Community Structure and Diversity of Endophytic Bacteria in Melon (Cucumis melo L.) Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Cultivable Endophytic Bacteria Isolation and Growth Conditions
2.3. Extraction of DNA
2.4. Amplification of 16S rDNA Gene Fragments
2.5. Library Construction and Sequencing
2.6. Sequencing Data Analysis
2.7. 16S Functional Gene Prediction
3. Results
3.1. Assessment of Sequencing Data
3.2. Counts of Endophytic Bacterial in Melon Seeds
3.3. Species Diversity Analysis for Melon Seed Endophytes
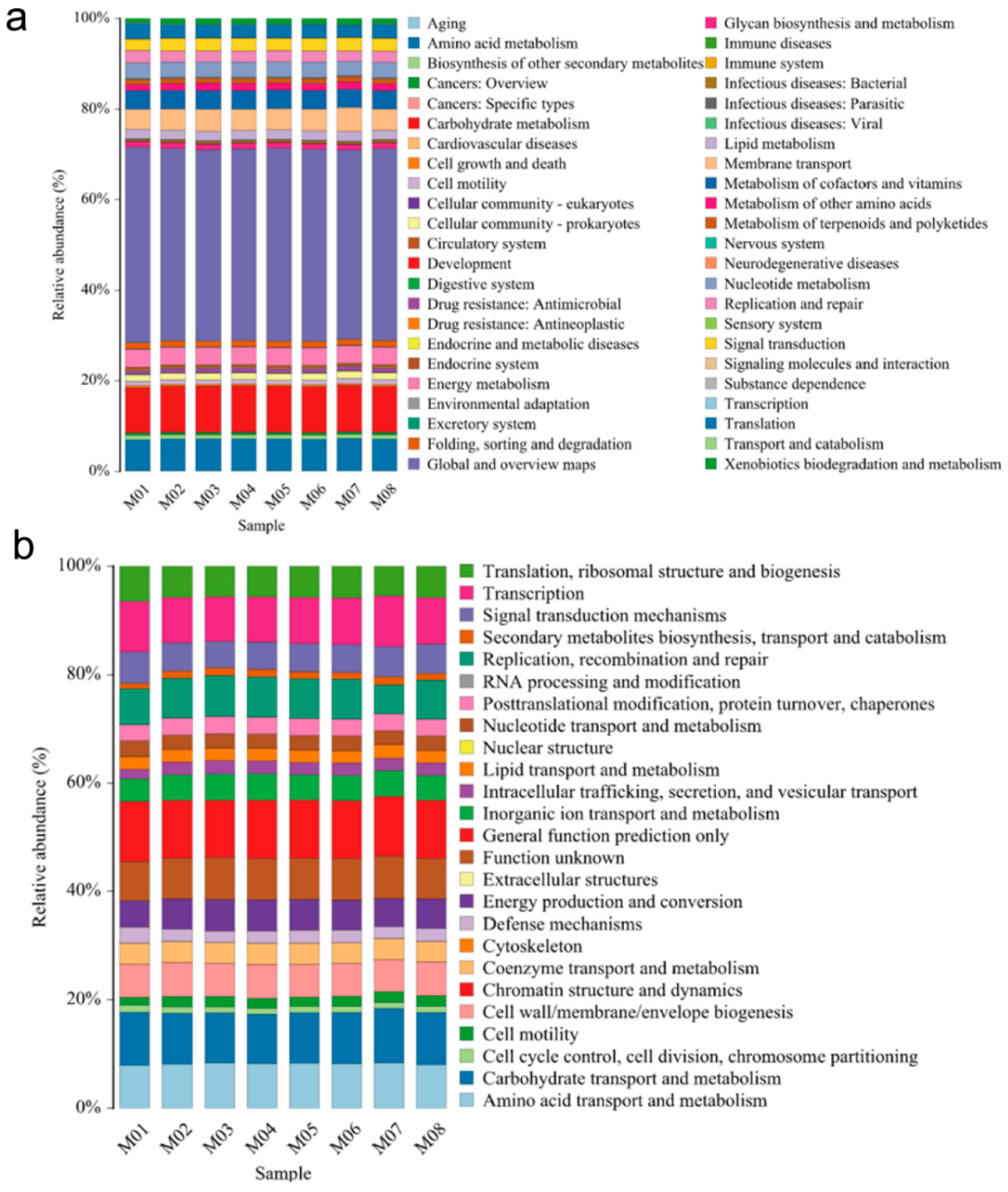
3.4. Prediction and Analysis of Functional Genes from Melon Seed Endophytes
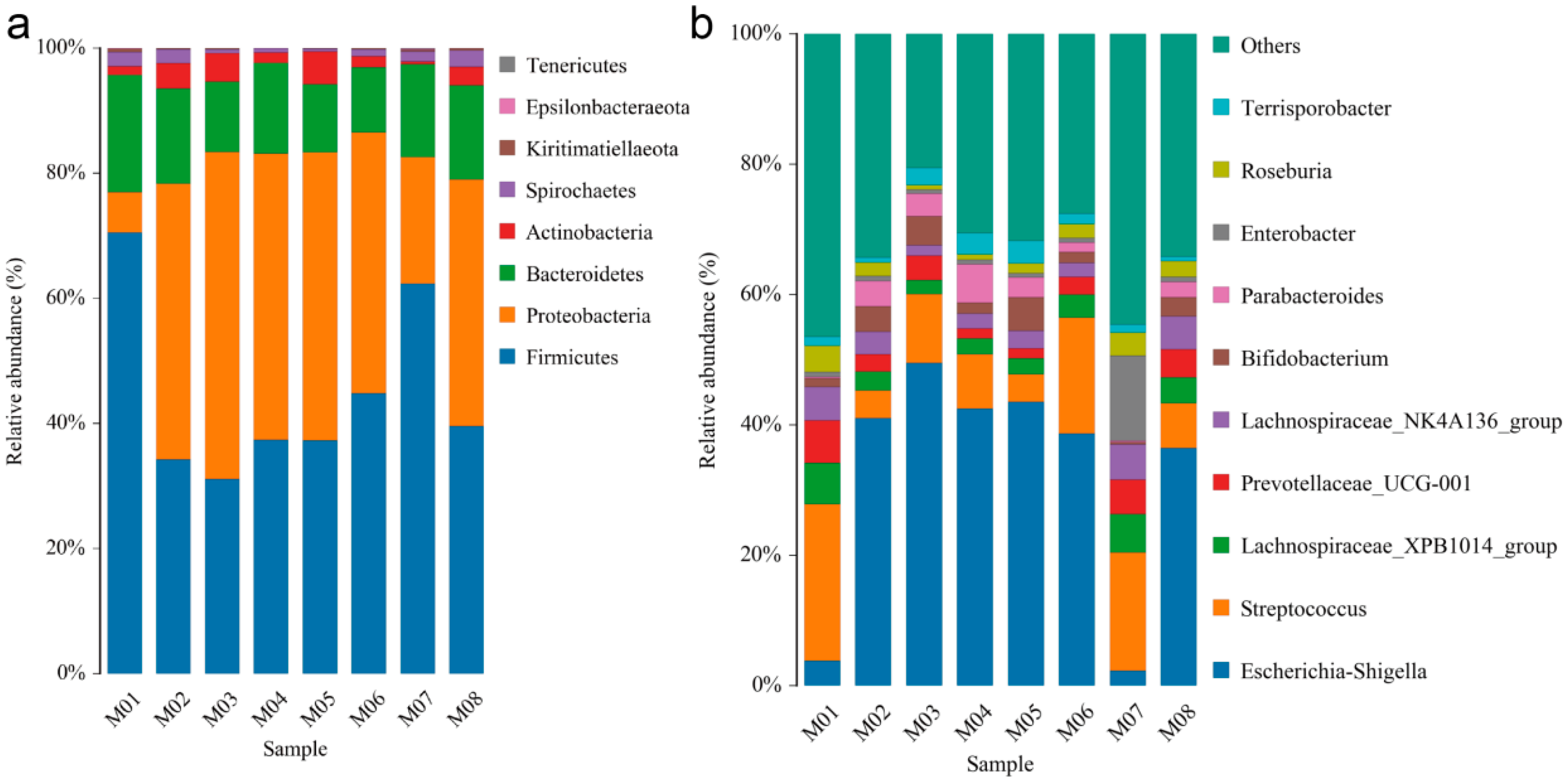
3.5. Structural Analysis of Endophytic Community in Melon Seeds
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chikh-Rouhou, H.; Abdedayem, W.; Solmaz, I.; Sari, N.; Garcés-Claver, A. Melon (Cucumis melo L.): Genomics and Breeding. In Smart Plant Breeding for Vegetable Crops in Post-Genomics Era; Springer Nature Singapore: Singapore, 2023; pp. 25–52. [Google Scholar]
- Klomchit, A.; Calderin, J.D.; Jaidee, W.; Watla-iad, K.; Brooks, S. Napthoquinones from Neocosmospora sp.—Antibiotic Activity against Acidovorax citrulli, the Causative Agent of Bacterial Fruit Blotch in Watermelon and Melon. J. Fungi. 2021, 7, 370. [Google Scholar]
- Burdman, S.; Walcott, R. Acidovorax citrulli: Generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 2012, 13, 805–815. [Google Scholar] [PubMed]
- Wang, H.; Qian, C.; Jiang, H.; Liu, S.; Yang, D.; Cui, J. Visible-Light-Driven Zinc Oxide Quantum Dots for the Management of Bacterial Fruit Blotch Disease and the Improvement of Melon Seedlings Growth. J. Agric. Food Chem. 2023, 71, 2773–2783. [Google Scholar] [CrossRef] [PubMed]
- Ning, X.F. Identification of resistance to Acidovorax citrulli among different melon accessions and detection of QTLs for bacterial fruit blotch and downy mildew resistance in melon. Ph.D. Thesis, Xinjiang University, Xinjiang, China, 2018. [Google Scholar]
- Bahar, O.; Kritzman, G.; Burdman, S. Bacterial fruit blotch of melon: Screens for disease tolerance and role of seed transmission in pathogenicity. Eur. J. Plant Pathol. 2009, 123, 71–83. [Google Scholar] [CrossRef]
