Genome-Wide Classification and Evolutionary Analysis of the KNOX Gene Family in Plants
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Identification of KNOX Gene
2.3. Multiple Sequence Alignment and Phylogenetic Analyses
2.4. Gene Duplication and Loss Inference
2.5. Collinearity Analysis and Replication Type Identification of KNOX Gene
2.6. Expression Pattern Analysis of the KNOX Gene
2.7. Nonsynonymous (Ka) and Synonymous (Ks) Substitution Rate Calculation Analysis
3. Results
3.1. Identifying the KNOX Gene in 118 Plants
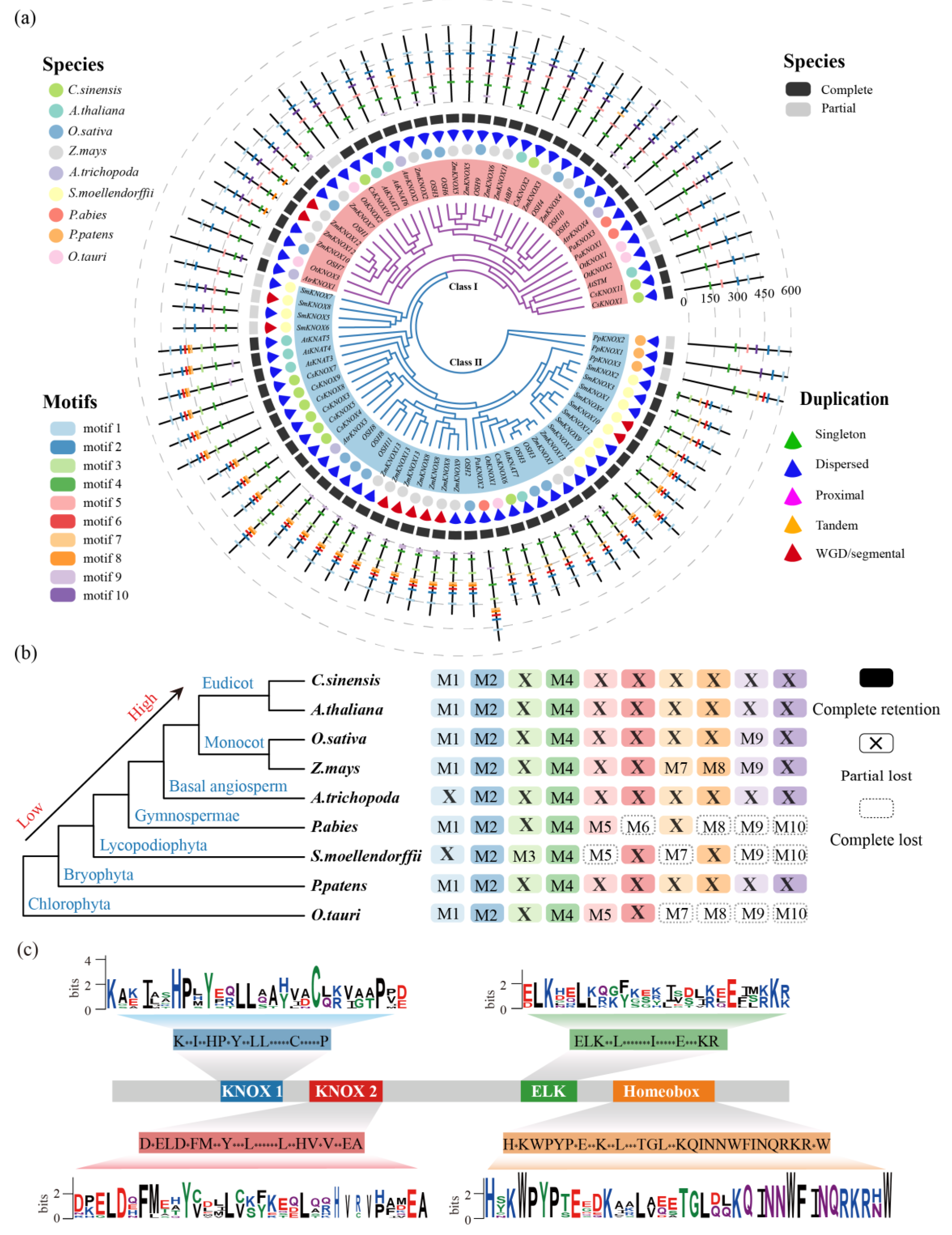
3.2. Comparative Analysis of the KNOX Gene in 118 Plants
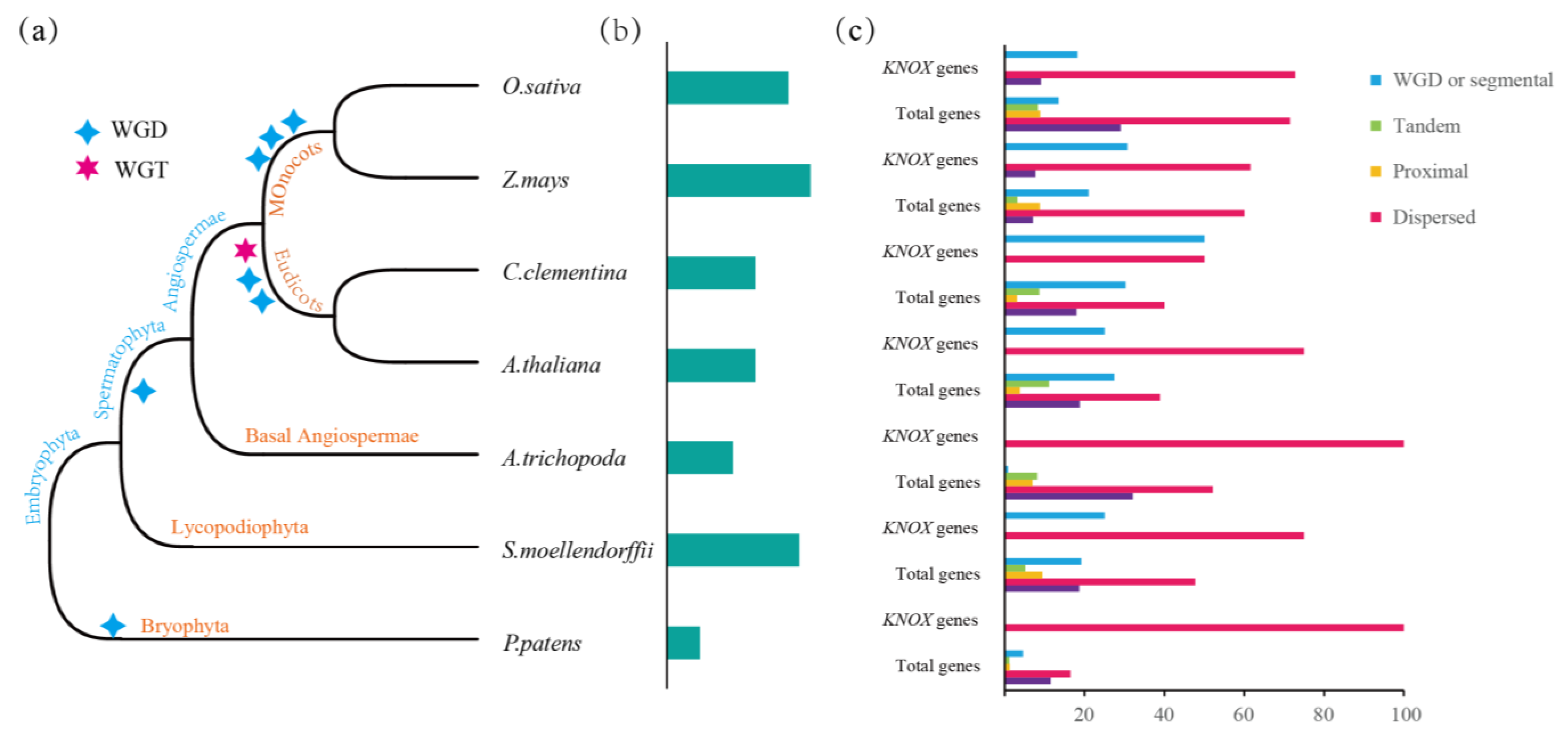
3.3. Gene Duplication and Loss Analysis of the KNOX Gene
3.4. Conserved Motifs Analysis of the KNOX Gene
3.5. Collinearity Analysis and Replication Type Identification of KNOX Gene
3.6. Expression Pattern Analysis of the KNOX Gene in Model Plants
3.7. Pan-Genome Analysis of the KNOX Gene Family in 17 Citrinae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Efroni, I.; Eshed, Y.; Lifschitz, E. Morphogenesis of simple and compound leaves: A critical review. Plant Cell 2010, 22, 1019–1032. [Google Scholar] [CrossRef]
- Tsuda, K.; Hake, S. Diverse functions of KNOX transcription factors in the diploid body plan of plants. Curr. Opin. Plant Biol. 2015, 27, 91–96. [Google Scholar] [CrossRef]
- Frangedakis, E.; Saint, D.; Moody, L.A.; Rabbinowitsch, E.; Langdale, J.A. Nonreciprocal complementation of KNOX gene function in land plants. New Phytol. 2017, 216, 591–604. [Google Scholar] [CrossRef]
- Qin, W.; Yin, Q.; Chen, J.; Zhao, X.; Yue, F.; He, J.; Yang, L.; Liu, L.; Zeng, Q.; Lu, F. The class II KNOX transcription factors KNAT3 and KNAT7 synergistically regulate monolignol biosynthesis in Arabidopsis. J. Exp. Bot. 2020, 71, 5469–5483. [Google Scholar] [CrossRef]
- Gao, J.; Yang, X.; Zhao, W.; Lang, T.; Samuelsson, T. Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar] [CrossRef]
- Serikawa, K.A.; Martinez-Laborda, A.; Zambryski, P. Three knotted1-like homeobox genes in Arabidopsis. Plant Mol. Biol. 1996, 32, 673–683. [Google Scholar] [CrossRef]
