Abstract
In this study, the biodiversity of endophytic bacteria of cultivated grape varieties from the vineyards of Primorsky Krai, Russia, was analyzed for the first time. Far Eastern grape varieties with a high level of stress resistance are a unique object of research as they are cultivated in cold and humid climates with a short summer season. Grapevine endophytic microorganisms are known as promising agents for the biological control of grapevine diseases and agricultural pests. Using genomic approaches, we analyzed the biodiversity of the endophytic bacteria and fungi in the most common grape varieties of Primorsky Krai, Russia: Vitis vinifera × Vitis amurensis cv. Adele (hybrid No. 82-41 F3), Vitis riparia × V. vinifera cv. Mukuzani (pedigree unknown), two cultivars Vitis labrusca × V. riparia cv. Alfa, and Vitis Elmer Swenson 2-7-13 cv. Prairie Star for the first time. The main representatives of the endophytic microorganisms included 16 classes of bacteria and 21 classes of fungi. The endophytic bacterial community was dominated by Gammaproteobacteria (31–59%), followed by Alphaproteobacteria (9–34%) and, to a lesser extent, by the classes Bacteroidia (9–22%) and Actinobacteria (6–19%). The dominant fungal class was Dothideomycetes (43–77%) in all samples analyzed, with the exception of the grapevine cv. Mukuzani from Makarevich, where Malasseziomycetes was the dominant fungal class. In the samples cv. Alfa and cv. Praire Star, the dominant classes were Tremellomycetes and Microbotriomycetes. A comparative analysis of the endophytic communities of the cultivated grape varieties and the wild grape V. amurensis Rupr. was also carried out. We found that 18–20% of the variance between the endophytic communities accounted for the differences between the cultivated and wild grapevines, while the factors “plant location” and “individual plants” accounted for 50–56% and 3–10% of the variance, respectively. The results of this study can be used to develop new means of biocontrol in vineyards to protect plants from abiotic stresses and pathogens.
Keywords:
grapes; metagenome; bacteria; fungi; endophytes; Far East of Russia; 16S; ITS1; Vitis amurensis; NGS 1. Introduction
Grapevine is among the oldest cultivated plant species, being one of the most highly consumed fruit crops in the world [1]. The grapevine belongs to the Vitaceae family, consisting of approximately 14 genera and 900 species [2]. The genus Vitis represents more than 7.5 million ha of cultivated surfaces in the world with 27 million tons of wine produced each year [3]. The grapevines consist of multiple compartments like roots, leaves, and fruits, each providing distinct habitats for microorganisms. The microbiota found within different plant compartments is influenced by the grape genotype, development, and local environment. However, the extent to which the microbiota in one compartment, such as the roots, shapes the microbiota in other parts like the leaves or fruits is not well known. Several factors, including the plant’s geographic location, genotype, and biotic/abiotic stresses, influence the composition and diversity of plant microbiota. Previous research has revealed that specific bacterial and fungal communities are retained within grapevines compartments, irrespective of grafting or the rootstock genotype used [4]. This suggests that the environmental conditions experienced by microorganisms in different parts of the plant play a crucial role.
The most serious problems faced in the cultivation of grapes are abiotic and biotic stresses, which lead to a decrease in grape yield and fruit quality [5,6,7]. Biocontrol is gaining popularity in viticulture as a means to reduce the use of chemical pesticides, which have negative effects on the environment and human safety [8,9]. The grapevine endophytic microbiome, consisting of bacteria and fungi that inhabit plant tissues without causing harm, holds great potential as a source of biocontrol agents. These endophytes can be found in both natural and managed ecosystems [10]. Endophytes have the ability to produce beneficial metabolites like phosphorous, iron, and nitrogen from the environment, as well as growth-regulating phytohormones that help plants withstand abiotic stresses [11]. Additionally, endophytic microorganisms occupy a similar ecological niche as many plant pathogens, making them suitable candidates for biocontrol agents. Exploiting these endophytes as biocontrol agents could be an effective strategy to reduce pesticide usage in vineyards [3].
For example, the grapevine endophytic bacteria Bacillus velezensis, Pseudomonas chlororaphis, and Serratia plymuthica constitute potential biocontrol agents against grapevine trunk diseases [5]. Furthermore, the endophytic strain Bacillus velezensis KOF112, isolated from the Japanese wine grape Vitis sp. cv. Koshu, inhibits the growth of several aggressive plant pathogens such as Botrytis cinerea (gray mold), Colletotrichum gloeosporioides (causing anthracnose), and Phytophthora infestans (some of the most aggressive and widespread plant pathogens) [12]. In addition to bacteria, endophytic fungi also play a role in promoting plant growth. Phialocephala fortinii, for instance, has been found to increase phosphorus accumulation and biomass in Vitis macrocarpon [13]. These endophytes have the potential to enhance the overall health and productivity of grape plants. Moreover, endophytic bacteria like Pseudomonas fluorescens RG11 have been found to promote plant growth and increase the levels of melatonin, an important plant hormone, in different grape cultivars [14]. This hormone is known to aid in plant growth and can be particularly useful in helping plants withstand salt stress conditions [15]. By utilizing melatonin-producing endophytes, it may be possible to develop strategies that support plant growth under challenging environmental conditions. Another notable example is Burkholderia phytofirmans PsJN, a rhizobacterium that promotes plant growth and induces resistance to gray mold in grapevines. Additionally, it triggers physiological changes that enhance the grapevine’s tolerance to low non-freezing temperatures. This bacterium shows promise as a biological agent for disease control and improving grapevine resilience to abiotic stresses [16]. Thus, endophytic microorganisms of grapes have a number of favorable properties for plant growth, also they have the potential to be utilized as biological agents in grape cultivation, contributing to sustainable agriculture practices and integrated plant production methods.
Vitis vinifera, a commonly cultivated grape species, is not able to survive harsh winters in regions with extremely low temperatures like northern China and Primorsky Krai in Russia. However, certain wild Vitis species such as Vitis amurensis and Vitis riparia have shown great tolerance to freezing conditions [7]. Despite this, the specific mechanisms responsible for grapevine cold tolerance are still largely unknown. Perhaps, in addition to the physiological and molecular features of grapes, endophytic microorganisms may be a key element in resistance to adverse environmental factors. The microbiome processes essential for vine growth and wine production exhibit distinct patterns linked to the location of the vineyard [17,18]. Currently, in the viticulture of Primorsky Krai, bred or imported grape varieties are used, capable of yielding a good harvest in conditions of a short summer period, high relative humidity, and cold winter. It is known that the grapevines have been extensively manipulated through centuries of cultivation and breeding, resulting in highly selected germplasms [19]. Thus, information about the microbiome of Far Eastern grape cultivars is an interesting object of research.
Understanding the composition of the microbiome of grapevine varieties grown in the vineyards of Primorsky Krai of Russia will make it possible to correct the biodiversity and species ratio of endophytic microorganisms, which may contribute to improving grapevine growth and production characteristics. Therefore, the aim of this work was to study the biodiversity of endophytic bacteria and fungi in the most common grape cultivars growing in the vineyards of the Primorsky Krai of Russia using next-generation sequencing (NGS).
2. Materials and Methods
2.1. Samples Collection and Pre-Treatment
Healthy and normally developed grapevine tissues were collected from two different vineyards in Primorsky Krai of Russia in July 2022. In particular, from each site, Makarevich (longitude 43.718600 and latitude 132.11040) and PRIM ORGANICA (longitude 44.218073 and latitude 132.475822), were collected young shoots with three leaves of two 10-year-old grapevines of two cultivars V. vinifera × V. amurensis cv. Adele (hybrid No. 82-41 F3), Vitis riparia × V. vinifera cv. Mukuzani (pedigree unknown) and two cultivars Vitis labrusca × V. riparia cv. Alfa (https://www.vivc.de/index.php?r=passport%2Fview&id=346, accessed on 18 October 2023), and Vitis Elmer Swenson 2-7-13 cv. Prairie Star (https://www.vivc.de/index.php?r=passport%2Fview&id=23087, accessed on 18 October 2023), respectively. In order to control weeds in the two vineyards, Makarevich and PRIM ORGANICA, a combination of manual weeding and mechanical processing of the aisles was employed. Additionally, approximately 5 kg of humus derived from cattle manure was used as a mineral fertilizer every two years during spring time. To prevent fungal diseases, particularly mildew, two fungicides were utilized in the PRIM ORGANICA vineyard: Thanos (containing famoxadone and cymoxanil) from DuPont, Wilmington, DE, USA, and Orden (containing copper chloride and cymoxanil) from August, Russia. These treatments were applied 3–4 times throughout the season, alternating between the two preparations. For defense against spider mites, the PRIM ORGANICA vineyard used the inorganic contact fungicide and acaricide Tiovit Jet (containing colloidal sulfur) from August, Russia, which exhibits high gas phase activity. In the Makarevich vineyard, three different treatments were employed to defend against spider mites, anthracnose, and mildew: Abiga Peak (containing copper chloroxide) from Abiga Peak, Russia, Ridomil Gold (containing ethylene bisdithiocarbamate and mefenoxam) from Syngenta, Switzerland, and Kurzat R (containing copper chloroxide) at a specified concentration. Insect protection was provided by using two insecticides: Fufanon (containing malathion) from BuFF Fufanon, Germany, and Actara (containing thiamethoxam) from Syngenta, China. These insecticides were employed to safeguard the vineyards against harmful insects. Overall, these vineyard processing techniques and the use of various fertilizers, fungicides, and insecticides help maintain the health and productivity of the vineyards by controlling weeds, preventing fungal diseases, and protecting against harmful insects.