- Wani, Z.A.; Ashraf, N.; Mohiuddin, T.; Riyaz-Ul-Hassan, S. Plant-endophyte symbiosis, an ecological perspective. Appl Microbiol Biotechnol. 2015, 99, 2955–2965. [Google Scholar] [PubMed]
- Thomas, P.; Sahu, P.K. Vertical transmission of diverse cultivation-recalcitrant endophytic bacteria elucidated using watermelon seed embryos. Front. Microbiol. 2021, 12, 635810. [Google Scholar]
- Mastretta, C.; Taghavi, S.; van der Lelie, D.; Mengoni, A.; Galardi, F.; Gonnelli, C.; Vangronsveld, J. Endophytic bacteria from seeds of Nicotiana tabacum can reduce cadmium phytotoxicity. Int. J. Phytoremed. 2009, 11, 251–267. [Google Scholar] [CrossRef]
- Puente, M.E.; Li, C.Y.; Bashan, Y. The development of cactus seedlings can be improved by the presence of endophytic bacteria in cactus seeds. Environ. Exp. Bot. 2009, 66, 402–408. [Google Scholar] [CrossRef]
- Verma, S.; Kingsley, K.; Irizarry, I.; Bergen, M.; Kharwar, R.N.; White, J. Seed-vectored endophytic bacteria modulate development of rice seedlings. J. Appl. Microbiol. 2017, 122, 1680–1691. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, E.M.; Raizada, M.N. Bacterial Seed Endophytes of Domesticated Cucurbits Antagonize Fungal and Oomycete Pathogens Including Powdery Mildew. Front. Microbiol. 2018, 9, 42. [Google Scholar]
- Fessehaie, A.; Walcott, R.R. Biological Control to Protect Watermelon Blossoms and Seed from Infection by Acidovorax avenae subsp. citrulli. Phytopathology 2005, 95, 413–419. [Google Scholar]
- Jiang, C.-H.; Wu, F.; Yu, Z.-Y.; Xie, P.; Ke, H.-J.; Li, H.-W.; Yu, Y.-Y.; Guo, J.-H. Study on screening and antagonistic mechanisms of Bacillus amyloliquefaciens 54 against bacterial fruit blotch (BFB) caused by Acidovorax avenae subsp. citrulli. Microbiol. Res. 2015, 170, 95–104. [Google Scholar] [CrossRef]
- de Melo, E.A.; Mariano, R.d.L.R.; Laranjeira, D.; dos Santos, L.A.; Gusmão, L.d.O.; de Souza, E.B. Efficacy of Yeast in the Biocontrol of Bacterial Fruit Blotch in Melon Plants. Trop. Plant Pathol. 2015, 40, 56–64. [Google Scholar] [CrossRef]
