1. Introduction
Kiwifruit holds a prominent position in the global fruit industry, owing to its distinctive flavor, exceptional nutritional value (high vitamin C content, rich in dietary fiber), and associated health advantages (positive action on cardiovascular diseases, lipid metabolism, inflammatory response, blood pressure problems, overweight, etc., while it is also preferred by vegans) [
1]. Kiwifruits are available in a diverse range of cultivars, distinguished by their green, gold, or red flesh as well as their size. Despite the existence of over 60 species within the
Actinidia genus, only a few have achieved notable economic significance, with the green-fleshed “Hayward” cultivar enjoying widespread cultivation worldwide [
2].
The cultivation and production of kiwifruits continue to expand. with leading global producers being China, Italy, New Zealand, and Greece. As many countries increase their kiwifruit production, the kiwifruit industry is emphasizing not only product quality (in terms of dry matter, total soluble solids, and firmness at harvest) but also the development of a market-oriented approach to maintain competitiveness [
3]. Furthermore, recognizing that the survival of the industry hinges on innovating new products, significant breeding programs have been launched to formalize the development of new cultivars with desirable characteristics (size, storability, increased firmness during storage and shipment, enhanced disease resistance, etc.), as well as less vulnerable to climate change (fewer chilling units requirements, etc.) [
3]. There is optimism that these endeavors will produce commercially accessible fruits with a variety of sizes, peel characteristics, flesh colors, flavors, nutraceutical properties, and textures to complement the “Hayward” cultivar. Considering the wide geographical distribution and diversity of wild kiwifruit populations, Ferguson [
4] anticipates that there will likely be numerous appealing traits that can be integrated into breeding programs.
To evaluate as well as to distinguish different kiwifruit cultivars, both traditional and modern approaches have been applied. Traditional approaches encompass the evaluation of morphological and biochemical traits, including fruit shape, size, color, chemical composition, and flowering time [
5]. For instance, Mavromatis [
6] distinguished two kiwifruit cultivars based on morphological characteristics such as fruit weight, length, and diameter. Lee [
7] and Latocha [
8] used biochemical markers such as total phenolic content and antioxidant activity. Nevertheless, it is important to note that these traditional methods have inherent limitations in terms of accuracy and influence exerted by environmental factors.
The use of molecular markers has gained momentum as a reliable technique to distinguish kiwifruit cultivars based on their genetic profiles [
6]. This approach has grown in popularity due to its high accuracy, reproducibility, and independence from environmental and agronomic factors. Various molecular markers, including simple sequence repeats (SSR), random amplified polymorphic DNA (RAPD), and inter-simple sequence repeats (ISSR), have been widely employed for this purpose [
9,
10,
11,
12,
13]. Among these options, ISSRs have garnered particular attention due to their high polymorphism levels, reproducibility, and cost-effectiveness in identifying and classifying kiwifruit cultivars [
12,
13]. Furthermore, inter-simple sequence repeat (ISSR) markers have been successfully used to ascertain the genetic fidelity among mother plants and their in vitro multiplied kiwifruit regenerates [
13].
“Arta Kiwifruit” is a clonal selection originating from the renowned “Hayward” cultivar, found as a random plant within a “Hayward” orchard by the farmer Mrs. Xylogianni Evanthia, in Arta City, Greece. This unique genotype was distinguished and was specifically selected for its exceptional ability to produce large-sized fruits, making it a valuable candidate for commercialization and promotion. Therefore, the primary objective of this study was to evaluate some physical and biochemical traits of the new kiwifruit genotype both at harvest and after storage. Additionally, the genetic difference between the widely cultivated “Hayward” cultivar and the new Greek kiwifruit selection “Arta Kiwifruit”, was also examined using molecular markers, to ensure the uniqueness of this new genetic material. Part of the evaluation process was also to compare the bioactive compounds of this genotype with those of the well-known and widely consumed “Hayward” cultivar.
3. Results and Discussion
The biochemical composition of kiwifruits at harvest reflects the accumulation of primary metabolites, such as sugars and organic acids, as well as secondary metabolites, including phenolic compounds and other antioxidants. These compounds play a significant role in determining the taste, texture, nutritional value, and potential health benefits of the fruit. During storage, various biochemical changes occur, which can affect the overall quality and consumer acceptance of the fruit. Thus, monitoring and comparing the changes in these compounds at harvest and during storage is crucial for assessing the biochemical dynamics of the new kiwifruit genotype.
Productivity also plays an important role in the selection of a new genotype, since this is the first desirable trait the farmer is looking for. The mean production per vine during the first year differed between the two genotypes with “Hayward” presenting a significantly higher yield than the new genotype (
Table 3). During the second year, there were not any significant differences between the genotypes, with the yield being approximately 22.5 Kg per vine for both genotypes.