- Vollbrecht, E.; Veit, B.; Sinha, N.; Hake, S. The developmental gene Knotted-1 is a member of a maize homeobox gene family. Nature 1991, 350, 241–243. [Google Scholar] [CrossRef]
- Kerstetter, R.; Vollbrecht, E.; Lowe, B.; Veit, B.; Yamaguchi, J.; Hake, S. Sequence analysis and expression patterns divide the maize knotted1-like homeobox genes into two classes. Plant Cell 1994, 6, 1877–1887. [Google Scholar]
- Wechter, W.P.; Farnham, M.W.; Smith, J.P.; Keinath, A.P. Identification of resistance to peppery leaf spot among Brassica juncea and Brassica rapa plant introductions. HortScience 2007, 42, 1140–1143. [Google Scholar] [CrossRef]
- Bhat, S.; Sarla, N. IIdentification and overcoming barriers between Brassica rapa (L. ) and B. nigra (L.) Koch crosses for the resynthesis of B. juncea (L.) Czern. Genet. Resour. Crop Evol. 2004, 51, 455–469. [Google Scholar]
- Cheng, F.; Sun, R.; Hou, X.; Zheng, H.; Zhang, F.; Zhang, Y.; Liu, B.; Liang, J.; Zhuang, M.; Liu, Y. Subgenome parallel selection is associated with morphotype diversification and convergent crop domestication in Brassica rapa and Brassica oleracea. Nat. Genet. 2016, 48, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Hay, A.; Tsiantis, M. The genetic basis for differences in leaf form between Arabidopsis thaliana and its wild relative Cardamine hirsuta. Nat. Genet. 2006, 38, 942–947. [Google Scholar] [CrossRef]
- Zeng, R.F.; Fu, L.M.; Deng, L.; Liu, M.F.; Gan, Z.M.; Zhou, H.; Hu, S.F.; Hu, C.G.; Zhang, J.Z. CiKN1 and CiKN6 are involved in leaf development in citrus by regulating CimiR164. Plant J. 2022, 110, 828–848. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Song, M.; Cheng, F.; Zhang, M.; Davoudi, M.; Chen, J.; Lou, Q. A SNP Mutation in Homeodomain-DDT (HD-DDT) Transcription Factor Results in Multiple Trichomes (mt) in Cucumber (Cucumis sativus L.). Genes 2021, 12, 1478. [Google Scholar] [CrossRef] [PubMed]
- Kabak, G.; Şehsuvars, S.; Turgut, S.; Gökdemir, Ş. Genome-wide analysis of Fragaria vesca three-amino-acid-loop-extension (TALE) genes. Biotechnol. Stud. 2021, 30, 79–85. [Google Scholar] [CrossRef]