- Edwards, J.; Santos-Medellín, C.; Sundaresan, V. Extraction and 16S rRNA sequence analysis of microbiomes associated with rice roots. Bio-Protocol 2018, 8, e2884. [Google Scholar] [CrossRef] [PubMed]
- Majeed, A.; Kaleem Abbasi, M.; Hameed, S.; Imran, A.; Rahim, N. Isolation and characterization of plant growth-promoting rhizobacteria from wheat rhizosphere and their effect on plant growth promotion. Front. Microbiol. 2015, 6, 198. [Google Scholar]
- Qaisrani, M.M.; Mirza, M.S.; Zaheer, A.; Malik, K.A. Isolation and identification by 16s rRNA sequence analysis of achromobacter, azospirillum and rhodococcus strains from the rhizosphere of maize and screening for the beneficial effect on plant growth. Pak. J. Agric. Sci. 2014, 51, 91–99. [Google Scholar]
- Oliveira, C.; Shakiba, E.; North, D.; McGraw, M.; Ballard, E.; Barrett-D’amico, M.; Glazko, G.; Rahmatallah, Y. 16S rRNA gene-based metagenomic analysis of rhizosphere soil bacteria in arkansas rice crop fields. Agronomy 2022, 12, 222. [Google Scholar] [CrossRef]
- Du, K.; Geng, Y.; Liu, L.; Gao, D.M. Community structure and diversity of endophytes of Bupleurum chinense DC. seeds. Biotic Resour. 2022, 44, 36–44. [Google Scholar]
- Yang, L.; Wu, Q.; Gao, Y.; Chen, Q.; Wang, Y.; Niu, X.; Weng, Q. Analysis of the composition of endophytic community of Dendrobium nobile Lindl. seeds based on high-throughput sequencing. Seed 2020, 39, 94–98. [Google Scholar]
- Gerna, D.; Clara, D.; Allwardt, D.; Mitter, B.; Roach, T. Tailored media are key to unlocking the diversity of endophytic bacteria in distinct compartments of germinating seeds. Microbiol. Spectr. 2022, 10, e00172-22. [Google Scholar]
- Sha, W.; Hong, D.; Che, Y.; Xue, Y.; Kong, Y.; Yi, X.; Liu, B. Differences in Root Endophytic Bacterial Communities of Chinese Cork Oak (Quercus variabilis) Seedlings in Different Growth Years. Forests 2023, 14, 1489. [Google Scholar] [CrossRef]
- Li, J.; Wang, S.; Yu, J.; Wang, L.; Zhou, S. A modified CTAB protocol for plant DNA extraction. Chin. Bull. Bot. 2013, 48, 72. [Google Scholar]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [PubMed]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Knights, D. BugBase predicts organism-level microbiome phenotypes. BioRxiv 2017, 133462, 1–19. [Google Scholar]
