Abstract
Cannabis sativa is gaining attention as an agronomically important crop in many countries around the world. The identification and control of leaf diseases in cannabis are very important for cannabis cultivators as leaves are the most economically important part of the cannabis plants. In 2022, several cannabis plants in cultivations showing olive leaf spot symptoms emerged from Chiang Rai province, Thailand. Preliminary studies indicated that the causal organism is Pseudocercospora sp. Species of Pseudocercospora are important plant pathogens that are now identified through morphological studies combined with DNA sequence data of Internal Transcribed Spacer (ITS), Actin (act), Translation Elongation Factor (tef), and RNA Polymerase II second largest subunit (rpb2) gene regions. We aimed to investigate and understand the emergence of olive leaf spot disease in cannabis plants in Chiang Rai province, Thailand, with a specific focus on the combined morpho-molecular identification of the pathogen. In our study, Pseudocercospora cannabina, the causal organism of olive leaf spot disease, was identified as the leaf spot-causing pathogen with both morphological and phylogenetic analyses. Our study is the first to provide molecular data for Ps. cannabina as the typenor Ps. cannabina isolates from previous studies have made molecular data available for this species. A pathogenicity test, re-isolation, and identification steps were performed to fulfill Koch’s postulates. This comprehensive approach enhances our understanding of the olive leaf spot disease and its causative agent in cannabis.
1. Introduction
Cannabis sativa L., also known as hemp or marijuana, has emerged as an agronomically important crop due to its assorted portfolio of uses [1]. It is also relatively easy to grow cannabis, as it has low demand for fertilizers, biocides, weed control, or crop rotation [2]. As a crop with both food and non-food products and a high yield, many counties have legalized the cultivation of cannabis for its potential contribution in improving agro-industrial fields such as agriculture, textiles, bio-composite, paper making, automotive, construction, bio-fuel, functional food, oil, cosmetics, personal care, and the pharmaceutical industry [3,4]. The legal cultivation of cannabis is carried out in over 40 countries, while it still remains illegal to cultivate or use cannabis products in many countries all over the world due to its narcotic potential [5]. The main cannabis cultivators in the world are the United States, China, and Canada, where it is carried out under strict government regulations. Cannabis farming has a good turnover value for the harvest. For example, in the USA alone, the value of hemp production in the open totaled USD 824 million in 2021 [6].
Historically, cannabis was introduced to Thailand and other countries in Southeast Asia from India [7], and has been used for medicinal purposes, as a food source, and as a source of fiber. The drug-containing genotype of cannabis is more well-known, as its active ingredients are used for medical and recreational purposes, for which it is only legally grown in a few countries of the world, including Thailand. On the other hand, industrial hemp (with THC levels below 0.3%) is used as a food source rich in proteins, omega-3 fatty acids, magnesium, and vitamins in the form of hemp seeds, oil, milk, cheese, protein powder [8], and as a source of strong and stiff and easily recyclable natural fiber [9].
Due to the legalization of cannabis cultivation in some countries and its potential impact on public health, ecological balance, agricultural productivity, and economic benefits, pathogens affecting the cultivation of cannabis and hemp and disease prevention measures have gained the attention of cannabis cultivators and researchers [10]. Both insect pests and fungal, bacterial, and viral pathogens can cause diseases in cannabis [11]. Common diseases causing major economic losses during cannabis cultivation include leaf spots, damping-off, root and crown rot, powdery mildew, bud rots, post-harvest molds, and dudding. Among them, diseases caused by fungal pathogens have risen as the most devastating [12,13].
To avoid the economic losses caused during cultivation and storage in post-harvest stages, it is important to correctly identify diseases and their causal organisms to establish prevention strategies [14]. Additionally, it is important that the end product be free of diseases because the recreational use of cannabis is highly regulated by the governments in all countries where it is grown legally, and for the beneficial and economical manufacturing of high-quality downstream products. This is crucial not only for meeting regulatory standards but also for safeguarding public health.