- Wang, L.; Yang, X.; Gao, Y.; Yang, S. Genome-Wide Identification and Characterization of TALE Superfamily Genes in Soybean (Glycine max L.). Int. J. Mol. Sci. 2021, 22, 4117. [Google Scholar] [CrossRef]
- Giannino, D.; Frugis, G.; Mele, G.; Nicolodi, C.; Chiappetta, A.; Bitonti, M.; Innocenti, A.; Mariotti, D. The Knotted-like homeobox genes (Knox) and cytokinins are involved in the differentiation and morphogenesis of lettuce leaves. In Proceedings of the Atti XVII Convegno Nazionale Della Societa’Italiana di Chimica Agraria, Portoferraio, Italy, 29 September–1 October 1999; pp. 181–189. [Google Scholar]
- Jia, P.; Zhang, C.; Xing, L.; Li, Y.; Shah, K.; Zuo, X.; Zhang, D.; An, N.; Han, M.; Ren, X. Genome-Wide Identification of the MdKNOX Gene Family and Characterization of Its Transcriptional Regulation in Malus domestica. Front. Plant Sci. 2020, 11, 128. [Google Scholar] [CrossRef]
- Di Giacomo, E.; Sestili, F.; Iannelli, M.A.; Testone, G.; Mariotti, D.; Frugis, G. Characterization of KNOX genes in Medicago truncatula. Plant Mol. Biol. 2008, 67, 135–150. [Google Scholar] [CrossRef]
- Testone, G.; Condello, E.; Verde, I.; Nicolodi, C.; Caboni, E.; Dettori, M.T.; Vendramin, E.; Bruno, L.; Bitonti, M.B.; Mele, G. The peach (Prunus persica L. Batsch) genome harbours 10 KNOX genes, which are differentially expressed in stem development, and the class 1 KNOPE1 regulates elongation and lignification during primary growth. J. Exp. Bot. 2012, 63, 5417–5435. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, P.; Cheng, B.; Zhang, Y.; Shen, Y.; Wang, X.; Zhang, Q.; Lou, Q.; Zhang, S.; Wang, B. Identification of TALE transcription factor family and expression patterns related to fruit chloroplast development in tomato (Solanum lycopersicum L.). Int. J. Mol. Sci. 2022, 23, 4507. [Google Scholar] [CrossRef]
- Bằng, C.P. Identification, classification and expression analysis of KNOX gene family in potato (Solanum tuberosum L.) by using bioinformatic methods. J. Sci. Technol. 2017, 3, 84–87. [Google Scholar]
- Song, N.; Liang, H.; An, Y.; Bai, S.; Ma, F.; Zhang, Z.; Li, H.; Zhou, Y.; Guo, G.; Song, C. Identification and Bioinformatics Analysis of KNOX Gene Family in Wheat (Triticum aestivum L.). Mol. Plant Breed. 2021, 12, 19. [Google Scholar] [CrossRef]