The plant material was collected at 11–12 a.m. on low-cloud days without precipitation, and the air temperature was 18–20 °C. Each plant material specimen was delivered to the laboratory within 3 h. Four biological replicates (two stems and two leaves) of each of the grapevine cultivars were collected and analyzed using a cultivation-independent approach (NGS).
To prepare the grapevine tissues for further analysis, each cultivar’s tissues (0.5 g) were cleaned using a specific procedure. First, the tissues were washed with soap under running water. Then, they were sequentially washed under sterile conditions using 75% ethanol for 2 min, 10% hydrogen peroxide for 1 min, and finally rinsed five times with sterile water. This process aimed to effectively sterilize the surface of the tissues [20,21]. To verify the success of this sterilization method, a sample of the last wash water (100 µL) was incubated on R2A and potato dextrose agar (PDA) plates. The purpose was to ensure that there was no bacterial or fungal colony growth on these plates, indicating the absence of any contamination from the outside (in vitro control of epiphytic microorganisms). This sterilization procedure is crucial for maintaining the integrity and purity of the grapevine tissues, preventing any unwanted microorganisms from interfering with subsequent analysis or experiments.
2.2. DNA Extraction and Illumina MiSeq Sequencing
The DNA for NGS was isolated using the method as described earlier [22]. The quality and quantity of the DNA was assessed using the NanoPhotometer P300 (IMPLEN, Munich, Germany).
The DNA samples were sent to Sintol (Moscow, Russia) for Illumina high-throughput sequencing. The libraries were prepared for sequencing following the protocol described in “16S Meta-genomic Sequencing Library Preparation” (Part # 15,044,223 Rev. B; Illumina (San Diego, CA, USA)). The bacterial 16S rRNA V4 region (515Fmod-806R) was amplified from all samples using plant primers modified for Vitis sp. 515F (5′GGTAATACGKAGGKKGCDAGC) and 806R (5′RTGGACTACCAGGGTATCTAA) [20]. The ITS1-ITS2 rDNA region of the fungi was amplified from all samples using primers ITS1f (5′CTTGGTCATTTAGAGGAAGTAA) and ITS2 (5′GCTGCGTTCTTCATCGATGC) [21]. Once the amplicons had been obtained, the libraries were purified and mixed in an equimolar ratio using the SequalPrep™ Normalization Plate Kit (ThermoFisher, Waltham, MA, USA, Cat # A10510-01). The resulting library pools were quality-checked using the Fragment Analyzer, and quantitative analysis was performed using qPCR. The library pool was sequenced on Illumina MiSeq (2 × 250 paired end) using the MiSeq Reagent Kit v2 (500 cycles). The FASTQ files were obtained using bcl2fastq conversion software v2.17.1.14 (Illumina). The phage PhiX library was used to control the sequencing parameters. The majority of the reads that belonged to the phage DNA were removed during the demultiplexing process.
Bacterial and fungal sequences of the endophytes were deposited into NCBI under the accession number PRJNA998468 and in the database of the Biotechnology laboratory at the Federal Scientific Center of the East Asia Terrestrial Biodiversity, Far Eastern Branch of the Russian Academy of Sciences, Russia (https://biosoil.ru/downloads/biotech/Vitis%20metagenom/, accessed on 18 October 2023).
2.3. Bioinformatics and Biostatistics
The samples used in the bioinformatic analysis are presented in Supplementary Materials Tables S1 and S2. Custom scripts based on the R and Bash languages were used to process the data obtained (https://github.com/niknit96/Aleynova_et.al.2023.10, accessed on 18 October 2023). The QIIME 2 [23] and DADA2 [24] programs were used to pre-process the raw data. Paired-end reads were merged and sorted to remove primers, remaining PhiX reads, and chimeric sequences. Taxonomic identification of sequences was performed using the QIIME 2 Scikit-learn algorithm using the SILVA 138 pre-trained classifier for 16S sequences (99% OTUs from V4 region of sequences) [25] and the UNITE pre-trained classifier for ITS sequences (99% OTUs from ITS1f/ITS2 region of sequences) [26].
The qiime2R [27], phyloseq [28], ggdendro [29], RColorBrewer [30], circlize [31], and tidyverse [32] libraries were used in the pre-filtering and data preparation. Amplicon se-quence variants were merged into genus-level taxonomic ranks. Mitochondria, chloro-plast, Viridiplantae, Metazoa, Rhizaria, Protista, Alveolata, and unidentified sequences were deleted from the obtained data. Taxa at the genus level were filtered on the basis of relative abundance > 0.1% for a plant. We merged the filtered genera into a group called “other” in the taxonomy bar plots at the class level. Also, “other” genus taxa were removed from the UpSet diagrams.
The tidygraph [33], ggraph [34], Netcommi [35], and SpiecEasi [36] R libraries were used to analyze the positive and negative associations between bacteria and fungi in the Vitis endophytic community. Bacterial and fungal genera that did not occur more than 3 times in at least 60% of the samples were removed to reduce sparsity and ensure reliable results. The genus-level taxa were normalized, transformed, and converted into an adjacency matrix based on covariance by the package SpiecEasi using Meinshausen and Bühlmann neighborhood selection to estimate the conditional dependence of each pair of genera. This approach is suitable for compositional amplicon data and, unlike correlation-based methods, prevents most spurious, indirect relationships from being included in the networks [36]. The adjacency matrix was then converted into a Tidygraph object with help the Netcommi package and visualized as a circular network using the ggraph library. The centrality of nodes was measured using Kleinberg’s hub centrality scores. The top 10% of taxa of genus level based on the values of Kleinberg’s hub centrality score were qualified as hubs.
The Shannon alpha diversity and Bray–Curtis beta diversity data were obtained using the Vegan package [37]. The Bray–Curtis dissimilarity data were transformed into an even sampling depth and converted into non-metric multidimensional scaling (NMDS). To analyze the alpha diversity data between groups, a pairwise Wilcoxon rank-sum test with false discovery rate correction was performed. The PERMANOVA test with 999 permutations was used to statistically validate the beta diversity data. For the graphical representation of the results, the ggplot2 [32] and ComplexHeatmap [38] R libraries were used.
3. Results
3.1. Illumina Next-Generation Sequencing Results
Using Illumina technology, all 16S rRNA V4 regions (16S) and ITS1-ITS2 rDNA regions (ITS1) of the DNA samples were successfully sequenced, and libraries were constructed. NGS produced a total of 3,285,397 16S and 743,375 ITS1 paired reads, respectively. After paired-end alignments, quality filtering, and the deletion of singletons and chimeric and non-bacteria or non-fungal sequences, a total of 1,990,927 bacterial 16S and 262,933 fungal ITS1 sequences were generated from 4 plant samples (4 samples from each plant). On average, 124,433 and 16,433 sequences, per grapevine sample, were obtained from the 16S and ITS1 regions (Supplementary Materials Tables S1 and S2).