Leaf diseases caused by fungi in cannabis include yellow leaf spot disease caused by Septoria cannabis and S. neocannabina, brown leaf spot caused by Phoma and Ascochyta species, white leaf spot caused by Diaporthe ganjae (=Phomopsis ganjae) [15], olive leaf spot caused by Pseudocercospora cannabina and Cercospora cannabis, Stemphylium Leaf and Stem Spot caused by Stemphylium botryosum and S. herbarum, black mildew caused by Schiffnerula cannabis, Black Dot caused by Epicoccum nigrum, and Pepper spot caused by Leptosphaerulina trifolii [16]. Among them, leaf spot disease caused by Pseudocercospora cannabina remains a high-priority disease [17]. Due to its ability to overwinter on infected planting materials and soil, this disease is very commonly observed in field-grown hemp. Ps. Cannabina infections can cause the complete defoliation of cannabis and hemp plants [18,19].
Pseudocercospora Speg. (Mycosphaerellaceae, Mycosphaerellales) contains many plant-pathogenic species causing leaf spots, fruit spots, and blights in a wide range of hosts dispersed in a vast geographical distribution area [20,21,22]. Their diversity is relatively higher in tropical and temperate areas, causing diseases in important agricultural crops, such as Sigatoka leaf disease in bananas, sooty spot disease in kiwi, leaf and fruit spot in citrus [23,24], and angular leaf spot disease in beans [25], leading to devastating economical losses. Quarantine regulations also consider Pseudocercospora ssp. As an important phytopathogenic fungi as it has the potential to cause devastating disease to many crops [20,26].
Pseudocercospora species are identified using various characteristics, such as their associated host plants, morphology, pathogenicity, and phylogenetic relationships generated by a multi-locus phylogeny [20,22,23,27,28]. Many phytopathogenic species of Pseudocercospora are suggested as host-specific from the results of multi-locus phylogeny and inoculation tests on specific plant hosts [20,22,23]. Therefore, DNA barcoding data of the causal agents of the disease play an important role in the correct identification of species of the genus Pseudocercospora when an unknown disease is observed in the field.
During a survey of cannabis fields in Chiang Rai province of Northern Thailand, Cannabis sativa leaves showing leaf spot symptoms were collected and brought to the laboratory. After preliminary morphological studies under microscopical observations, the causal organism observed on the leaf spots was identified to be a Pseudocercospora sp.
Even though the phylogenetic analyses revealed that this species could be a novel one, we concluded that this is Pseudocercospora cannabina, considering the host specificity of this genus, the unavailability of sequence data for this species until our study, and morphological similarities present between our isolates and the previously described morphological characteristics of the type Ps. cannabina.
Even though Ps. cannabina is reported and identified as the pathogen associated with olive leaf spot disease in cannabis all around the world, the identification is performed only with the observation of field symptoms and morphological characteristics of the isolated pathogen. Although these steps are important for identification, they are not highly accurate. Most countries have introduced molecular-based diagnosis techniques for rapid diagnosis or epidemiological studies to grasp the origin of the pathogen. Our study aims to solve this issue by providing DNA sequence data of multiple gene regions, contributing to a more accurate identification and comprehensive understanding of Ps. cannabina as a pathogen of cannabis. This study reveals the phylogenetic position within the genus Pseudocercospora, indicating several species’ barcodes, such as tef and rpb2 gene regions. Also, we describe the symptoms in detail and the morphology based on the current taxonomical criteria. This information will be helpful for field diagnoses in the cropping fields and for quarantine purposes.
We conducted etiological studies, including a pathogenicity test, detailed morphological observations, and multi-locus phylogenetic analyses, to reveal the fundamental information needed for accurate identification and disease control. Illustrations and detailed descriptions are provided.
2. Materials and Methods
2.1. Sample Collection and Isolation
Samples of leaf spot in Cannabis sativa were collected from a plantation area located in Chiang Rai Province, northern Thailand, on 5th February, 2022. Twenty symptomatic leaves were randomly collected from this plantation. Leaf samples were kept in sterile zip-lock plastic bags and carried to the laboratory within 24 h of collection. Single spore isolation was carried out as described by To-Anun et al. (2011) [29]. The fungal specimens with desired structures (i.e., conidiomata, conidiophore, conidiogenous cells, and conidia) were mounted on lactic acid and photographs were taken using the Axiovision Zeiss Scope-A1 microscope fitted with a Canon EOS 6D digital camera. The morphological measurements were carried out using the Tarosoft (R) Image Frame Work program. The specimens were deposited in the Sustainable Development of Biological Resources Laboratory (SDBR) at Chiang Mai University, Chiang Mai, Thailand.