- Yao, J.; Zhang, S.; Wu, N.; Li, X.; Ahmad, B.; Wu, J.; Guo, R.; Wang, X. KNOX transcription factor VvHB63 affects grape seed development by interacting with protein VvHB06. Plant Sci. 2023, 330, 111665. [Google Scholar] [CrossRef] [PubMed]
- Scofield, S.; Murray, J.A. KNOX gene function in plant stem cell niches. Plant Mol. Biol. 2006, 60, 929–946. [Google Scholar] [CrossRef]
- Song, J.-M.; Xie, W.-Z.; Wang, S.; Guo, Y.-X.; Koo, D.-H.; Kudrna, D.; Gong, C.; Huang, Y.; Feng, J.-W.; Zhang, W. Two gap-free reference genomes and a global view of the centromere architecture in rice. Mol. Plant 2021, 14, 1757–1767. [Google Scholar] [CrossRef]
- Yang, T.; Jiao, Y.; Wang, Y. Stem cell basis of shoot branching. Plant Cell Physiol. 2023, 64, 291–296. [Google Scholar] [CrossRef]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of pluripotency pathways regulates stem cell maintenance in the Arabidopsis shoot meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, S.; Chen, C.-Y.; Li, C.; Shan, W.; Lu, W.; Cui, Y.; Liu, X.; Wu, K. Arabidopsis BREVIPEDICELLUS interacts with the SWI2/SNF2 chromatin remodeling ATPase BRAHMA to regulate KNAT2 and KNAT6 expression in control of inflorescence architecture. PLoS Genet. 2015, 11, e1005125. [Google Scholar] [CrossRef]
- Tabata, R.; Ikezaki, M.; Fujibe, T.; Aida, M.; Tian, C.-E.; Ueno, Y.; Yamamoto, K.T.; Machida, Y.; Nakamura, K.; Ishiguro, S. Arabidopsis auxin response factor6 and 8 regulate jasmonic acid biosynthesis and floral organ development via repression of class 1 KNOX genes. Plant Cell Physiol. 2010, 51, 164–175. [Google Scholar] [CrossRef]
- Scofield, S.; Dewitte, W.; Murray, J.A. STM sustains stem cell function in the Arabidopsis shoot apical meristem and controls KNOX gene expression independently of the transcriptional repressor AS1. Plant Signal. Behav. 2014, 9, e28934. [Google Scholar] [CrossRef]