The fruit physical parameters of the “Hayward” and “Arta Kiwifruit” genotypes are summarized in
Table 3. Notably, the mean fruit weight of “Arta Kiwifruit” was significantly higher compared to “Hayward” during both years of experimentation both at harvest and after the storage period. Additionally, fruits of “Arta Kiwifruit” exhibited larger lengths compared to “Hayward” during both years. Sotiropoulos et al. [
20] also described a genotype of higher fruit weight than “Hayward”, which due to this characteristic was considered commercially interesting. Thus, what truly distinguishes “Arta Kiwifruit” is its large fruit size, providing substantial marketable advantages in terms of fruit appearance. These desirable characteristics played a pivotal role in the selection of “Arta Kiwifruit”. It is worth noting that the final fruit size of “Hayward” can be increased through various horticultural practices, including the application of plant growth regulators such as thidiazuron, forchlorfenuron, gibberellic acid, or dichlorophenoxyacetic acid [
21]. “Arta Kiwifruit” produces large fruits without the need for chemical treatments, which is a significant advantage, as both the production costs remain low, and the overall kiwifruit cultivation becomes more sustainable (less use of pesticides and plant growth regulators), while at the same time extra fruit quality is achieved (FAO Codex Alimentarius, Standard for Kiwifruit, CXS 338-2020, adopted in 2020,
https://www.fao.org/fao-who-codexalimentarius/committees/committee/related-standards/jp/ accessed on 16 November 2023).
In addition to fruit size, factors such as firmness, soluble sugars (TSS), organic acids, and dry matter accumulation play crucial roles in determining fruit quality and the ability to be stored effectively [
22]. In the present study, the fruit dry matter did not differ between genotypes and was beyond the lower allowable limit for harvest (15%) (FAO Codex Alimentarius, Standard for Kiwifruit, CXS 338-2020, adopted in 2020,
https://www.fao.org/fao-who-codexalimentarius/committees/committee/related-standards/jp/ accessed on 16 November 2023) (
Table 3). Similarly, the fruit firmness at harvest ranged from 21.3 N to 38.9 N, with “Arta Kiwifruit” exhibiting higher firmness during the first year. After storage, no significant difference was observed between genotypes. Generally, the commercially approved fruit firmness can vary depending on the specific cultivar and market preferences. Previous studies have indicated that fruit ripeness, as assessed through firmness measurements, can significantly influence consumer preferences [
23]. Following the findings of Stec et al. [
23], Jaeger et al. [
3] concluded that commercial Actinidia fruits with firmness ranging from 0.6 to 0.8 Kg (approximately 5.9 N to 7.8 N) were significantly more acceptable to consumers. Furthermore, the life storage of kiwifruit is affected by the extent to which the flesh softens [
24] and this was obvious in the present study, as both genotypes presented a significant reduction in fruit firmness during both years. According to Krupa [
25], kiwifruits are considered ripe and ready to be consumed when the firmness of the flesh measures below 9.81 N.
No significant difference was observed between genotypes in terms of pH, TA, and TSS: TA ratio, as shown in
Table 4. Both “Hayward” and “Arta Kiwifruit” fruits were harvested at the commercial maturity stage, which is determined by reaching the minimum marketing value of 6.2 °Brix TSS content [
26]. The TSS content ranged from 6.8 to 7.9 °Brix, with “Hayward” fruits having a statistically significantly higher amount during the first year. After storage, TSS content was doubled, ranging on average from 12.6 to 13.8 °Brix, which is generally considered acceptable by consumers [
27,
28], with “Hayward” fruits of the second year presenting higher TSS than those of “Arta Kiwifruit”. Furthermore, both “Hayward” and “Arta Kiwifruit” are green-fleshed kiwifruits, sharing a similar hue angle and they do not exhibit significant differences in terms of flesh color parameters (
Table 4). After the storage period though, significant reductions of all color parameters were determined, in both genotypes, indicative of either chlorophyll breakdown and/or just simply pigment concentration due to water loss, as was evident by the slight fruit weight loss.
According to Nishiyama et al. [
29], the
Actinidia genus exhibits significant variation in fruit total chlorophyll content, ranging from trace amounts to 4.4 ± 0.8 mg 100 g
−1 FW in a hybrid variety of
A. arguta ×
deliciosa. In this study, the total chlorophyll content ranged from 14.7 μg g
−1 FW in “Hayward” to 11.7 μg g
−1 FW in “Arta Kiwifruit” (
Table 5). Similar to previous research by Drummond [
30], kiwifruits contain both chlorophylls a and b, with varying levels and ratios between different cultivars. In the current study, the “Hayward” fruits had a significantly higher total Chla content at harvest (9.1 and 8.5 μg g
−1 FW during the first and second year, respectively), compared to “Arta Kiwifruit” (7.3 and 7.5 μg g
−1 FW, respectively). No significant differences were observed in Chlb content for both genotypes, as well as in the total Chls content at harvest. These findings align with previous studies conducted by Nishiyama et al. [
29], Drummond [
30], and Pilkington et al. [
31].
Generally, immature fruits commonly contain chlorophylls, but their levels decline rapidly as the fruit matures and ripens. Consequently, very few fruits retain their green color when ripe [
30,
31]. However, mature green kiwifruits of the
Actinidia deliciosa species are an exception, as they are characterized by high concentrations of chlorophyll in the fruit flesh. In this study, the total chlorophyll content, as well as Chla and Chlb levels, decreased during the maturation process in storage conditions in “Hayward” fruits, in accordance with Robertson [
32]. Based on the present trial the chlorophyll content of the fruit after the storage period was approximately 10 μg g
−1 fruit flesh, which is consistent with the findings of Pilkington et al. [
31], indicating that both green-fleshed kiwifruit genotypes have one of the highest concentrations of chlorophyll, second only to avocados [
33].