3.2. The Biodiversity of Bacterial Endophytes from Grapevine Cultivars in Primorsky Krai of Russia
After the bioinformatic quality control procedures, a total of 1,990,927 16S sequences in 16 samples and 172 taxa of genus level with a relative representation above 0.1% were identified, which belong to 18 taxa of class level. The endophytic bacterial community composition was mostly matched by class Gammaproteobacteria (28–59%) followed by class Alphaproteobacteria (8–35%) and, to a lesser extent, by classes Bacteroidia (6–22%) and Actinobacteria (5–21%) (Figure 1a). The biodiversity of the endophytic bacteria in grapevines cv. PrairieStar and cv. Alfa from the vineyard PRIM ORGANICA was most similar in the percentage of bacterial classes, as were the cv. Adele and cv. Mukuzani from the vineyard Makarevich (Figure 1a). The leaf and stem tissues of the grapevine were similar in terms of the bacterial biodiversity within the plant based on UPGMA clustering.
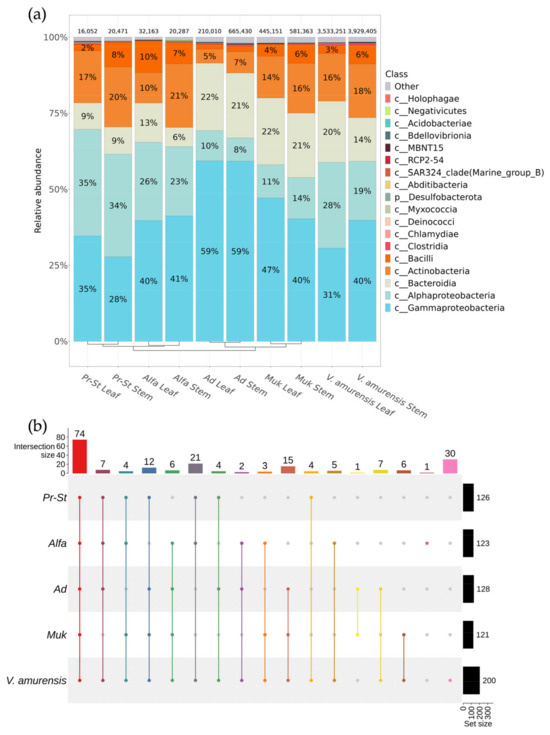
Figure 1.
Comparative analysis of the bacterial endophytic community composition of grapevine cultivars and wild grape Vitis amurensis growing in Primorsky Krai of Russia according to genomic approach (NGS). The composition of endophytic bacteria of grapevine variates Pr-St—Vitis Elmer Swenson 2-7-13 cv. Prairie Star from commercial vineyard PRIM ORGANICA; Alfa—Vitis labrusca × Vitis riparia cv. Alfa from PRIM ORGANICA; Ad—Vitis vinifera × V. amurensis cv. Adele from commercial vineyard Makarevich; Muk—V. riparia × V. vinifera cv. Mukuzani; V. amurensis—the sum of data for all V. amurensis grapevines obtained early [39]: (a) class-level taxonomical bar plots for the bacteria endophytic community depends on the variates of grapevines, and the sum of data for all V. amurensis grapevines obtained early [39]; (b) genus-level UpSet diagrams depicting overlapping taxa of NGS in different cultivars of grapevines. For each biocompartment, taxa were filtered based on relative abundance > 0.1%. Taxa with relative frequencies < 0.1% were removed from the UpSet plot. Number of sequences are shown above taxonomical bar plots. For clustering in bar plots, we used unweighted pair group method with arithmetic mean (UPGMA).
We also compared the biodiversity of the endophytic bacteria of grape varieties with the bacterial endophytic community of wild grapes V. amurensis; the data were obtained in previous research [39]. According to the analysis, in all samples of cultivated grapes, except for the cv. Adele, there was a similar percentage of classes of endophytic bacteria. In the grapevine cv. Adele, the percentage of the main Gammaproteobacteria class increased up to 59% and the percentage of the Alphaproteobacteria reduction class decreased up to 8–10% (Figure 1a).
According to the analysis, the endophytic bacterial community in the studied grape cultivars was represented by 121–128 genera of bacteria, while the community of endophytic bacteria in wild grape V. amurensis was represented by 200 genera (Figure 1b). Among them, 74 taxa were common for all grapevines (Figure 1b and Supplementary Materials Table S3). At the same time, 30 genera of endophytic bacteria were unique for the wild V. amurensis grape. The representation of the endophytic genera in the grape varieties collected at the Makarevich vineyard was similar to the grape samples from PRIM ORGANICA. Each sample of the studied grape variety contained 4–7 overlapping genera with the wild V. amurensis grape (Figure 1b). The grapevine cv. Alpha from PRIM ORGANICA had one unique genus, Eikenella, of endophytic bacteria (Figure 1b and Supplementary Materials Table S3).
The most common taxa for all analyzed grapevines were the taxa Comamonadaceae, Sphingomonas and Methylobacterium-Methylorubrum (Figure 2). For the grapes cv. Adel and cv. Mukuzani from the Makarevich vineyard, a large percentage were the bacterial genera Aquabacterium and Spirosoma, as well as Asinibacterium and Ralstonia being represented in these grape varieties and in the wild grape V. amurensis and absent in the grapevines from PRIM ORGANICA (Figure 2). The representation of endophytic bacteria Abiotrophia and Neisseria was 4 and 9% in the grape cv. Alpha from PRIM ORGANICA, while the representation of these bacterial genera in other varieties and the wild grape V. amurensis was less than 0.1% (Figure 2).
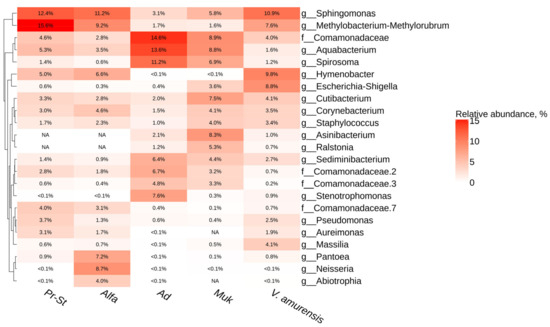
Figure 2.
Genus-level relative endophytic bacteria abundance heat map of significant taxa according to next-generation sequencing (NGS) in grapevines cultivars and wild grape Vitis amurensis. Pr-St—Vitis Elmer Swenson 2-7-13 cv. Prairie Star from commercial vineyard PRIM ORGANICA; Alfa—Vitis labrusca × Vitis riparia cv. Alfa from PRIM ORGANICA; Ad—Vitis vinifera × V. amurensis cv. Adele from commercial vineyard Makarevich; Muk—V. riparia × V. vinifera cv. Mukuzani; V. amurensis—the sum of data for all V. amurensis grapevines obtained early [39]. The top 10 most abundant taxa from each grapevine are displayed. White squares (NA) represent the absence of taxa.
3.3. The Fungal and Fungi-like Endophytic Microorganisms from Grapevine Varietes in Primorsky Krai of Russia
A total of 262,933 ITS1 reads were used for the phyla description for fungi and fungi-like endophytes of the vine varieties. According to metagenomic analysis of the ITS1 sequences, 146 taxa of genus level with a relative representation above 0.1% were represented in the community of fungi and fungi-like endophytes in different grapevines. These genera belonged to 24 taxa of class level in the analyzed grapevines (Figure 3a). The dominant class of fungi was Dothideomycetes (22–79%) in all analyzed samples except the grapevine cv. Mukuzani and the stem tissue from cv. Adel from Makarevich. In the grapes of the Mukuzani variety, the class Malasseziomycetes was dominant. In addition, the presence of Oomycetes (6–23%) and Agaricomycetes (4–14%) was found in the samples collected at the Makarevich vineyard. In the samples of grapes from PRIM ORGANICA, the predominating classes were Tremellomycetes and Microbotryomycetes (Figure 3a). The leaf and stem tissues of cv. Prairie Star and cv. Alfa were similar in terms of their intra-plant fungal biodiversity based on UPGMA clustering, while the stem tissue in cv. Adel was similar to the leaf and stem tissues in cv. Mukuzani.