- .Shani, E.; Burko, Y.; Ben-Yaakov, L.; Berger, Y.; Amsellem, Z.; Goldshmidt, A.; Ori, N. Stage-specific regulation of Solanum lycopersicum leaf maturation by class 1 KNOTTED1-LIKE HOMEOBOX proteins. Plant Cell 2009, 21, 3078–3092. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Han, L.; Li, G.; Chai, M.; Fu, C.; Cheng, X.; Wen, J.; Tang, Y.; Wang, Z.-Y. STM/BP-like KNOXI is uncoupled from ARP in the regulation of compound leaf development in Medicago truncatula. Plant Cell 2014, 26, 1464–1479. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Zhao, Y.; Wang, J.; Lu, M.-Z. The transcription factor KNAT2/6b mediates changes in plant architecture in response to drought via down-regulating GA20ox1 in Populus alba × P. glandulosa. J. Exp. Bot. 2021, 72, 5625–5637. [Google Scholar] [CrossRef]
- Sufyan, M.; Daraz, U.; Hyder, S.; Zulfiqar, U.; Iqbal, R.; Eldin, S.M.; Rafiq, F.; Mahmood, N.; Shahzad, K.; Uzair, M. An overview of genome engineering in plants, including its scope, technologies, progress and grand challenges. Funct. Integr. Genom. 2023, 23, 119. [Google Scholar] [CrossRef]
- Pautot, V.; Dockx, J.; Hamant, O.; Kronenberger, J.; Grandjean, O.; Jublot, D.; Traas, J. KNAT2: Evidence for a link between knotted-like genes and carpel development. Plant Cell 2001, 13, 1719–1734. [Google Scholar] [CrossRef]
- Scofield, S.; Dewitte, W.; Murray, J.A. The KNOX gene SHOOT MERISTEMLESS is required for the development of reproductive meristematic tissues in Arabidopsis. Plant J. 2007, 50, 767–781. [Google Scholar] [CrossRef]
- Roth, O.; Alvarez, J.P.; Levy, M.; Bowman, J.L.; Ori, N.; Shani, E. The KNOXI transcription factor SHOOT MERISTEMLESS regulates floral fate in Arabidopsis. Plant Cell 2018, 30, 1309–1321. [Google Scholar] [CrossRef]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016, 45, gkw982. [Google Scholar] [CrossRef]
- Liu, H.; Wang, X.; Liu, S.; Huang, Y.; Guo, Y.-X.; Xie, W.-Z.; Liu, H.; Qamar, M.T.U.; Xu, Q.; Chen, L.-L. Citrus Pan-Genome to Breeding Database (CPBD): A comprehensive genome database for citrus breeding. Mol. Plant 2022, 15, 1503–1505. [Google Scholar] [CrossRef]
- Sato, Y.; Fukuda, Y.; Hirano, H.Y. Mutations that cause amino acid substitutions at the invariant positions in homeodomain of OSH3 KNOX protein suggest artificial selection during rice domestication. Genes Genet. Syst. 2001, 76, 381–392. [Google Scholar] [CrossRef][Green Version]