The ratio of Chla to Chlb fluctuated between 1.5 and 1.7 at harvest, and between 1.5 and 1.8 after storage (
Table 5). “Hayward” exhibited a significantly higher content of total carotenoids compared to “Arta Kiwifruit” at harvest, while the concentration of carotenoids was similar in both genotypes after the storage period (
Table 5).
The level of soluble sugars found in kiwifruit is an important indicator of ripeness. As the fruit ripens, the concentration of sugars increases significantly, while the starch content decreases. This transformation is accompanied by changes in fruit texture as well. Once the fruit reaches a state of ripeness suitable for consumption, the sugars contribute to the sweet taste. However, this sweetness is balanced by the acid composition of the fruit, ensuring a harmonious flavor profile [
34,
35].
The main sugars found in
Actinidia fruits are glucose, fructose, and sucrose [
30].
Table 6 displays the sugar composition within the “Hayward” and “Arta Kiwifruit” genotypes, revealing nearly equal levels of glucose at harvest, while “Hayward” presented higher levels of fructose (both years) and total sugars (during the second year only), which increased significantly (in both genotypes) after the storage period. Similar findings have been reported by Perez et al. [
35] and Sanz et al. [
36]. Notably, “Hayward” fruits exhibited significantly higher concentrations of fructose, sucrose, glucose, and total sugars as well as sweetness index after the storage period, during both years of the study. No significant differences were observed for starch though. Nishiyama et al. [
34] noted that the levels of total sugars and their ratios vary not only with maturity but also across different kiwifruit cultivars, justifying the present results.
Both genotypes exhibited a near doubling of fructose and glucose concentrations after storage, while sucrose and inositol concentrations remained at similar levels to that found at harvest (
Table 6). The concentration of total sugars in “Hayward” was near 52–56 mg g
−1 FW at harvest and increased to 126–139 mg g
−1 FW after storage. Similarly, in “Arta Kiwifruit”, the total sugar concentration was between 42–44 mg g
−1 FW at harvest during the two-year experimentation period and increased up to 94–98 mg g
−1 FW after storage. These findings represent an almost 147% increase in total sugars for “Hayward” kiwifruits after storage and a nearly 120% increase for “Arta Kiwifruit”. The present results are within the broad range reported by Drummond [
30] (11.7 g 100 g
−1 FW) and those reported by Nishiyama et al. [
34], ranging from 6.15 to 10.47 g 100 g
−1 FW. Other studies have also reported total sugar values ranging from 7.7 g 100 g
−1 FW [
35] to 10.8 g 100 g
−1 FW [
36,
37]. On a per-fruit consumption basis, one fruit of “Hayward” offers 13.31 g of sugars while one fruit of “Arta Kiwifruit” offers 11.86 g of sugars (mean of the two-year study). This is of utmost importance for a specific category of people who seek fruits of low sugar content. On the other hand, though, calculating the sweetness index (SI) of the fruit of the two genotypes based on the method of Denaxa et al. [
14], “Hayward” is sweeter (SI = 20.7–23.3) than “Arta Kiwifruit” (SI = 15.6–16.2) after the storage period, which may be an advantage for people who have a preference for sweet fruits.
Kiwifruit also contains various organic acids that contribute to the fruit’s overall sensory attributes, particularly in terms of the balance between sweetness and acidity. These organic acids play essential roles in both health and metabolism. Although ascorbic acid is the primary organic acid with notable health benefits in kiwifruit, other acids may be present in high quantities too. The levels and proportions of organic acids in kiwifruit depend on the fruit’s maturity, while their distribution within the fruit is not uniform [
30].
As shown in
Table 6, there were significant differences observed between the two genotypes in terms of organic acid concentrations, both at harvest and after storage. “Hayward” fruits exhibited significantly higher levels of ascorbic acid, ranging from 6.1 to 6.9 mg g
−1 FW (at harvest, during the two-year study) and from 8.8–10.4 mg g
−1 FW (after the storage period, during the two-year study). Park et al. [
38] also noted a significant difference in ascorbic acid content among various kiwifruit cultivars. It is worth noting that the measured values of ascorbic acid in our study exceeded those reported in previous ones [
24,
30,
34,
39]. According to Esti et al. [
40], factors such as genotype, ripening stage, storage conditions, and the analytical technique employed, can influence the ascorbic acid content of kiwifruit. These same authors also demonstrated that ascorbic acid content in kiwifruit samples from
Actinidia chinensis (Planch) var. chinensis genotypes exceeded the average content found in
Actinidia deliciosa (A. Chev) “Hayward”.
Furthermore, since reductions in ascorbic acid may indicate a loss of nutritional value [
41], fluctuations in ascorbic acid content during storage and ripening are critical factors influencing consumer preferences. Tavarini et al. [
42] suggested that the timing of harvest significantly impacts the ascorbic acid content of the “Hayward” cultivar, with more mature fruits exhibiting lower levels. However, when these fruits were stored for 6 months and subsequently ripened, the ascorbic acid content either remained stable or increased compared to that determined at harvest. A similar pattern was observed in the current study, where both genotypes showed an approximately 46% increase in ascorbic acid concentration after storage. This finding is consistent with a study on gold kiwifruit conducted in New Zealand, which reported a substantial 17% increase in ascorbic acid content after 20 weeks of storage, with no significant changes in moisture content [
30]. The per fruit consumption quantity of ascorbic acid (based on mean fruit weight determined after storage, as presented in
Table 3) indicates that a single “Arta Kiwifruit” fruit offers approximately 1.03 g of ascorbic acid versus 0.96 g of a single “Hayward” fruit (based on the mean value of fruit weight and of ascorbic acid concentration determined during the two-year study).