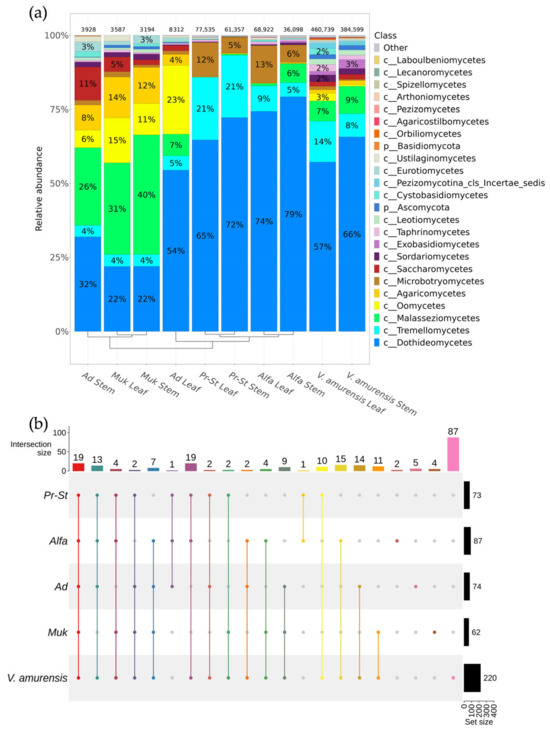
Figure 3.
Comparative analysis of the fungal and fungi-like endophytic community grapevine cultivars and wild grape Vitis amurensis growing in Primorsky Krai of Russia according to genomic approach (NGS). The composition of endophytic fungi and fungi-like microorganisms of grapevine variates Pr-St—Vitis Elmer Swenson 2-7-13 cv. Prairie Star from commercial vineyard PRIM ORGANICA; Alfa—Vitis labrusca × Vitis riparia cv. Alfa from PRIM ORGANICA; Ad—Vitis vinifera × V. amurensis cv. Adele from commercial vineyard Makarevich; Muk—V. riparia × V. vinifera cv. Mukuzani; V.amurensis—the sum of data for all V. amurensis grapevines obtained early [39]: (a) class-level taxonomical bar plots for the fungal and fungi-like endophytic community depend on the variates of grapevines, and the sum of data for all V. amurensis grapevines obtained early [39]; (b) genus-level UpSet diagrams depicting overlapping taxa of NGS in different cultivars of grapevines. For each biocompartment, taxa were filtered based on relative abundance > 0.1%. Taxa with relative frequencies < 0.1% were removed from the UpSet plot. Number of sequences are shown above taxonomical bar plots. For clustering in the bar plots, we used UPGMA.
The representation of the genera of fungi and fungi-like microorganisms varied from 62 to 87 in the cultivated grape varieties, while the representation of genera in the wild grape V. amurensis was 220 genera (Figure 3b and Supplementary Materials Table S4). Among them, 19 taxa were found in all grapevines. The largest number of taxa of genus level were present in the grapevine cv. Alfa from PRIM ORGANICA (87 genera). The 87 fungi and fungi-like genera were unique for wild grape V. amurensis, 2 for grapevine cv. Alfa from PRIM ORGANICA, and 4–5 for each cultivar of grape from the Makarevich vineyard (Figure 3b and Supplementary Materials Table S4).
The most dominant taxa for the grape cv. Adel and cv. Mukuzani from the Makarevich vineyard were Malassezia, Aureobasidium, and Plasmopara (Figure 4). For the grapevines from PRIM ORGANICA, the predominant genera were Alternaria, Aureobasidium, and Cladosporium (Figure 4). The genera Kalmanozyma and Pseudopithomyces were representative for the grapevine cv. PrairiStar, cv. Alfa from PRIM ORGANICA, and wild grape V. amurensis, and absent in the grapevine from Makarevich (Figure 4).
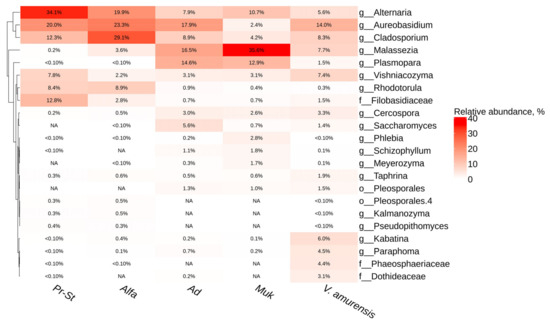
Figure 4.
Genus-level relative endophytic fungi and fungi-like microorganism abundance heat maps of significant taxa according to next-generation sequencing (NGS) in grapevine cultivars and wild grape Vitis amurensis. Pr-St—Vitis Elmer Swenson 2-7-13 cv. Prairie Star from commercial vineyard PRIM ORGANICA; Alfa—Vitis labrusca × Vitis riparia cv. Alfa from PRIM ORGANICA; Ad—Vitis vinifera × V. amurensis cv. Adele from commercial vineyard Makarevich; Muk—V. riparia × V. vinifera cv. Mukuzani; V.amurensis—the sum of data for all V. amurensis grapevines obtained early [39]. The top 10 most abundant taxa from each grapevine are displayed. White squares (NA) represent the absence of taxa.
3.4. A Comparative Analysis of the Endophytic Microbial Communities from Cultivated and Wild Grapevine Varietes
A comparative analysis of the endophytic communities of the cultivated grapevine varieties analyzed in this study with the previously studied endophytic biodiversity in wild grape V. amurensis growing in the Far East of Russia was carried out. A total of 7,462,656 16S and 845,338 ITS1 paired reads were obtained from 50 samples of wild grape V. amurensis (Supplementary Materials Tables S1 and S2).
Figure 5a,b shows the results of the analysis of the alpha and beta bacterial endophytic diversity, respectively. The grapevine cultivar samples are not statistically different based on alpha diversity compared to the samples of the wild grape V. amurensis (Figure 5a).
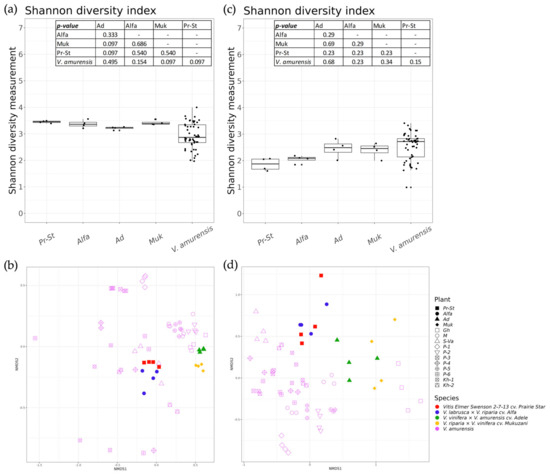
Figure 5.
A comparison of endophytic bacterial, fungi, and fungi-like microorganism communities of grapevine cultivars and wild grape Vitis amurensis. Pr-St—Vitis Elmer Swenson 2-7-13 cv. Prairie Star from commercial vineyard PRIM ORGANICA; Alfa—Vitis labrusca × Vitis riparia cv. Alfa from PRIM ORGANICA; Ad—Vitis vinifera × V. amurensis cv. Adele from commercial vineyard Makarevich; Muk—V. riparia × V. vinifera cv. Mukuzani; V.amurensis—the sum of data for all V. amurensis grapevines obtained early [39]: (a) Shannon’s alpha diversity boxplot of bacterial communities; (b) Bray–Curtis beta diversity NMDS plot of bacterial communities; (c) Shannon’s alpha diversity boxplot of fungi and fungi-like microorganism communities; (d) Bray–Curtis beta diversity NMDS plot of fungi and fungi-like microorganism communities.
The beta diversity was examined using non-metric multidimensional scaling (NMDS) ordination to compare the significant differences and understand the clustering of samples between groups (Figure 5b). The NMDS ordination showed that samples of the grape cultivars were located in separate small clusters, while the V. amurensis samples were more distributed. Samples from one vineyard are closer to each other according to the NMDS ordination plot (Figure 5b). The PERMANOVA test demonstrated that a significant proportion of the variance between bacterial communities is explained when comparing grape varieties with V. amurensis samples (20% of variance, p < 0.001). The factors “location of plants” and “individual plants” explain 50% and 10% of variance between the endophytic microbiomes, respectively (p < 0.001) (Figure 5b and Supplementary Materials Table S5).
Figure 5c,d shows the results of the alpha and beta diversity analysis of the fungi and fungal-like endophytic biodiversity of different grape samples. The alpha diversity of the grape variety samples was not statistically different among each other and from the wild grape V. amurensis samples. Also, the Shannon diversity index median value was slightly lower in the Prairie Star and Alfa grape varietes from PRIM ORGANICA than the median value in V. amurensis, whereas the medians were similar in the V. amurensis samples and the grapes Adel and Mukuzani from the Makarevich vineyard (Figure 5c). NMDS ordination showed that samples of grape cultivars from one vineyard are closer to each other (Figure 5d). The PERMANOVA test demonstrated that the grape cultivars and V. amurensis samples were significantly different based on beta diversity (18% of variance, p < 0.001). The “location of plants” and “individual plants” factors explain 56% and 3% of the variance between endophytic mycobiomes, respectively (p < 0.001) (Figure 5d and Supplementary Materials Table S6).