- Roswell, M.; Dushoff, J.; Winfree, R. A conceptual guide to measuring species diversity. Oikos 2021, 130, 321–338. [Google Scholar] [CrossRef]
- Hemme, C.L.; Deng, Y.; Gentry, T.J.; Fields, M.W.; Wu, L.; Barua, S.; Barry, K.; Tringe, S.G.; Watson, D.B.; He, Z.; et al. Metagenomic insights into evolution of a heavy metal-contaminated groundwater microbial community. ISME J. 2010, 4, 660–672. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed]
- Saikkonen, K.; Wali, P.R.; Helander, M. Genetic compatibility determines endophyte-grass combinations. PLoS ONE 2010, 5, e11395. [Google Scholar] [CrossRef]
- Hume, D.; Schmid, J.; Rolston, M.; Vijayan, P.; Hickey, M. Effect of climatic conditions on endophyte and seed viability in stored ryegrass seed. Seed Sci. Technol. 2011, 39, 481–489. [Google Scholar] [CrossRef]
- Welty, R.E. Influence of moisture content, temperature, and length of storage on seed germination and survival of endophytic fungi in seeds of tall fescue and perennial ryegrass. Phytopathology 1987, 77, 893. [Google Scholar] [CrossRef]
- Zhang, Z.; Wang, X.; Han, S.; Liu, C.; Liu, F. Effect of two seaweed polysaccharides on intestinal microbiota in mice evaluated by illumina PE250 sequencing. Int. J. Biol. Macromol. 2018, 112, 796–802. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic profiling of the community structure, diversity, and nutrient pathways of bacterial endophytes in maize plant. Antonie Van Leeuwenhoek 2020, 113, 1559–1571. [Google Scholar] [PubMed]
- Purushotham, N.; Jones, E.; Monk, J.; Ridgway, H. Community Structure, Diversity and Potential of Endophytic Bacteria in the Primitive New Zealand Medicinal Plant Pseudowintera colorata. Plants 2020, 9, 156. [Google Scholar] [CrossRef]
- Park, K.; Paul, D.; Kim, E.; Kloepper, J.W. Hyaluronic acid of Streptococcus sp. as a potent elicitor for induction of systemic resistance against plant diseases. World J. Microbiol. Biotechnol. 2007, 24, 1153–1158. [Google Scholar] [CrossRef]