Several studies have demonstrated that kiwifruit cultivars grown under identical geographic and climatic conditions can exhibit significant variations in their content of bioactive compounds [
43,
44].
Table 7 presents the results of the total phenolic compounds and antioxidant activity analysis in the studied kiwifruit genotypes. It can be observed that there were few significant differences in the measured total phenols or other phenolic compounds between the two genotypes at harvest, while their levels can be considered high [
42,
45]. “Hayward” fruits exhibited higher total phenol (first year) and total flavonoid (second year) content at harvest, while the opposite stood for total flavanols (second year). After storage though, “Hayward” fruits presented higher total flavonoid content (first year) while “Arta Kiwifruit” fruits had higher o-diphenol and total flavanol (second year) concentrations. “Hayward” fruits exhibited the highest level of antioxidant activity at harvest (first year), and higher antioxidant activity (ABTS assay during the second year). After storage, they exhibited the highest antioxidant activity (during the first year), but this was lower (based on the DPPH assay) during the second year. In general, fruits of both genotypes experienced a reduction of their antioxidant capacity after storage, during both years according to the literature [
27,
28,
42]. A correlation analysis between antioxidant capacity, measured by all three assays (both at harvest and after storage), and various possible bioactive kiwifruit metabolites (total phenols,
o-diphenols, flavanols, flavonoids, ascorbic acid, and carotenoids) revealed significant, positive relationships (correlation coefficient ranged between 0.32 and 0.54) in accordance with [
46,
47]. This indicates that any factor that could negatively influence the content of these bioactive compounds could deteriorate the nutritional value of the fruit.
Kiwifruit is not only rich in ascorbic acid, carbohydrates, and bioactive compounds but in proteins too [
48]. Actinidin, a cysteine protease, makes up approximately 50% of the soluble protein content in kiwifruit [
49]. The total protein concentration in kiwifruit though, is influenced by the cultivar, fruit developmental stage, and environmental conditions [
48,
50]. In the present study, the total protein content was found to be similar in both genotypes at harvest and after storage, as shown in
Table 7. After storage though, it seemed that the protein content decreased in “Hayward” (first year) and “Arta Kiwifruit” fruits (second year), indicative of possible protease activity [
48].
Kiwifruit contains significant amounts of free amino acids, which undergo significant changes as the fruit ripens [
51]. In this study, we observed a decrease in some amino acids during storage, as fruit continued to mature (
Table 8). Similarly, studies conducted by Drummond [
30] and MacRae and Redgwell [
51] indicated a reduction in most of the free amino acids during maturation, compared to immature fruits. Nonetheless, there were also some amino acids that increased after storage, such as glycine, γ-aminobutyric acid, and tryptophan (in both genotypes, during both study years). Furthermore, the major components identified in both genotypes in the present study were aspartic acid, glutamic acid, and glutamine. Drummond [
30], however, found that arginine and γ-amino butyric acid (GABA) were the predominant amino acids, accounting for 36% of the total free amino acids, whereas MacRae and Redgwell [
51] reported asparagine, glutamine, and arginine/GABA as the major amino acids in the whole fruit. Another study also reported the presence of tryptophan and indole amines [
52], as detected in the present study.
As shown in
Table 8, alanine, arginine, glutamic acid, phenylalanine, threonine, and tryptophan were significantly higher in “Arta Kiwifruit” compared to “Hayward” at harvest of the first year, while valine concentration was higher in “Hayward” fruits during both study years. After storage, “Arta Kiwifruit” exhibited significantly higher concentrations of alanine, arginine, asparagine, γ-aminobutyric acid, glutamic acid, glutamine, glycine, leucine, phenylalanine, and tyrosine compared to “Hayward” fruits during the first year and of asparagine, γ-aminobutyric acid, glutamic acid, and glutamine, during the second year.
PCA was used to examine the differences between the genotypes based on fruit quality parameters measured at harvest and after the storage period, separately per study year.
