – | – | – | – | 5 | 1 | – | – | 6 |
| Enterobacter cloacae | 1 | – | – | 1 | – | 9 | – | – | – | 11 |
| Enterobacter kobei | – | – | – | – | – | 6 | – | 1 | – | 7 |
| Enterobacter ludwigii | – | – | – | 2 | – | 5 | – | – | – | 7 |
| Enterobacter xiangfangensis | – | 2 | – | – | – | 2 | 1 | – | – | 5 |
| Klebsiella aerogenes | 1 | – | 3 | – | – | 2 | – | 1 | 1 | 8 |
| Klebsiella oxytoca | 2 | 1 | 3 | – | 2 | 6 | 2 | – | 4 | 20 |
| Klebsiella variicola | – | – | – | – | – | 1 | – | – | – | 1 |
| Kosakonia kowanii | – | – | 1 | – | 1 | 3 | – | 1 | – | 6 |
| Kluyvera cryocrescens | – | – | – | – | – | 1 | – | – | – | 1 |
| Leclercia adecarboxylata | – | – | – | – | – | 1 | – | – | – | 1 |
| Pantoea agglomerans | 5 | 4 | 2 | 2 | 16 | 12 | 4 | 2 | 1 | 48 |
| Pantoea ananatis | – | 2 | – | – | 2 | 6 | – | – | – | 10 |
| Pantoea anthophila | 2 | – | 2 | 1 | – | 6 | 2 | 4 | 1 | 18 |
| Pantoea vagans | – | – | – | – | 3 | 2 | 5 | |||
| Pseudescherichia vulneris | – | 1 | – | – | – | – | – | – | – | 1 |
| Rahnella aquatilis | – | 1 | – | – | – | – | – | – | – | 1 |
| Raoultella ornithinolytica | – | 3 | – | – | – | 2 | – | – | – | 5 |
| Raoultella terrigena | – | – | – | – | – | 3 | – | 1 | 1 | 5 |
| Serratia ficaria | – | – | – | – | – | 4 | 3 | 1 | 8 | |
| Serratia fonticola | – | – | – | – | – | 1 | – | – | – | 1 |
| Serratia grimesii | 2 | – | – | 1 | – | 4 | – | – | 7 | |
| Serratia liquefaciens | – | – | – | 1 | – | 5 | – | – | – | 6 |
| Serratia marcescens | – | – | – | – | 2 | – | – | – | – | 2 |
| Serratia ureilytica | – | – | – | 1 | – | – | – | 1 | 1 | 3 |
| Total | 13 | 15 | 12 | 11 | 21 | 93 | 20 | 17 | 10 | 213 |
| Microbial Community | Diversity Index | Fruits (n = 45) | Vegetables (n = 40) | p-Value |
| Bacteria in leaf tissue | Richness | 258.8 (66.2) * | 221.3 (67.8) | 0.0119 |
| Shannon’s Index | 4.52 (1.21) | 4.27 (1.31) | 0.3647 | |
| Corrected evenness | 0.517 (0.139) | 0.503 (0.15) | 0.6623 | |
| Fungi in leaf tissue | Richness | 21.0 (8.0) | 22.0 (8.9) | 0.5962 |
| Shannon’s Index | 2.30 (0.83) | 2.24 (1.00) | 0.7802 | |
| Corrected evenness | 0.492 (0.162) | 0.470 (0.192) | 0.5736 | |
| Bacteria in pre-enrichment | Richness | 163.4 (49.1) | 306.9 (93.3) | 0.0001 |
| Shannon’s Index | 3.47 (1.21) | 4.59 (0.87) | 0.0001 | |
| Corrected evenness | 0.421 (0.144) | 0.494 (0.089) | 0.0075 |
| Microbial Community | Diversity Indice | Inner Leaves | Outer Leaves | Difference +/− 95% Confidence | p-Value |
|---|---|---|---|---|---|
| Bacteria in leaf tissue | Richness | 187 | 239 | 52.4 +/− 52.08 | 0.0486 |
| Shannon’s Index | 3.74 | 4.77 | 1.03 +/− 0.96 | 0.0374 | |
| Corrected evenness | 0.448 | 0.550 | 0.102 +/− 0.108 | 0.0635 | |
| Fungi in leaf tissue | Richness | 18.5 | 27.5 | 9.09 +/− 6.80 | 0.0139 |
| Shannon’s Index | 2.31 | 2.80 | 0.49 +/− 0.61 | 0.1032 | |
| Corrected evenness | 0.477 | 0.564 | 0.086 +/− 0.115 | 0.1245 | |
| Bacteria in pre-enrichment | Richness | 193 | 283 | 90.0 +/− 80.6 | 0.0321 |
| Shannon’s Index | 3.27 | 4.18 | 0.90 +/− 0.92 | 0.0560 | |
| Corrected evenness | 0.381 | 0.443 | 0.062 +/− 0.112 | 0.2452 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Artimová, R.; Játiová, M.; Baumgartnerová, J.; Lipková, N.; Petrová, J.; Maková, J.; Javoreková, S.; Hleba, L.; Medová, J.; Medo, J. Microbial Communities on Samples of Commercially Available Fresh-Consumed Leafy Vegetables and Small Berries. Horticulturae 2023, 9, 150. https://doi.org/10.3390/horticulturae9020150
Artimová R, Játiová M, Baumgartnerová J, Lipková N, Petrová J, Maková J, Javoreková S, Hleba L, Medová J, Medo J. Microbial Communities on Samples of Commercially Available Fresh-Consumed Leafy Vegetables and Small Berries. Horticulturae. 2023; 9(2):150. https://doi.org/10.3390/horticulturae9020150
Chicago/Turabian StyleArtimová, Renata, Michaela Játiová, Juliána Baumgartnerová, Nikola Lipková, Jana Petrová, Jana Maková, Soňa Javoreková, Lukáš Hleba, Janka Medová, and Juraj Medo. 2023. "Microbial Communities on Samples of Commercially Available Fresh-Consumed Leafy Vegetables and Small Berries" Horticulturae 9, no. 2: 150. https://doi.org/10.3390/horticulturae9020150
APA StyleArtimová, R., Játiová, M., Baumgartnerová, J., Lipková, N., Petrová, J., Maková, J., Javoreková, S., Hleba, L., Medová, J., & Medo, J. (2023). Microbial Communities on Samples of Commercially Available Fresh-Consumed Leafy Vegetables and Small Berries. Horticulturae, 9(2), 150. https://doi.org/10.3390/horticulturae9020150