2.2. DNA Extraction and PCR Amplification and Sequencing
The genomic DNA from the fungal mycelia was extracted using the DNA Extraction Mini Kit (FAVORGEN, Ping Tung, Taiwan) following the manufacturer’s protocol. The internal transcribed spacer (ITS1, 5.8S, ITS2), the partial actin (act), translation elongation factor 1-alpha (tef1), and RNA polymerase second largest subunit (rpb2) genes were amplified by the polymerase chain reaction (PCR) using ITS4/ITS5 primers [30], ACT-512F/ACT-783R primers [31], EF1-668/EF1-1251 primers [31,32], and RPB2-5F2/RPB2-7cR primers [33], respectively. PCR reactions were performed in a 25 µL reaction mixture containing 1.0 µL DNA template, 1.0 µL each forward and reverse primer, 12.5 µL 2X Quick Taq® HS DyeMix (TOYOBO, Japan), and 9.5 µL deionized water. The amplification program for all four genes was performed in separate PCR reactions and consisted of an initial denaturation step at 95 °C for 5 min followed by 35 cycles of denaturation at 95 °C for 30 s, an annealing step at 52 °C for 45 s (ITS), 55 °C for 1 min (act) and 56 °C for 1 min (rbp2), and an extension step at 72 °C for 10 min on a GeneMax thermal cycler (Hangzhou Bioer Technology Co., Ltd. (BIOER), Zhejiang, China). PCR products were checked on 1% agarose gel electrophoresis. The purified PCR products were sent to the 1st Base Company (Kembangan, Malaysia). The obtained nucleotide sequences were deposited in GenBank (accession numbers available in Table 1).

Table 1.
GenBank accession numbers of the Pseudocercospora species isolates used for the phylogenetic analysis. Culture collection numbers with “*” designate the type. The isolates obtained in this study are in bold.
2.3. Phylogenetic Analyses
The DNA sequences generated were assembled and aligned with 63 sequences of Pseudocercospora species retrieved from previous studies by Nakashima et al. (2016) [23], Videira et al. (2017) [28], and Chen et al. (2022) [22] using MEGA X software package [34] (Table 1). This matrix was aligned by using MAFFT (Multiple Alignment using Fast Fourier Transform) online version [35] and edited manually. Maximum-likelihood (ML) analysis was used in this study to estimate the phylogenetic relationship of the samples. ModelTest-NG [36] was used to estimate the best substitution model for each gene for ML analysis, and ML analysis was performed using RAxML-NG [37]. Branch strengths were tested by a bootstrap analysis of 100 replications [38]. ML analysis was performed with the evolutional models set as TN93ef + I + G4 for the ITS and act regions, and TN93+G4 for tef1 and rpb2 regions. Trochophora simplex (CBS 124744) was selected as an outgroup in all analyses and trees were viewed by using FigTree v. 1.4.2 (Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK).
2.4. Pathogenicity Test
To confirm Koch’s postulates, pathogenicity tests were conducted using the newly isolated strain from Cannabis sativa, following the methodology outlined by Sautua et al. (2020) [39]. For each isolate, spore suspensions (106 conidia/mL) were sprayed onto C. sativa leaves. Spore suspensions were prepared by combining sterile water with 1 mL of Tween 20 to aid in the dispersion of conidia onto 35-day-old colonies. As a control, leaves were sprayed with sterile distilled water and Tween 20. Following inoculation, the plants were placed in wet plastic boxes under sterile conditions and closely monitored on a daily basis for symptom detection. Once symptoms appeared on the inoculated leaves, the causal organism was reisolated. The reisolated strains were then compared to the original isolates to confirm the fulfillment of Koch’s postulates.
3. Results
Field observations of the cannabis plants showed olive leaf spot symptoms with circular to elliptical spots with a grey center and dark brown to black margins (Figure 1A–E). Severely affected plants showed signs of defoliation. The morphological characteristics and measurements were similar to the type specimen of Ps. cannabina described in Wakefield, 1917 [40].