- Seipke, R.F.; Kaltenpoth, M.; Hutchings, M.I. Streptomyces as symbionts: An emerging and widespread theme? FEMS Microbiol. Rev. 2012, 36, 862–876. [Google Scholar] [CrossRef]
- Brandl, M.T. Plant lesions promote the rapid multiplication of Escherichia coli O157: H7 on postharvest lettuce. Appl. Environ. Microbiol. 2008, 74, 5285–5289. [Google Scholar] [CrossRef]
- Simko, I.; Zhou, Y.; Brandl, M.T. Downy mildew disease promotes the colonization of romaine lettuce by Escherichia coli O157: H7 and Salmonella enterica. BMC Microbiol. 2015, 15, 19. [Google Scholar] [CrossRef]
- Muralidharan, J.; Galiè, S.; Hernández-Alonso, P.; Bulló, M.; Salas-Salvadó, J. Plant-based fat, dietary patterns rich in vegetable fat and gut microbiota modulation. Front. Nutr. 2019, 6, 157. [Google Scholar]
- Ajmeer, A.J.; Selvarajan, R.; Mearns, K.; Pandian, J. Unveiling the Bacterial Communities and Its Potential Agricultural Applications from Organic Manure (Panchagavya) Using Targeted Amplicon Analysis. Pol. J. Environ. Stud. 2013, 6, 1–9. [Google Scholar] [CrossRef]
- Mushtaq, S.; Shafiq, M.; Tariq, M.R.; Sami, A.; Nawaz-Ul-Rehman, M.S.; Bhatti, M.H.T.; Haider, M.S.; Sadiq, S.; Abbas, M.T.; Hussain, M.; et al. Interaction between bacterial endophytes and host plants. Front. Plant Sci. 2023, 13, 1092105. [Google Scholar] [PubMed]

| Sample ID | Collection Time (Year) | Collection Site | Seed Accession Code |
|---|---|---|---|
| M01 | 2019 | Tuokexun County | MR-1 |
| M02 | 2013 | Changji Hui Autonomous Prefecture | MR-1 |
| M03 | 2011 | Changji Hui Autonomous Prefecture | MR-1 |
| M04 | 2019 | Tuokexun County | HH |
| M05 | 2017 | Changji Hui Autonomous Prefecture | HH |
| M06 | 2011 | Changji Hui Autonomous Prefecture | HH |
| M07 | 2019 | Tuokexun County | MR-1 (♀) × HH (♂) |
| M08 | 2019 | Tuokexun County | HH (♀) × MR-1 (♂) |
| Sample ID | PE Reads | Raw Tags | Clean Tags | Effective Tags | AvgLen (bp) | GC (%) | Q20 (%) | Q30 (%) | Effective (%) |
|---|---|---|---|---|---|---|---|---|---|
| M01 | 110,497 | 107,104 | 103,204 | 88,082 | 402 | 52.74 | 98.09 | 96.08 | 79.71 |