3.5. Analysis of Associations of Endophytic Bacteria and Fungi in the Vitis Microbiome
Using the microbiome network method, we analyzed the effect of endophytic communities of bacteria and fungi on each other in visually healthy Vitis plants, based on the data presented in Supplementary Materials Tables S1 and S2.
According to the analysis of the microbiome networks, the largest number of associations are observed between bacteria (196) (Figure 6a,b). The number of positive associations is ~three times higher than the number of negative ones (154 “+” vs. 42 “−”). Moreover, the number of positive and negative edges between bacteria and fungi is approximately the same (11 “+” and 12 “−”) and amounts to ~12% of the total number of associations. On the other hand, fungi are significantly less connected to each other compared to bacteria, and have approximately the same number of positive and negative associations with each other (8 “+” and 6 “−”) (Figure 6b).
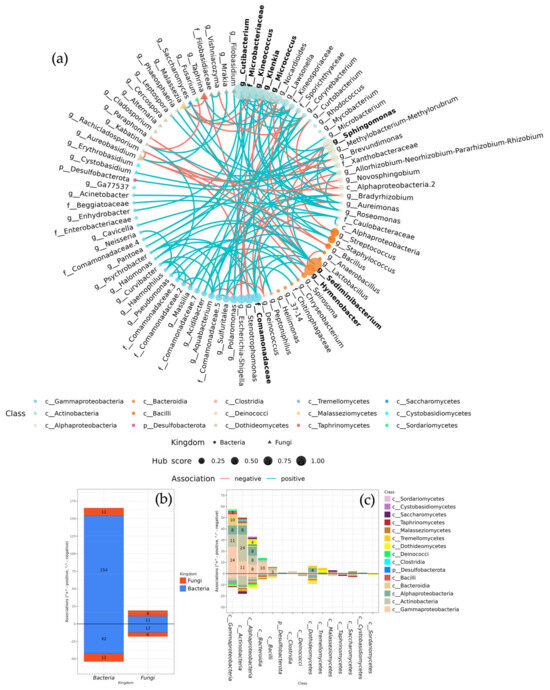
Figure 6.
Positive and negative associations of endophytic bacteria and fungi in the Vitis microbiome. (a) Interdomain ecological networks of the bacterial–fungal associations; (b) summary bar plot of kingdom-level bacteria-fungi associations; (c) summary bar plot of class-level bacteria–fungi associations. The top 10% taxa of genus level based on the values of Kleinberg’s hub centrality score were qualified as hubs. In (a), hub taxa are shown in bold.
Among bacteria, three classes are distinguished in descending order according to the number of positive and negative connections between the taxa at the genus level, Gammaproteobacteria, Actinobacteria, and Alphaproteobacteria, and among fungi—the class Dothideomycetes (Figure 6c). Gamaproteobacteria and Actinobacteria have the highest number of positive intergeneric relationships within their class. The genus-level taxa of the class Actinobacteria have the largest number of negative relationships compared to other classes.
We then discovered hub taxa in the Vitis endophytic microbial community, which have an important influence on the connectivity between the bacterial and fungal taxa (Figure 6a). The class Actinobacteria is characterized by the largest number of genus-level taxa that we marked as hubs, viz Cutibacterium, Microbacteriaceae, Kineococcus, Klenkia, and Micrococcus. The Bacteroidia class has two hubs: Sediminibacterium and Hymenobacter. Alphaproteobacteria and Gammaproteobacteria are characterized by Sphingomonas and Comamonadaceae hubs, respectively. No hubs were found in the fungi.
4. Discussion
The environmental conditions in any wine-growing area can affect the quality of the grapes and lead to variations in the taste and aroma of the wine, even for the same grape varieties [40]. New research has suggested that specific microbial communities associated with V. vinifera may be a key element of the terroir since microbial processes essential to grape growing and wine production show spatial patterns linked to the vineyard site [41]. Also, to reduce plant pathogens and the negative impact of agricultural practices on the environment, biological control is an increasingly successful and widespread strategy. Beneficial microbes, especially endophytic bacteria and fungi, are used to counteract plant pathogens and limit the use of agrochemicals [42].
This study aimed at characterizing the endophytic microbiome of the most common grape varieties of Primorsky Krai, Russia. In general, the endophytic bacterial community composition was presented by the classes Gammaproteobacteria, Alphaproteobacteria, Bacteroidia, and Actinobacteria. The general biodiversity profile of endophytic bacteria correlated with the data from previous studies [20,40]. The most dominant taxon for the grapevines cvs. Adel and Mukuzani from Makarevich was the taxon Comamonadaceae (15–26%) (Figure 2). It is known that the family Comamonadaceae, which are the dominant denitrifiers, might play a primary role in improving the denitrification performance of the environment [43]. Perhaps an increase in the percentage of this taxon of endophytic bacteria indicates an excessive amount of nitrates in the soil of the Makarevich vineyard. Also, in the samples of the grapevine cvs. Adel and Mukuzani, endophytic bacteria Aquabacterium (9–14%) and Spirosoma (7–11%) were found in large numbers (Figure 2). It has been shown early that Aquabacterium sp. A7Y can produce endo-chitosanase AqCoA, which degrades the fungal cell walls, and the resulting oligosaccharides are promising weapons to protect plants from fungal disease [44]. Also, it was shown that Spirosoma sp. are radiation-resistant [45,46] and propanil-degrading bacteria [47]. The community of endophytic bacteria was represented in a significant percentage by the genera Sphingomonas and Methylobacterium-Methylorubrum in the samples of grape cvs. Prairie Star and Alpha from the vineyard PRIM ORGANICA and in the wild grape V. amurensis (Figure 2). The presence of Sphingomonas and Methylobacterium-Methylorubrum bacteria in wines can affect their sensory qualities due to the production of volatile organic compounds, and these compounds contribute to the unique characteristics of the wines [48].
The dominant classes of fungi were Dothideomycetes and Malasseziomycetes in all analyzed samples, which correlated with the endophytic community of fungi in grapes from a vineyard in China [49]. In samples of grapes from the Makarevich vineyard, we identified characteristic dominant genera typically linked with wine and grapes (Malassezia) [49] along with several pathogenic fungi-like genera (Plasmopara) (Figure 4). It is known that powdery mildew, caused by Plasmopara viticola, is a severe disease that leads to significant grape harvest losses worldwide. However, certain American and Asian Vitis species exhibit varying levels of resistance to P. viticola. Some species like V. rupestris show moderate resistance, while others like V. rubra, V. candicans, V. amurensis, V. riparia, V. cinerea, or Muscadinia rotundifolia display high resistance [50,51,52]. Surprisingly, P. viticola can exist within the internal tissues of certain grape species or varieties without causing visible symptoms of powdery mildew. To control fungal-like organisms such as oomycetes (to which P. viticola belongs), the regular application of fungicides is necessary to prevent damage and economic losses [53]. However, the excessive use of fungicides can lead to the development of P. viticola strains that are resistant to these chemicals [54,55]. In the Makarevich vineyard, the presence of Plasmopara in the grape samples could indicate that either the analyzed grape varieties are resistant to this oomycete or that the presence of P. viticola in the tissues does not necessarily result in the manifestation of powdery mildew symptoms. It is likely that continuous chemical treatments against oomycetes at the Makarevich vineyard contribute to a reduction in the number of P. viticola within the grape tissues, preventing the appearance of symptoms. However, these treatments may not completely eliminate the pathogen, allowing it to persist within grape tissues as an endophyte. The near absence of Plasmopara representation in the samples collected at the PRIM ORGANICA vineyard indicates a more effective treatment against this oomycete.