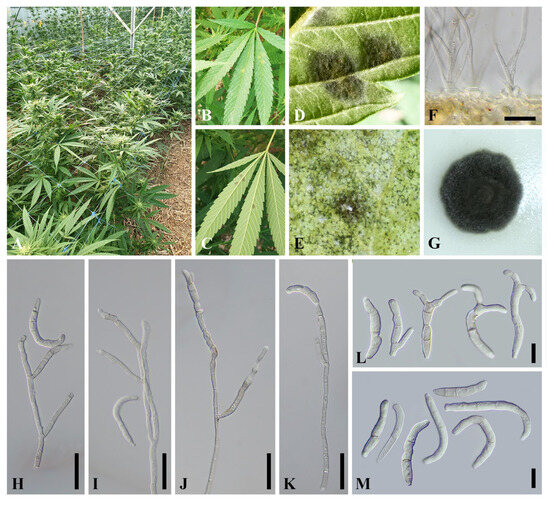
Figure 1.
Natural symptoms of olive leaf spot on Cannabis sativa L. (A–C). Conidial masses on the lower leaf surface (D,E); cross section (F); colony on PDA after 15 days at 25–30 °C (G); conidiophores and conidiogenous cells (H–K); conidia (L,M); scale bars: F, H–K = 50 µm; L–M = 20 µm.
3.1. Taxonomy
Taxonomic Description
Pseudocercospora cannabina (Wakef.) Deighton, Mycological Papers 140:141 (1976) [MB#321527] (Figure 1).
Description in vivo—Leaf spots amphigenous, on the upper surface, at first inconspicuous to slightly discolored, vein limited, later pale yellowish, irregular to subcircular, 2–6 mm diam., without distinct margin, finally covering the whole leaf, on the lower surface, indistinct, pale yellowish, covered with sooty conidial masses. Caespituli hypophyllous, effuse, sooty, vein-limited, greyish to pale brown patches, velutinous. Mycelium internal, hyphae septate, branched, brown to dark brown. Stromata rudimentary or poorly developed, forming substomatal hyphal aggregations. Conidiophores emerged from stromata or branched from superficial hyphae, 1–5(–8) in a loose fascicle, olivaceous brown or brown, darker than conidia or concolored, 9–15-septate, 0–3 times branched near the middle, straight to slightly sinuous, not geniculate, short or well developed, 75–115 × 4–6 μm; conidiogenous cells integrated, apical, proliferating percurrently or sympodially, conspicuously constricted at the middle part caused by percurrent proliferation, conically truncated at the apex, conidial loci inconspicuous. Conidia holoblastic, solitary, filiform to obclavato-cylindric, straight to curved, smooth, pale olivaceous to very pale brown, guttulate, 2–5-septate, sometimes constricted at the septa, uniform or irregular in width, broadly rounded at the apex, obconically truncated to truncated at the base, 35–60 × 3.5–5 μm; hilum unthickened, and not darkened.
Culture characteristics: Colonies on PDA reaching 2 cm in diameter after 25 days at room temperature; moderate aerial mycelium, circular, growth effuse with elevated colony center, grey to dark in outer region, entire margin, aerial mycelium dense.
Specimens examined: Thailand, Chiang Rai, Muang, on leaves of Cannabis sativa L., 5 February 2022, N. Tamakaew, CRC188, living culture SDBR-CMU372; ibid., Chiang Rai, Muang, on leaves of Cannabis sativa L., 5 February 2022, N. Tamakaew, CRC189, living culture SDBR-CMU373.
3.2. Pathogenicity Test
Within the 24 h incubation period, the penetration of germ tubes or hyphae of Ps. cannabina into leaf tissue could be observed. Under the compound microscope, hyphae invaded through the stomata and were growing inside of the leaf tissue (Figure 2). In contrast, no symptoms were observed on the control leaves sprayed with sterile distilled water. The leaves developed leaf spot symptoms 5 days post-inoculation. The reisolated pathogen from the inoculated leaves showed a similar morphology to the isolate used for the pathogenicity test, and the DNA sequence data for the ITS gene region obtained from the reisolated samples were also identical to the isolate (SDBR-CMU373) used for the pathogenicity test.
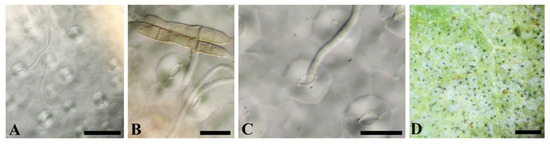
Figure 2.