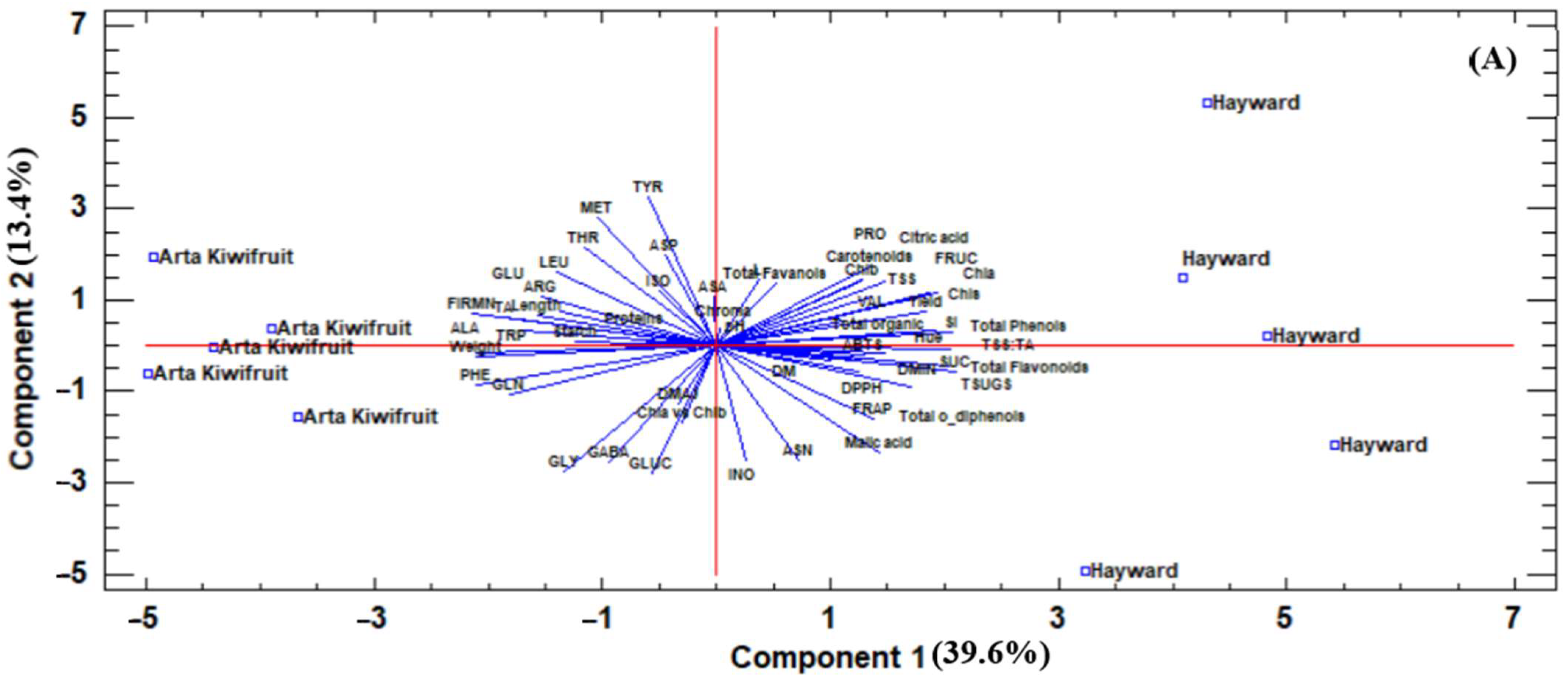
As can be seen both during harvest and after the storage period of the first year (
Figure 1A,B), the two genotypes separated as “Hayward” were located at the right side of the principal component (PC) 1, while “Arta Kiwifruit” was located at the left side of PC1 at harvest, while the exact opposite stood after storage. At harvest, “Hayward” was characterized by a high concentration of phenolic compounds, carotenoids, sugars, and antioxidant capacity (based on DPPH and FRAP assays), while “Arta Kiwifruit” was characterized by high fruit weight, firmness, length, and starch, among others.
After storage (
Figure 1B) “Hayward” was characterized by high sugar concentration, antioxidant activity (by FRAP and ABTS assays), and TSS, among others, while “Arta Kiwifruit” was characterized by high fruit weight, some amino acids, and pH.
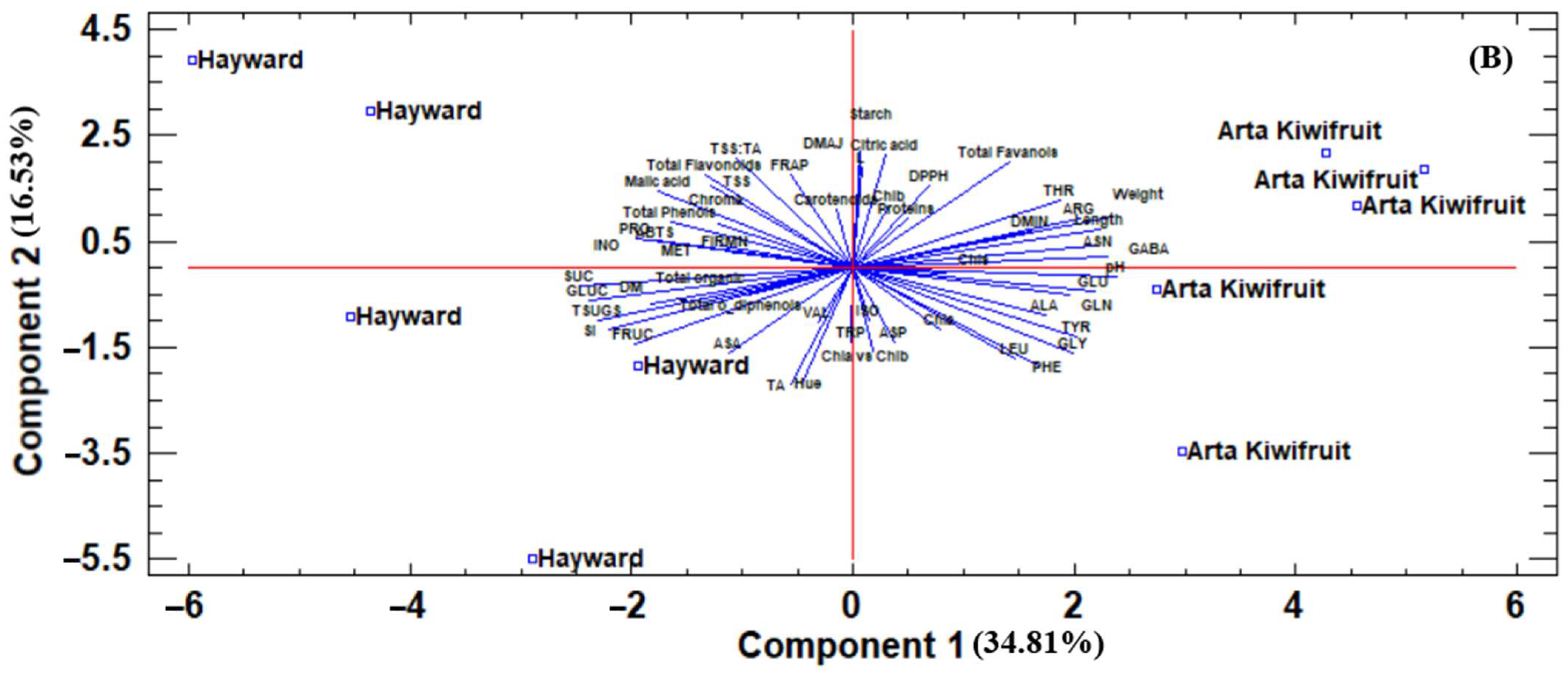
Similarly, as before, the two genotypes were clearly separated during the second year, both at harvest (