Also, for the grapevines from PRIM ORGANICA, the predominant genera were Alternaria, Aureobasidium, and Cladosporium. Notably, Alternaria is considered one of the main mycobiota populations of grapes at harvest [56,57]. Additionally, the pathogenic fungus Alternaria alternata is known to cause significant post-harvest losses in grapes. This issue has been extensively documented in various reports [58]. Perhaps the analyzed grape samples contained other Alternaria species that occupied the same ecological niches without causing crop losses. Also, contrasting findings indicate that several species of Alternaria can control the growth of different pathogens such as Rhizoctonia solani, Fusarium oxysporum, B. cinerea, and Pseudomonas aeruginosa [59,60]. As in our study, Aureobasidium was the predominant genus also in grapes in other studies [61,62]. It was shown that Aureobasidium pullulans had enzyme activity, such as pectinases, xylanases and cellulases, which encourages future studies regarding their application in winemaking, in particular, in improving the color extraction, technological parameters, and antioxidant activity of wine [63]. In addition, it is known that the Cladosporium sp. is indirectly involved in food spoilage because it produces mycotoxins [64], and their effect on wine quality is due to grape damage. Also, the fungus Cladosporium can withstand a high sugar content and low moisture [49]. Thus, the presence of dominant endophytic fungi of the genera Alternaria, Aureobasidium, and Cladosporium is an indicator that the selected grape varieties are grown in conditions of high humidity, and also that they can be used for winemaking. Also, samples of varieties from the Makarevich vineyard contained unique fungi genera such as Resinicium, Ustilago, Ascotricha (cv. Adele), Daedalea, and Nothophoma (cv. Mukuzani) (Supplementary Materials Table S3). It is known that fungi of the genus Resinicium is a worldwide genus of corticoid wood-inhabiting fungi [65], and Ustilago is the causative agent of corn smut disease and the culprit of considerable losses in grain yields [66]. Also, a unique taxon, Bipolaris, was found in the samples of the cultivar Alpha from PRIM ORGANICA (Supplementary Materials Table S3). Bipolaris sorokiniana (teleomorph, Cochliobolus sativus) is a wheat pathogen [67]. Perhaps the presence of these taxa is due to the proximity of cereal agricultural crops. Purnomo et al. [68] reported that the fungus Daedalea dickinsii has the capability to degrade dichlorodiphenyltrichloroethane (DDT, organochlorine pesticides) via a Fenton reaction. The presence of this taxon in the samples collected at the Makarevich vineyard may be indirect evidence of the presence of DDT in the soil, but this assumption needs to be checked.
According to the comparative analysis of cultivated grape varieties and wild grape V. amurensis endophytic communities, it was found that 30 and 87 unique genera of endophytic bacteria and fungi are represented in wild grapes, respectively (Figure 1b and Figure 3b and Supplementary Materials Tables S3 and S4). The microbial community found in grapes can vary greatly, mainly due to external factors like the environment, location, and specific characteristics of grape varieties [69]. Previous studies have demonstrated that the types and quantities of endophytic bacteria and fungi living inside wild V. amurensis grapes in the Far East of Russia differ significantly depending on the plant’s organs and weather conditions [20,21]. Leaves and stems tend to have the highest number of these microorganisms. The presence of an average temperature around 15 °C and ample precipitation seem to contribute to both the diversity and abundance of endophytic bacteria and fungi in V. amurensis. Conversely, hot and dry weather can lead to a significant decrease in the number of these microorganisms. Additionally, the use of various chemical treatments can also have a significant impact on the composition of the grape’s endophytic community. The lowest representation of taxa of endophytic bacteria and fungi compared with the wild grape V. amurensis is explained by the annual treatment of vineyards with chemicals against grape pathogens. However, in addition to pathogenic microorganisms, the wild grape V. amurensis may contain endophytic microorganisms that help the vine to withstand abiotic and biotic stresses. Therefore, the study of endophytes unique to wild grapes is an interesting task that requires future research.
Understanding the structure and nature of associations in the microbiome of healthy grapevines is important for the development of methods to make both wild and cultivated forms of Vitis more resistant to biotic stresses. The analysis of microbial associations between bacteria and fungi in visually healthy grapevine plant communities revealed mainly positive associations between bacteria–bacteria interactions and only a relatively small number of bacteria–fungi and fungi–fungi interactions. The classes Gammaproteobacteria, Actinobacteria, and Alphaproteobacteria had the strongest influence on the connectivity of the microbiome network. In microbial communities, hub taxa, characterized by high hub scores, are important as their removal can significantly affect the connectivity of the network [70,71]. Notably, the Actinobacteria class has the highest number of hub-taxa in our data, namely Cutibacterium, Microbacteriaceae, Kineococcus, Klenkia, and Micrococcus. Actinobacteria are known to be actively involved in stimulating plant growth and increasing disease resistance via beneficial interactions with the plant organism [72]. Further research is needed to explore the intriguing study of endophytic bacterial hubs of healthy grapes in the context of their role in microbiome networks associated with Vitis diseases.
5. Conclusions
This research focused on examining the diversity of endophytic bacteria and fungi found in cultivated grape varieties grown in the vineyards of Primorsky Krai, Russia. It was the first study of its kind in this region. According to our findings, around 18–20% of the variation in endophytic communities can be attributed to the disparities between cultivated and wild grapevines. Additionally, plant location and individual plants contribute to 50–56% and 3–10% of the variation, respectively. This suggests that factors like the environment in which the plants are grown and the specific characteristics of each plant play a significant role in shaping the diversity of the endophytic communities in grapevines. In addition, a comprehensive analysis was undertaken to investigate the microbial composition and inherent relationships within the grapevine microbiome. This research has significant potential to advance the use of endophytic microorganisms to enhance grapevine productivity and improve the resilience of cultivated Vitis species to various biotic and abiotic stresses.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae9121257/s1: Table S1: 16S data samples used in analysis; Table S2: ITS data samples used in analysis; Table S3: Intersections in 16S metagenome data of cultivars and V. amurensis plant samples; Table S4: Intersections in ITS metagenome data of cultivars and V. amurensis plant samples; Table S5: PERMANOVA results (16s data); Table S6: PERMANOVA results (ITS data).
Author Contributions
O.A.A. and K.V.K. performed the research design, data analysis, paper preparation, and experimental process. O.A.A., A.R.S. and P.A.C. collected the material. O.A.A., A.A.A., A.R.S., A.A.D., A.A.B. and N.N.N. performed the isolation of DNA for NGS. N.N.N. and Z.V.O. performed the bioinformatic analysis and visualization. A.S.D. was responsible for the writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the Russian Science Foundation (grant number 22–74–10001, https://rscf.ru/project/22-74-10001, accessed on 18 October 2023).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article and Supplementary Materials.