Pathogenicity test. (A–C) Fungal hyphae penetrated through Cannabis sativa stomata on the lower leaf surface after 24 h; (D) the reproduction of the symptoms with emerged conidiophores and conidia on the lower leaf surface 5 DPI. Scale bars: (A,B) = 20 µL; (C) = 50 µL; (D) = 1 cm.
3.3. Phylogeny
The two isolates of Pseudocercospora species on C. sativa had identical sequences on the loci analyzed in this study. Therefore, the sequences obtained from one isolate (SDBR-CMU372 = RC238) were included in these analyses. The sequencing results of all regions were combined and aligned in a data matrix of 65 OTU, consisting of Pseudocercospora and Trochophora species (Table 1.). The final alignment contained a total of 1826 characters consisting of four regional sequences, ITS: 495 sites, act: 233 sites, tef1: 422 sites, and rpb2: 676 sites, including alignment gaps. The ML tree is shown in Figure 3. The sequences of the isolate analyzed in this study formed a clade with hitherto known species: Ps. diplusodonii on Diplusodon sp., Ps. ershadii on Diospyros lotus, Ps. kobayashiana on Diospyros kaki, and Ps. liquidambaricola on Liquidambar formosana. The clade was weakly supported by bootstrap analyses, but it was distinguishable from closely related taxa and recognized as an independent species (Figure 3).
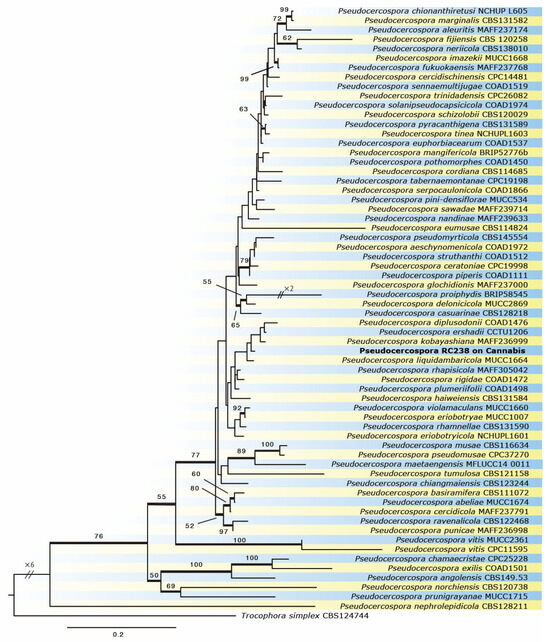
Figure 3.
Phylogram of Pseudocercospora resulting from a maximum likelihood analysis, based on a combined matrix of ITS, act, rpb2, and tef1. Numbers above the branches indicate ML bootstraps (ML BS ≥ 50%).
4. Discussion
The causal organism of olive leaf spot, Pseudocercospora cannabina, is an important pathogen in cannabis or hemp. It has been previously reported in China, India, Korea, Poland, Australia, and the USA [41]. Even though it is reported as a high-priority pathogen causing olive leaf spot disease in Cannabis sativa, in almost all studies mentioned, the identification was only carried out using morphological characteristics, which is not highly accurate. For the plant genus Cannabis, an alternative species of Cercosporoid fungi, Cercospora cannabis Hara & Fukui, is known [42,43]. According to Chupp (1954) [42], C. cannabina (=Ps. cannabina) is distinguishable by having colored conidia and branched conidiophores. Morphologically, the causal organism of leaf spots on C. sativa in Thailand is identical to the original description of C. cannabina by Wakefield (1917) [40]. From these results, our isolates were identified as Ps. cannabina. Additionally, sequence data originating from the type material of Ps. cannabina are not available and they were collected from Uganda, quite far from Thailand, where the disease was observed in our study. As mentioned above, Ps. cannabina is widely distributed around the world. Therefore, further studies are required for a more detailed understanding of the distribution and intraspecies diversity of this species.
Novel species introduced in this genus are nowadays verified by data involving host associations, morphological characters, and phylogenetic relationships using multi-locus data using DNA sequences [20]. Even though these species are important as phytopathogens, many species in Pseudocercospora lack sequence data from the type material. This causes difficulties in the identification and rapid diagnosis of plant disease since accurately distinguishing through just morphological observations can be challenging and sequence data can play a very important role in the accurate identification of Pseudocercospora species. Hence, adding sequence data to resolve this data gap is very important. Chen et al. 2022 [22] designated ITS as the barcode for Pseudocercospora, and species delineation can be undertaken further with additional gene regions act, rpb2, and tef1.