| M02 | 109,904 | 106,416 | 101,709 | 81,593 | 407 | 53.39 | 98.08 | 96.14 | 74.24 |
| M03 | 110,266 | 106,766 | 102,162 | 85,381 | 409 | 53.77 | 98.0 | 96.0 | 77.43 |
| M04 | 110,322 | 106,644 | 102,059 | 83,760 | 406 | 53.15 | 98.0 | 96.01 | 75.92 |
| M05 | 109,543 | 106,095 | 101,520 | 81,915 | 406 | 53.57 | 98.01 | 96.01 | 74.78 |
| M06 | 110,024 | 106,507 | 101,966 | 84,956 | 407 | 53.24 | 98.07 | 96.12 | 77.22 |
| M07 | 109,877 | 106,631 | 102,539 | 87,134 | 403 | 52.88 | 98.12 | 96.15 | 79.3 |
| M08 | 110,088 | 106,537 | 101,894 | 85,243 | 406 | 53.33 | 98.07 | 96.08 | 77.43 |
| Sample ID | OTU | ACE | Chao1 | Simpson | Shannon | Coverage |
|---|---|---|---|---|---|---|
| M01 | 469 | 471.6902 | 472.75 | 0.0686 | 4.273 | 0.9998 |
| M02 | 454 | 462.1405 | 467.3158 | 0.1771 | 3.3365 | 0.9996 |
| M03 | 447 | 464.2901 | 471.7 | 0.2618 | 2.6377 | 0.9995 |
| M04 | 458 | 470.477 | 487.5263 | 0.1934 | 3.1417 | 0.9995 |
| M05 | 457 | 467.9612 | 475.913 | 0.1979 | 3.1515 | 0.9996 |
| M06 | 458 | 464.6417 | 468.5556 | 0.1847 | 3.1388 | 0.9997 |
| M07 | 474 | 476.1013 | 478 | 0.0585 | 4.2468 | 0.9999 |
| M08 | 464 | 471.6609 | 476.8333 | 0.1442 | 3.5721 | 0.9997 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zeng, K.; Li, Y.; Wang, Z.; Du, Y.; Fan, M.; Xie, L. Community Structure and Diversity of Endophytic Bacteria in Melon (Cucumis melo L.) Seeds. Horticulturae 2023, 9, 1195. https://doi.org/10.3390/horticulturae9111195
Zeng K, Li Y, Wang Z, Du Y, Fan M, Xie L. Community Structure and Diversity of Endophytic Bacteria in Melon (Cucumis melo L.) Seeds. Horticulturae. 2023; 9(11):1195. https://doi.org/10.3390/horticulturae9111195
Chicago/Turabian StyleZeng, Kai, Yuandong Li, Zhou Wang, Yongkang Du, Mingqiang Fan, and Liqiong Xie. 2023. "Community Structure and Diversity of Endophytic Bacteria in Melon (Cucumis melo L.) Seeds" Horticulturae 9, no. 11: 1195. https://doi.org/10.3390/horticulturae9111195
APA StyleZeng, K., Li, Y., Wang, Z., Du, Y., Fan, M., & Xie, L. (2023). Community Structure and Diversity of Endophytic Bacteria in Melon (Cucumis melo L.) Seeds. Horticulturae, 9(11), 1195. https://doi.org/10.3390/horticulturae9111195