Acknowledgments
We would like to thank Sergei Makarevich and Alexandr and Anna Storozhenko for their primary contribution to the sample collection for the present study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Felber, A.C.; Orlandelli, R.C.; Rhoden, S.A.; Garcia, A.; Costa, A.T.; Azevedo, J.L.; Pamphile, J.A. Bioprospecting Foliar Endophytic Fungi of Vitis Labrusca Linnaeus, Bordô and Concord Cv. Ann. Microbiol. 2016, 66, 765–775. [Google Scholar] [CrossRef]
- Soejima, A.; Wen, J. Phylogenetic Analysis of the Grape Family (Vitaceae) Based on Three Chloroplast Markers. Am. J. Bot. 2006, 93, 278–287. [Google Scholar] [CrossRef]
- Compant, S.; Brader, G.; Muzammil, S.; Sessitsch, A.; Lebrihi, A.; Mathieu, F. Use of Beneficial Bacteria and Their Secondary Metabolites to Control Grapevine Pathogen Diseases. BioControl 2013, 58, 435–455. [Google Scholar] [CrossRef]
- Swift, J.F.; Hall, M.E.; Harris, Z.N.; Kwasniewski, M.T.; Miller, A.J. Grapevine Microbiota Reflect Diversity among Compartments and Complex Interactions within and among Root and Shoot Systems. Microorganisms 2021, 9, 92. [Google Scholar] [CrossRef] [PubMed]
- Bustamante, M.I.; Elfar, K.; Eskalen, A. Evaluation of the Antifungal Activity of Endophytic and Rhizospheric Bacteria against Grapevine Trunk Pathogens. Microorganisms 2022, 10, 2035. [Google Scholar] [CrossRef]
- Jiao, S.; Zeng, F.; Huang, Y.; Zhang, L.; Mao, J.; Chen, B. Physiological, Biochemical and Molecular Responses Associated with Drought Tolerance in Grafted Grapevine. BMC Plant Biol. 2023, 23, 110. [Google Scholar] [CrossRef]
- Ren, C.; Fan, P.; Li, S.; Liang, Z. Advances in Understanding Cold Tolerance in Grapevine. Plant Physiol. 2023, 192, 1733–1746. [Google Scholar] [CrossRef]
- Pertot, I.; Caffi, T.; Rossi, V.; Mugnai, L.; Hoffmann, C.; Grando, M.S.; Gary, C.; Lafond, D.; Duso, C.; Thiery, D.; et al. A Critical Review of Plant Protection Tools for Reducing Pesticide Use on Grapevine and New Perspectives for the Implementation of IPM in Viticulture. Crop Prot. 2017, 97, 70–84. [Google Scholar] [CrossRef]
- Carvalho, F.P. Pesticides, Environment, and Food Safety. Food Energy Secur. 2017, 6, 48–60. [Google Scholar] [CrossRef]
- Knapp, D.G.; Lázár, A.; Molnár, A.; Vajna, B.; Karácsony, Z.; Váczy, K.Z.; Kovács, G.M. Above-Ground Parts of White Grapevine Vitis vinifera cv. Furmint Share Core Members of the Fungal Microbiome. Environ. Microbiol. Rep. 2021, 13, 509–520. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Kiselev, K.V. Interaction of Plants and Endophytic Microorganisms: Molecular Aspects, Biological Functions, Community Composition, and Practical Applications. Plants 2023, 12, 714. [Google Scholar] [CrossRef] [PubMed]
- Hamaoka, K.; Aoki, Y.; Suzuki, S. Isolation and Characterization of Endophyte Bacillus velezensis KOF112 from Grapevine Shoot Xylem as Biological Control Agent for Fungal Diseases. Plants 2021, 10, 1815. [Google Scholar] [CrossRef] [PubMed]
- Mikheev, V.S.; Struchkova, I.V.; Ageyeva, M.N.; Brilkina, A.A.; Berezina, E.V. The Role of Phialocephala fortinii in Improving Plants’ Phosphorus Nutrition: New Puzzle Pieces. J. Fungi 2022, 8, 1225. [Google Scholar] [CrossRef]
- Ma, Y.; Jiao, J.; Fan, X.; Sun, H.; Zhang, Y.; Jiang, J.; Liu, C. Endophytic Bacterium Pseudomonas fluorescens RG11 May Transform Tryptophan to Melatonin and Promote Endogenous Melatonin Levels in the Roots of Four Grape Cultivars. Front. Plant Sci. 2017, 7, 2608. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant Growth-Promoting Bacteria That Confer Resistance to Water Stress in Tomatoes and Peppers. Plant Sci. 2004, 166, 525–530. [Google Scholar] [CrossRef]
- Theocharis, A.; Bordiec, S.; Fernandez, O.; Paquis, S.; Dhondt-Cordelier, S.; Baillieul, F.; Clément, C.; Barka, E.A. Burkholderia phytofirmans PsJN Primes Vitis Vinifera L. and Confers a Better Tolerance to Low Nonfreezing Temperatures. MPMI 2012, 25, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among Wine Grape Microbiome, Metabolome, and Fermentation Behavior Suggest Microbial Contribution to Regional Wine Characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef]
- Novello, G.; Gamalero, E.; Bona, E.; Boatti, L.; Mignone, F.; Massa, N.; Cesaro, P.; Lingua, G.; Berta, G. The Rhizosphere Bacterial Microbiota of Vitis vinifera cv. Pinot Noir in an Integrated Pest Management Vineyard. Front. Microbiol. 2017, 8, 1528. [Google Scholar] [CrossRef] [PubMed]
- Pacifico, D.; Squartini, A.; Crucitti, D.; Barizza, E.; Lo Schiavo, F.; Muresu, R.; Carimi, F.; Zottini, M. The Role of the Endophytic Microbiome in the Grapevine Response to Environmental Triggers. Front. Plant Sci. 2019, 10, 1256. [Google Scholar] [CrossRef] [PubMed]
- Aleynova, O.A.; Nityagovsky, N.N.; Dubrovina, A.S.; Kiselev, K.V. The Biodiversity of Grapevine Bacterial Endophytes of Vitis amurensis Rupr. Plants 2022, 11, 1128. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Nityagovsky, N.N.; Suprun, A.R.; Ananev, A.A.; Dubrovina, A.S.; Kiselev, K.V. The Diversity of Fungal Endophytes from Wild Grape Vitis amurensis Rupr. Plants 2022, 11, 2897. [Google Scholar] [CrossRef] [PubMed]
- Kiselev, K.V.; Nityagovsky, N.N.; Aleynova, O.A. A Method of DNA Extraction from Plants for Metagenomic Analysis Based on the Example of Grape Vitis amurensis Rupr. Appl. Biochem. Microbiol. 2023, 59, 361–367. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Abarenkov, K.; Zirk, A.; Piirmann, T.; Pöhönen, R.; Ivanov, F.; Nilsson, R.H.; Kõljalg, U. UNITE QIIME Release for Eukaryotes; Version 29.11.2022; UNITE Community: London, UK, 2022. [Google Scholar]
- Bisanz, J. qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. 2018. Available online: https://github.com/jbisanz/qiime2R (accessed on 18 October 2023).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- De Vries, A.; Ripley, B.D. ggdendro: Create Dendrograms and Tree Diagrams Using “ggplot2”. 2023. Available online: https://andrie.github.io/ggdendro/ (accessed on 18 October 2023).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes. 2022. Available online: https://cran.r-project.org/web/packages/RColorBrewer/ (accessed on 18 October 2023).
- Gu, Z. Circlize: Circular Visualization. 2022. Available online: https://cran.r-project.org/web/packages/circlize/ (accessed on 18 October 2023).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Pedersen, T.L. Tidygraph: A Tidy API for Graph Manipulation. 2023. Available online: https://tidygraph.data-imaginist.com/ (accessed on 18 October 2023).
- Pedersen, T.L. ggraph: An Implementation of Grammar of Graphics for Graphs and Networks. 2022. Available online: https://ggraph.data-imaginist.com/ (accessed on 18 October 2023).
- Peschel, S.; Müller, C.L.; von Mutius, E.; Boulesteix, A.-L.; Depner, M. NetCoMi: Network Construction and Comparison for Microbiome Data in R. Brief. Bioinform. 2021, 22, bbaa290. [Google Scholar] [CrossRef]
- Kurtz, Z.D.; Müller, C.L.; Miraldi, E.R.; Littman, D.R.; Blaser, M.J.; Bonneau, R.A. Sparse and Compositionally Robust Inference of Microbial Ecological Networks. PLoS Comput. Biol. 2015, 11, e1004226. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://CRAN.R-project.org/package=vegan (accessed on 18 October 2023).