In the pathogenicity test, similar symptoms observed in the field were reproduced and the causal organism re-isolated. From these results, the pathogen causing leaf spots on C. sativa was confirmed to be Ps. cannabina from morphological comparisons and sequence data fulfilling Koch’s postulates.
In Thailand, many species from Pseudocercospora have been reported but none from Cannabis [44]. Our study is the first to report olive leaf spot disease in Cannabis from Thailand. Even though leaf spot diseases are commonly reported in Cannabis around the world, in-depth studies have yet to be conducted and published. Moreover, this study is the first to use both morphological and molecular data to identify and confirm the pathogenicity of Ps. cannabina causing leaf spot disease on Cannabis sativa. The data obtained in this study will contribute to the rapid diagnosis and control of the disease.
Author Contributions
Conceptualization, R.C. and C.N.; methodology, D.L.H., S.H., P.W. and C.K.; validation, D.L.H., C.N., S.H., N.T. and N.N.; formal analysis, D.L.H. and C.N.; investigation, D.L.H., C.N., S.H., P.W., N.T., N.N., C.K., T.P. and R.C.; writing—original draft preparation, D.L.H.; writing—review and editing, C.N., S.H., P.W. and R.C.; supervision, T.P. and R.C.; project administration, R.C.; funding acquisition, C.N. and R.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding. The APC was funded by Mie University, Mie, Japan.
Data Availability Statement
All sequence data are available in NCBI GenBank following the accession numbers in the manuscript.
Acknowledgments
This study was partially supported by the Post-Doctoral Fellowship 2022 for Reinventing Chiang Mai University.
Conflicts of Interest
Author Chaorai Kanchanomai was employed by the company Chaofah Vineyard, Chiang Rai 57210, Thailand. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Salentijn, E.M.J.; Zhang, Q.; Amaducci, S.; Yang, M.; Trindade, L.M. New developments in fiber hemp (Cannabis sativa L.) breeding. Ind. Crops Prod. 2015, 68, 32–41. [Google Scholar] [CrossRef]
- Struik, P.C.; Amaducci, S.; Bullard, M.J.; Stutterheim, N.C. Agronomy of fibre hemp (Cannabis sativa L.) in Europe. Ind. Crops Prod. 2000, 11, 107–118. [Google Scholar] [CrossRef]
- Tsaliki, E.; Kalivas, A.; Jankauskiene, Z.; Irakli, M.; Cook, C.; Grigoriadis, I.; Panoras, I.; Vasilakoglou, I.; Dhima, K. Fibre and Seed Productivity of Industrial Hemp (Cannabis sativa L.) Varieties under Mediterranean Conditions. Agronomy 2021, 11, 171. [Google Scholar] [CrossRef]
- Rupasinghe, V.H.P.; Davis, A.; Kumar, S.K.; Murray, B.; Zheljazkov, V.Z. Industrial hemp (Cannabis sativa subsp. sativa) as an emerging source for value-added functional food ingredients and nutraceuticals. Molecules 2020, 25, 4078. [Google Scholar] [CrossRef]
- Gwinn, K.D.; Hansen, Z.; Kelly, H.; Ownley, B.H. Diseases of Cannabis sativa Caused by Diverse Fusarium Species. Front Agronomy 2022, 3, 796062. [Google Scholar] [CrossRef]
- Honig, L. National Hemp Report Agricultural Statistics Board Briefing, USDA. 2022. Available online: www.nass.usda.gov (accessed on 25 May 2023).