Figure 2A) and after the storage period (
Figure 2B). At harvest, “Hayward” was located at the right side of PC1 exhibiting high sugar concentration, high TSS:TA ratio, and high antioxidant capacity (based on FRAP and ABTS assays). “Arta Kiwifruit”, on the other hand, was located at the left side of PC1 presenting high fruit weight and dimensions, high antioxidant capacity (based on DPPH assay), and a high concentration of some amino acids such as threonine, tryptophan, glutamic acid, etc.
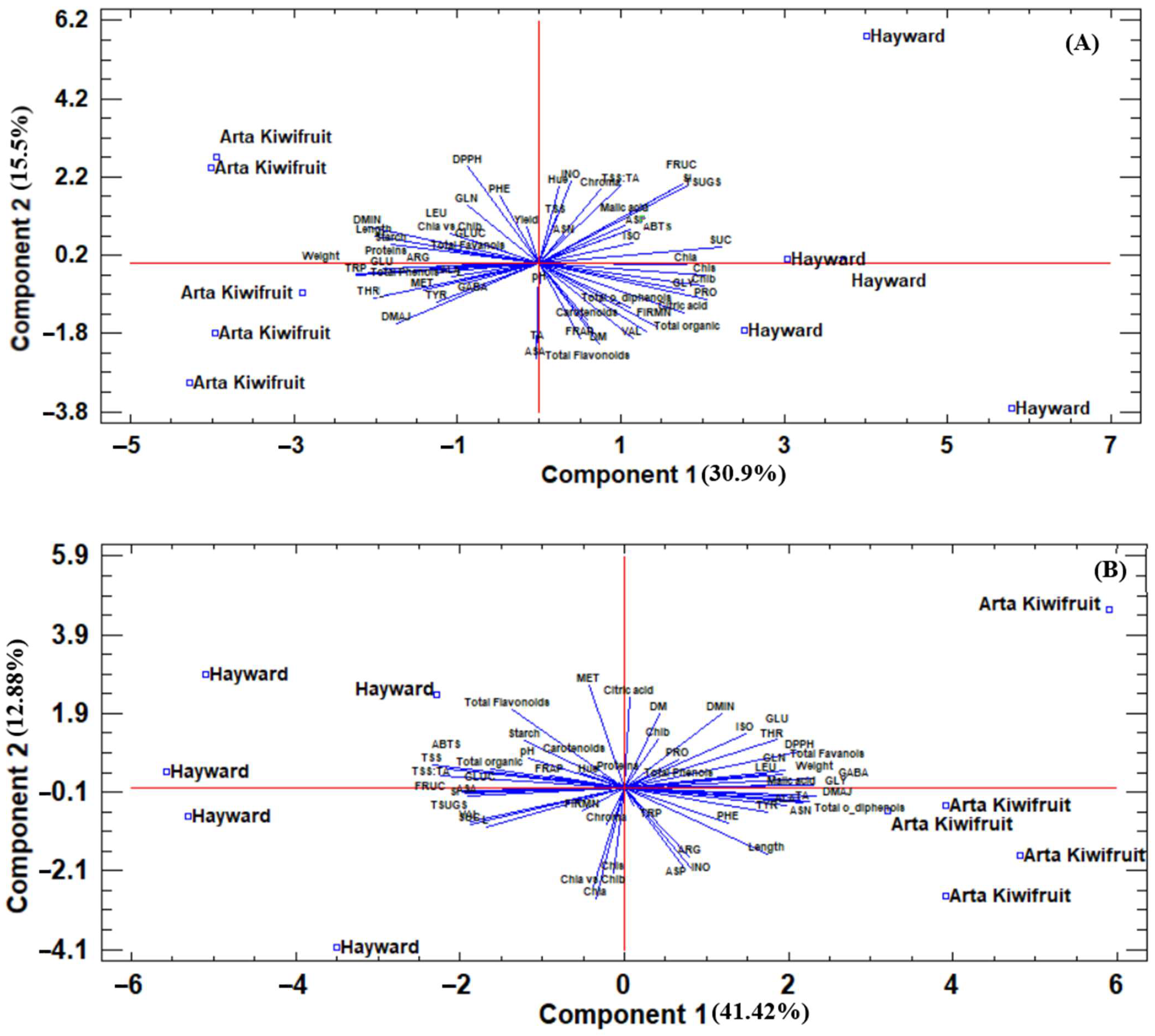
After the storage period, this clear separation still existed, as “Hayward” was located on the left side of PC1 and on the right the “Arta Kiwifruit” (
Figure 2B). “Hayward” was characterized by high TSS and TSS:TA ratio, total sugars, and antioxidant capacity (by FRAP and ABTS assays), while “Arta Kiwifruit” was characterized by high fruit weight, total flavanol, and
o-diphenol concentrations, and antioxidant capacity (by DPPH assay), among others.