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Aleynova, O.A.; Nityagovsky, N.N.; Ananev, A.A.; Suprun, A.R.; Ogneva, Z.V.; Dneprovskaya, A.A.; Beresh, A.A.; Tyunin, A.P.; Dubrovina, A.S.; Kiselev, K.V. The Endophytic Microbiome of Wild Grapevines Vitis amurensis Rupr. and Vitis coignetiae Pulliat Growing in the Russian Far East. Plants 2023, 12, 2952. [Google Scholar] [CrossRef]
- Hamaoka, K.; Aoki, Y.; Takahashi, S.; Enoki, S.; Yamamoto, K.; Tanaka, K.; Suzuki, S. Diversity of Endophytic Bacterial Microbiota in Grapevine Shoot Xylems Varies Depending on Wine Grape-Growing Region, Cultivar, and Shoot Growth Stage. Sci. Rep. 2022, 12, 15772. [Google Scholar] [CrossRef]
- Nanetti, E.; Palladino, G.; Scicchitano, D.; Trapella, G.; Cinti, N.; Fabbrini, M.; Cozzi, A.; Accetta, G.; Tassini, C.; Iannaccone, L.; et al. Composition and Biodiversity of Soil and Root-Associated Microbiome in Vitis vinifera Cultivar Lambrusco Distinguish the Microbial Terroir of the Lambrusco DOC Protected Designation of Origin Area on a Local Scale. Front. Microbiol. 2023, 14, 1108036. [Google Scholar] [CrossRef]
- Nigris, S.; Baldan, E.; Tondello, A.; Zanella, F.; Vitulo, N.; Favaro, G.; Guidolin, V.; Bordin, N.; Telatin, A.; Barizza, E.; et al. Biocontrol Traits of Bacillus licheniformis GL174, a Culturable Endophyte of Vitis vinifera cv. Glera. BMC Microbiol. 2018, 18, 133. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, W.; Li, C.; Wang, H.; Wang, H.; Ling, Y.; Yan, G.; Chang, Y. Effects of Antibiotics on Corncob Supported Solid-Phase Denitrification: Denitrification and Antibiotics Removal Performance, Mechanism, and Antibiotic Resistance Genes. J. Environ. Sci. 2023, 130, 24–36. [Google Scholar] [CrossRef]
- Wang, Y.; Li, D.; Liu, M.; Xia, C.; Fan, Q.; Li, X.; Lan, Z.; Shi, G.; Dong, W.; Li, Z.; et al. Preparation of Active Chitooligosaccharides with a Novel Chitosanase AqCoA and Their Application in Fungal Disease Protection. J. Agric. Food Chem. 2021, 69, 3351–3361. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Jung, J.-H.; Kim, M.-K.; Choe, H.N.; Seong, C.N.; Lim, S. Spirosoma taeanense sp. nov., a Radiation Resistant Bacterium Isolated from a Coastal Sand Dune. Antonie Van Leeuwenhoek 2021, 114, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Maeng, S.; Damdintogtokh, T.; Zhang, J.; Kim, M.-K.; Srinivasan, S.; Kim, M.K. Spirosoma profusum sp. nov., and Spirosoma validum sp. nov., Radiation-Resistant Bacteria Isolated from Soil in South Korea. Antonie Van Leeuwenhoek 2021, 114, 1155–1164. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, X.-Y.; Su, X.-J.; Hu, Q.; Jiang, J.-D. Spirosoma sordidisoli sp. nov., a Propanil-Degrading Bacterium Isolated from a Herbicide-Contaminated Soil. Antonie Van Leeuwenhoek 2019, 112, 1523–1532. [Google Scholar] [CrossRef]
- Wassermann, B.; Korsten, L.; Berg, G. Plant Health and Sound Vibration: Analyzing Implications of the Microbiome in Grape Wine Leaves. Pathogens 2021, 10, 63. [Google Scholar] [CrossRef]
- Zhu, L.; Li, T.; Xu, X.; Shi, X.; Wang, B. Succession of Fungal Communities at Different Developmental Stages of Cabernet Sauvignon Grapes From an Organic Vineyard in Xinjiang. Front. Microbiol. 2021, 12, 718261. [Google Scholar] [CrossRef] [PubMed]
- Olmo, H.P. Vinifera rotundifolia Hybrids as Wine Grapes. Am. J. Enol. Vitic. 1971, 22, 87–91. [Google Scholar] [CrossRef]
- Staudt, G.; Kassemeyer, H. Evaluation of Downy Mildew Resistance in Various Accessions of Wild Vitis Species. Vitis 1995, 34, 225–228. [Google Scholar]
- Díez-Navajas, A.M.; Wiedemann-Merdinoglu, S.; Greif, C.; Merdinoglu, D. Nonhost Versus Host Resistance to the Grapevine Downy Mildew, Plasmopara viticola, Studied at the Tissue Level. Phytopathology 2008, 98, 776–780. [Google Scholar] [CrossRef] [PubMed]
- Massi, F.; Torriani, S.F.F.; Borghi, L.; Toffolatti, S.L. Fungicide Resistance Evolution and Detection in Plant Pathogens: Plasmopara viticola as a Case Study. Microorganisms 2021, 9, 119. [Google Scholar] [CrossRef]
- Toffolatti, S.L.; Serrati, L.; Sierotzki, H.; Gisi, U.; Vercesi, A. Assessment of QoI Resistance in Plasmopara viticola Oospores. Pest Manag. Sci. 2007, 63, 194–201. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.; Kong, F.; van der Lee, T.; Wang, Z.; Zhang, H. Detection and Characterization of Carboxylic Acid Amide-Resistant Plasmopara viticola in China Using a TaqMan-MGB Real-Time PCR. Plant Dis. 2020, 104, 2338–2345. [Google Scholar] [CrossRef]
- Tournas, V.H.; Katsoudas, E. Mould and Yeast Flora in Fresh Berries, Grapes and Citrus Fruits. Int. J. Food Microbiol. 2005, 105, 11–17. [Google Scholar] [CrossRef]
- Ding, Y.; Wei, R.; Wang, L.; Wang, W.; Wang, H.; Li, H. Exploring the Ecological Characteristics of Natural Microbial Communities along the Continuum from Grape Berries to Winemaking. Food Res. Int. 2023, 167, 112718. [Google Scholar] [CrossRef]
- Shi, J.; Huang, D.; Du, Y.; Zhu, S.; Hussain, Z.; Haider, M.S.; Anwar, R. Effects of Exogenous Nitric Oxide Treatment on Grape Berries Against Botrytis cinerea and Alternaria alternata Related Enzymes and Metabolites. Plant Dis. 2023, 107, 1510–1521. [Google Scholar] [CrossRef]
- Yu, X.; Shentu, X.; Dong, S.; Hao, P.; Bian, Y.; Ma, Z. Application of Metabolites of Alternaria alternata 31 in Preventing and Treating Rhizoctonia solani, Fusarium oxysporium, and Botrytis cinerea. CN Patent 102,204,570 A, 5 October 2011. [Google Scholar]
- Ortega, H.E.; Torres-Mendoza, D.; Cubilla-Rios, L. Patents on Endophytic Fungi for Agriculture and Bio- and Phytoremediation Applications. Microorganisms 2020, 8, 1237. [Google Scholar] [CrossRef] [PubMed]
- Grube, M.; Schmid, F.; Berg, G. Black Fungi and Associated Bacterial Communities in the Phyllosphere of Grapevine. Fungal Biol. 2011, 115, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Lu, Y.; Chi, Z.; Liu, G.-L.; Jiang, H.; Hu, Z.; Chi, Z.-M. Genome Editing of Different Strains of Aureobasidium melanogenum Using an Efficient Cre/Loxp Site-Specific Recombination System. Fungal Biol. 2019, 123, 723–731. [Google Scholar] [CrossRef]
- Longhi, S.J.; Martín, M.C.; Merín, M.G.; Morata De Ambrosini, V.I. Yeast Multi-Enzymatic Systems for Improving Colour Extraction, Technological Parameters and Antioxidant Activity of Wine. Food Technol. Biotechnol. 2022, 60, 556–570. [Google Scholar] [CrossRef]
- Martins, G.; Vallance, J.; Mercier, A.; Albertin, W.; Stamatopoulos, P.; Rey, P.; Lonvaud, A.; Masneuf-Pomarède, I. Influence of the Farming System on the Epiphytic Yeasts and Yeast-like Fungi Colonizing Grape Berries during the Ripening Process. Int. J. Food Microbiol. 2014, 177, 21–28. [Google Scholar] [CrossRef]
- Yu, J.; Wang, X.-W.; Liu, S.-L.; Shen, S.; Zhou, L.-W. Taxonomy and Phylogeny of Resinicium sensu lato from Asia-Pacific Revealing a New Genus and Five New Species (Hymenochaetales, Basidiomycota). IMA Fungus 2021, 12, 19. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G. Plant Pathology, 4th ed.; Academic Press: London, UK, 1997; Volume 144, p. 146. [Google Scholar]
- Al-Sadi, A.M. Bipolaris Sorokiniana-Induced Black Point, Common Root Rot, and Spot Blotch Diseases of Wheat: A Review. Front. Cell. Infect. Microbiol. 2021, 11, 584899. [Google Scholar] [CrossRef]
- Purnomo, A.S.; Kamei, I.; Kondo, R. Degradation of 1,1,1-Trichloro-2,2-Bis (4-Chlorophenyl) Ethane (DDT) by Brown-Rot Fungi. J. Biosci. Bioeng. 2008, 105, 614–621. [Google Scholar] [CrossRef]
- Soliman, S.S.M.; Greenwood, J.S.; Bombarely, A.; Mueller, L.A.; Tsao, R.; Mosser, D.D.; Raizada, M.N. An Endophyte Constructs Fungicide-Containing Extracellular Barriers for Its Host Plant. Curr. Biol. 2015, 25, 2570–2576. [Google Scholar] [CrossRef]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–Microbiome Interactions: From Community Assembly to Plant Health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Palaniyandi, S.A.; Yang, S.H.; Zhang, L.; Suh, J.-W. Effects of Actinobacteria on Plant Disease Suppression and Growth Promotion. Appl. Microbiol. Biotechnol. 2013, 97, 9621–9636. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).