- Martin, M.A. Ethnobotanical Aspects of Cannabis in Southeast Asia. In Cannabis and Culture; Rubin, V., Ed.; Mouton Publishers: Berlin, Germany, 1975; pp. 63–76. ISBN 9027976694. [Google Scholar]
- Cerino, P.; Buonerba, C.; Cannazza, G.; D’Auria, J.; Ottoni, E.; Fulgione, A.; Di Stasio, A.; Pierri, B.; Gallo, A. A Review of Hemp as Food and Nutritional Supplement. Cannabis Cannabinoid Res. 2021, 6, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Dhakal, H.N.; Zhang, Z. The use of hemp fibers as reinforcements in composites. In Biofiber Reinforcements in Composite Materials; Woodhead Publishing: Sawston, UK, 2015; pp. 86–103. ISBN 9781782421221. [Google Scholar] [CrossRef]
- Punja, Z.K. Emerging diseases of Cannabis sativa and sustainable management. Pest Manag. Sci. 2021, 9, 3857–3870. [Google Scholar] [CrossRef]
- Finley, S.J.; Javan, C.T.; Green, R.L. Bridging Disciplines: Applications of Forensic Science and Industrial Hemp. Front. Microbiol. 2022, 13, 760374. [Google Scholar] [CrossRef] [PubMed]
- Punja, Z.K.; Collyer, D.; Scott, C.; Lung, S.; Holmes, J.; Sutton, D. Pathogens and Molds Affecting Production and Quality of Cannabis sativa L. Front. Plant Sci. 2019, 10, 1120. [Google Scholar] [CrossRef] [PubMed]
- Jerushalmi, S.; Maymon, M.; Dombrovsky, A. Fungal Pathogens Affecting the Production and Quality of Medical Cannabis in Israel. Plants 2020, 9, 882. [Google Scholar] [CrossRef]
- Manawasinghe, I.S.; Phillips, A.J.L.; Xu, J.; Balasuriya, A.; Hyde, K.D.; Stępień, Ł.; Harischandra, D.L.; Karunarathna, A.; Yan, J.; Weerasinghe, J.; et al. Defining a species in fungal plant pathology: Beyond the species level. Fungal Divers. 2021, 109, 267–282. [Google Scholar] [CrossRef]
- McPartland, J.M.; Hillig, K.W. White Leaf Spot. J. Ind. Hemp 2006, 11, 43–50. [Google Scholar] [CrossRef]
- Bakro, F.; Wielgusz, K.; Bunalski, M.; Jedryczka, M. An overview of pathogen and insect threats to fibre and oilseed hemp (Cannabis sativa L.) and methods for their biocontrol. In Proceedings of the Meeting 17th IOBC Working Group Meeting on Integrated Control in Oilseeds (ICOC), International Organization for Biological and Integrated Control of Noxious Animals and Plants, West Palearctic Regional Section (IOBC-WPRS), Zagreb, Croatia, 18–20 September 2018; Volume 136, pp. 9–20. [Google Scholar]
- Santo, D. Australian Industrial Hemp Industry Pesticide Review and Evaluation Program. AgriFutures Australia. Publication No. 22-060. 2022. Available online: https://agrifutures.com.au/wp-content/uploads/2022/06/22-060.pdf (accessed on 25 May 2023).
- Rodriguez, G.; Kibler, A.; Campbell, P.; Punja, Z.K. Fungal diseases of Cannabis sativa in British Columbia, Canada. In Proceedings of the American Phytopathological Society Annual Meeting, Pasadena, CA, USA, 1–5 August 2015. Poster 529-P. [Google Scholar]
- Ward-Gauthier, N.; Beale, J.; Amsden, B.; Dixon, E. Greenhouse hemp in Kentucky exhibits many common diseases. In Proceedings of the American Phytopathological Society Annual Meeting, Pasadena, CA, USA, 1–5 August 2015. Poster 502-P. [Google Scholar]
- Crous, P.W.; Braun, U.; Hunter, G.C.; Wingfield, M.J.; Verkley, G.J.M.; Shin, H.-D.; Nakashima, C.; Groenewald, J.Z. Phylogenetic lineages in Pseudocercospora. Stud. Mycol. 2013, 75, 37–114. [Google Scholar] [CrossRef] [PubMed]
- Wanasinghe, D.N.; Phukhamsakda, C.; Hyde, K.D.; Jeewon, R.; Lee, H.B.; Jones, E.B.G.; Tibpromma, S.; Tennakoon, D.S.; Dissanayake, A.J.; Jayasiri, S.C.; et al. Fungal diversity notes 709–839: Taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Divers. 2018, 89, 1–236. [Google Scholar] [CrossRef]