The phenotypic description of the two genotypes revealed differences concerning shoot, leaf, and fruit morphological indexes, as shown in
Table 9. “Hayward” exhibited strong coloration of the growing tip of the young shoots as well as strong coloration of the upper side of the petiole. “Arta Kiwifruit”, on the other hand, presented higher fruit weight, while its fruit was characterized by an oblong shape and rounded stalk end. On the contrary, “Hayward” fruit was lighter and elliptic with a flat stylar end, but sweeter than that of “Arta Kiwifruit”.
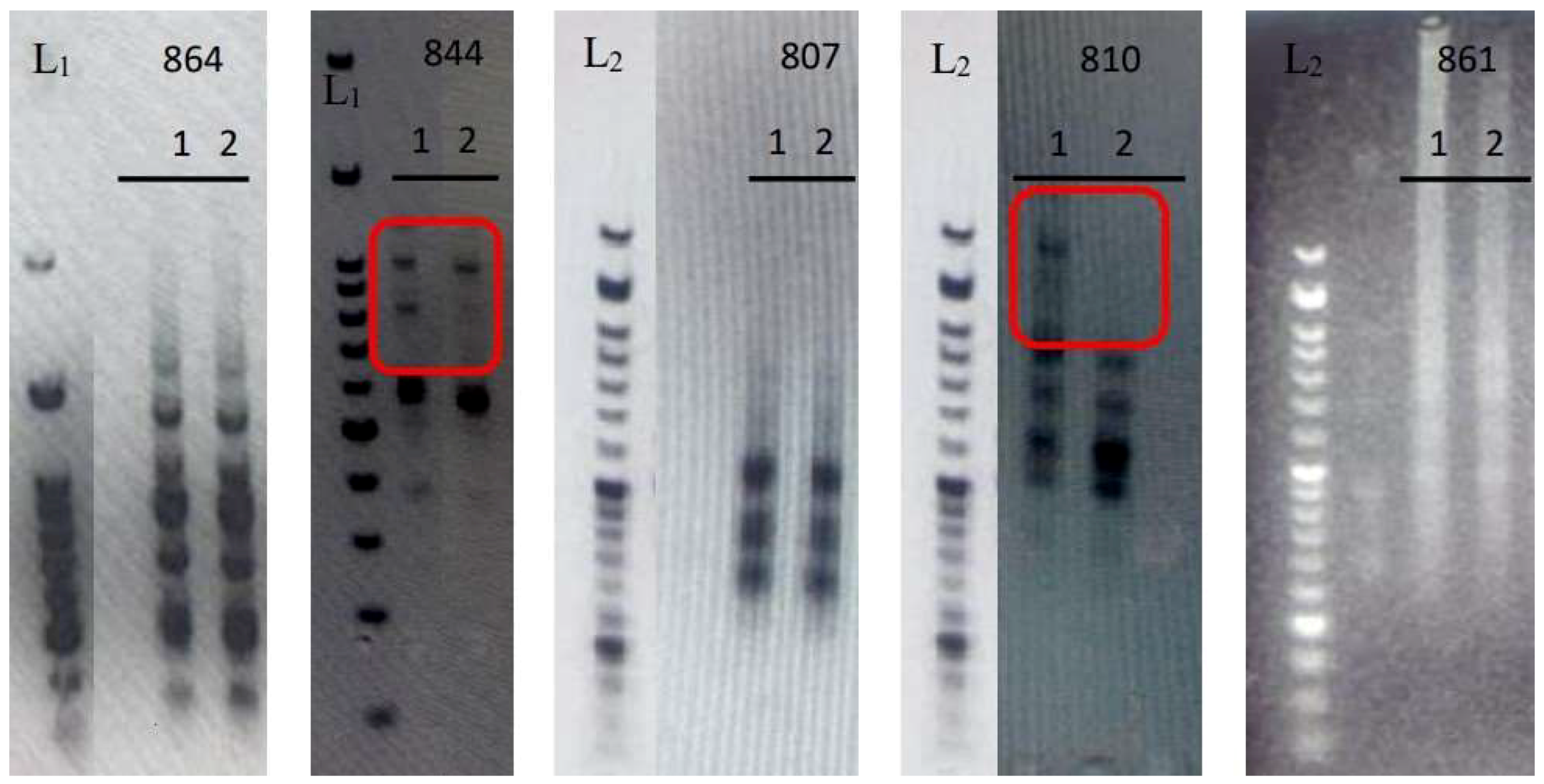
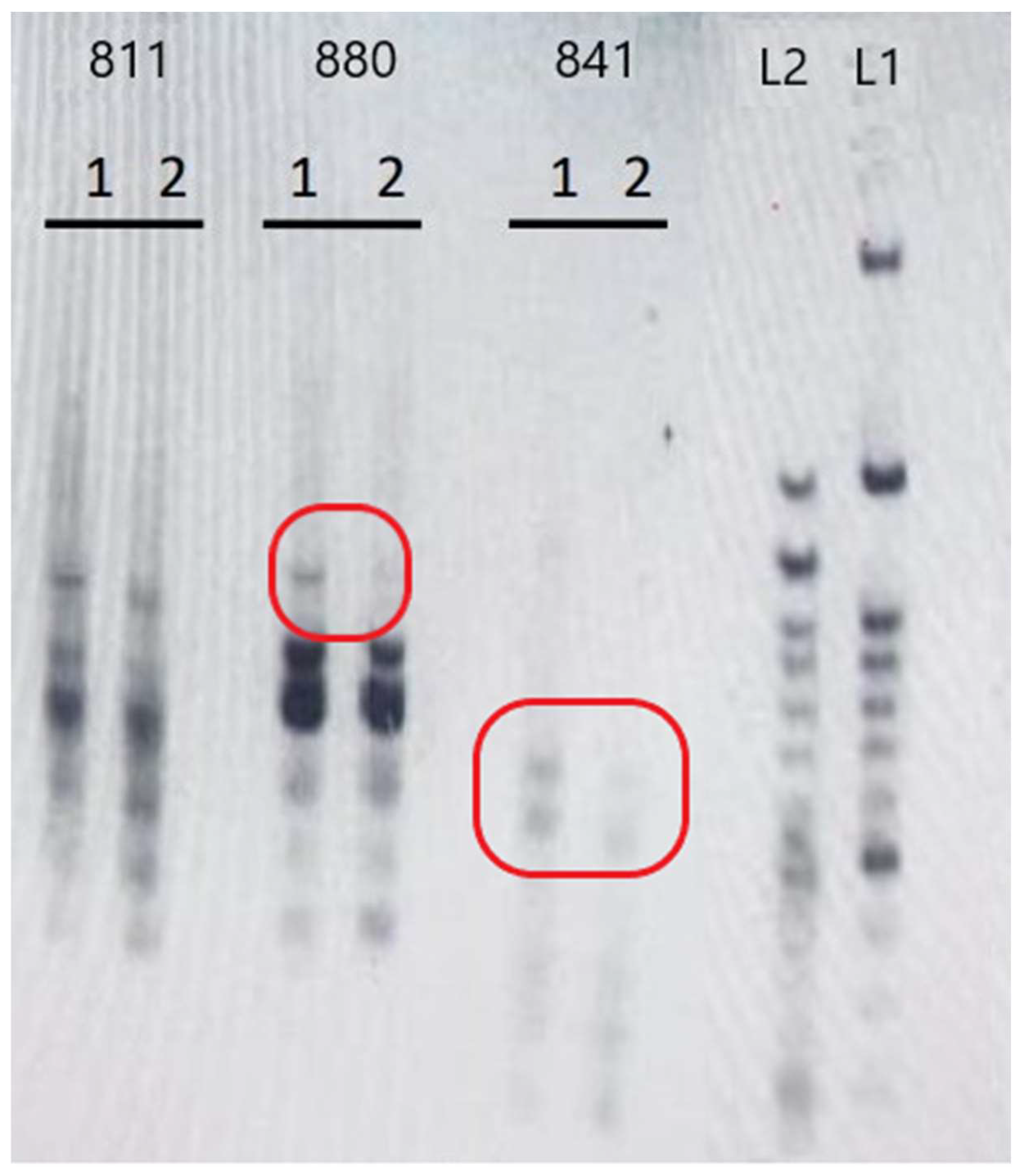
The extracted DNA exhibited sufficient quality and quantity for genetic analysis. The electrophoresis results of the PCR products are presented in
Figure 1. The framed figures and arrows highlight the observed genetic differences between the two genotypes. The two genotypes, i.e., the “Hayward” cultivar and the Greek genotype “Arta Kiwifruit”, were sufficiently distinguished by UBC-844 and UBC-810 ISSR markers. Different amplification bands were observed between the two genotypes, as indicated by the electrophoresis results (
Figure 3 and
Figure 4). ISSRs have proven effective in distinguishing kiwifruit genotypes [
53] and assessing the genetic stability of micropropagated Actinidia plants [
13], thereby solidifying their pivotal role as a valuable molecular tool in kiwifruit breeding programs.