- Chen, Q.; Bakhshi, M.; Balci, Y.; Broders, K.; Cheewangkoon, R.; Chen, S.; Fan, X.; Gramaje, D.; Halleen, F.; Jung, M.H.; et al. Genera of phytopathogenic fungi: GOPHY 4. Stud. Mycol. 2022, 101, 417–564. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, C.; Motohashi, K.; Chen, C.Y.; Groenewald, J.Z.; Crous, P.W. Species diversity of Pseudocercospora from Far East Asia. Mycol. Prog. 2016, 15, 1093–1117. [Google Scholar] [CrossRef]
- Aslake, M.; Sintayehu, A.; Alem, T.; Abera, M. Distribution and Status of Citrus Leaf and Fruit Spot (Pseudocercospora angolensis) Disease in North Western Ethiopia. Open J. Ecol. 2022, 12, 733–741. [Google Scholar] [CrossRef]
- Silva, K.J.D.E.; De Souza, E.A.; Sartorato, A.; De Souza, F. Pathogenic Variability of Isolates of Pseudocercospora griseola, the Cause of Common Bean Angular Leaf Spot, and its Implications for Resistance Breeding. J. Phytopathol. 2008, 156, 602–606. [Google Scholar] [CrossRef]
- Evans, H.C. The genus Mycosphaerella and its anamorphs Cercoseptoria, Dothistroma and Lecanosticta on pines. Mycol. Pap. 1984, 153, 1–102. [Google Scholar]
- Braun, U.; Nakashima, C.; Crous, P.W. Cercosporoid fungi (Mycosphaerellaceae) 1. Species on other fungi, Pteridophyta and Gymnospermae. IMA Fungus 2013, 4, 265–345. [Google Scholar] [CrossRef]
- Videira, S.I.R.; Groenewald, J.Z.; Nakashima, C.; Braun, U.; Barreto, R.W.; de Wit, P.J.G.M.; Crous, P.W. Mycosphaerellaceae—Chaos or clarity? Stud. Mycol. 2017, 87, 257–421. [Google Scholar] [CrossRef] [PubMed]
- To-Anun, C.; Hidayat, I.; Meeboon, J. Genus Cercospora in Thailand: Taxonomy and Phylogeny (with a dichotomous key to species). Plant Pathol. Quar. 2011, 1, 11–87. [Google Scholar] [CrossRef]
- White, T.J.; Burns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Carbone, I.; Kohn, L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 1999, 91, 553–556. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic Relationships Among Ascomycetes: Evidence from an RNA Polymerse II Subunit. Mol. Biol. Evol. 1998, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Stamatakis, A. RAxML-NG: A fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Sautua, F.J.; Doyle, V.P.; Price, P.P.; Porfiri, A.; Fernandez, P.; Scandiani, M.M.; Carmona, M.A. Fungicide resistance in Cercospora species causing Cercospora leaf blight and purple seed stain of soybean in Argentina. Plant Pathol. 2020, 69, 1678–1694. [Google Scholar] [CrossRef]
- Wakefield, E.M. Fungi Exotici: XXIII. Bull. Misc. Inf. R. Bot. Gard. Kew. 1917, 1917, 308–314. [Google Scholar] [CrossRef]
- Farr, D.F.; Rossman, A.Y.; Castlebury, L.A. United States National Fungus Collections Fungus-Host Dataset. Ag Data Commons. 2021. Available online: https://data.nal.usda.gov/dataset/united-states-national-fungus-collections-fungus-host-dataset (accessed on 13 November 2022).
- Chupp, C. A monograph of the Fungus genus Cercospora. by the Author Plant Pathology Department, Cornell University, Ithaca, N. Y. AIBS Bull. 1954, 4, 11. [Google Scholar] [CrossRef][Green Version]
- Crous, P.W.; Braun, U. Mycosphaerella and its Anamorphs 1: Names Published in Cercospora and Passalora; CBS Biodiversity Series 1; CBS Fungal Biodiversity Centre: Utrecht, The Netherlands, 2003; ISBN 90-70351-49-8. [Google Scholar]
- Chen, Y.J.; Jayawardena, R.S.; Bhunjun, C.S.; Harishchandra, D.L.; Hyde, K.D. Pseudocercospora dypsidis sp. Nov. (Mycosphaerellaceae) on Dypsis lutescens leaves in Thailand. Phytotaxa 2020, 474, 218–234. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).