- Yu, G.; Lam, T.T.-Y.; Zhu, H.; Guan, Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol. Biol. Evol. 2018, 35, 3041–3043. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Durand, D.; Farach, M. NOTUNG: A program for dating gene duplications and optimizing gene family trees. J. Comput. Biol. 2000, 7, 429–447. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Nam, J.; dePamphilis, C.W.; Ma, H.; Nei, M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol. Biol. Evol. 2003, 20, 1435–1447. [Google Scholar] [CrossRef]
- De La Torre, A.R.; Piot, A.; Liu, B.; Wilhite, B.; Weiss, M.; Porth, I. Functional and morphological evolution in gymnosperms: A portrait of implicated gene families. Evol. Appl. 2020, 13, 210–227. [Google Scholar] [CrossRef]
- Song, X.; Wang, J.; Ma, X.; Li, Y.; Lei, T.; Wang, L.; Ge, W.; Guo, D.; Wang, Z.; Li, C.; et al. Origination, expansion, evolutionary trajectory, and expression bias of AP2/ERF superfamily in Brassica napus. Front. Plant Sci. 2016, 7, 1186. [Google Scholar] [CrossRef]
- Zhang, H.; Jin, J.; Jin, L.; Li, Z.; Xu, G.; Wang, R.; Zhang, J.; Zhai, N.; Chen, Q.; Liu, P.; et al. Identification and analysis of the chloride channel gene family members in tobacco (Nicotiana tabacum). Gene 2018, 676, 56–64. [Google Scholar] [CrossRef]
- Sheridan, P.O.; Raguideau, S.; Quince, C.; Holden, J.; Zhang, L.; Williams, T.A.; Gubry, C. Gene duplication drives genome expansion in a major lineage of Thaumarchaeota. Nat. Commun. 2020, 11, 5494. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.-Q.; Ouyang, Y.; Tang, Z.-Y.; Lao, C.-C.; Zhang, Y.-Y.; Cheng, C.-S.; Zhou, H. Review on the development and applications of medicinal plant genomes. Front. Plant Sci. 2021, 12, 791219. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Wang, D.; Cheng, Z.; Wang, Y.; Jiao, Y. A near-complete assembly of an Arabidopsis thaliana genome. Mol. Plant 2022, 15, 1247–1250. [Google Scholar] [CrossRef] [PubMed]
- Cenci, A.; Concepción-Hernández, M.; Guignon, V.; Angenon, G.; Rouard, M. Genome-wide classification and phylogenetic analyses of the GDSL-type esterase/lipase (GELP) family in flowering plants. Int. J. Mol. Sci. 2022, 23, 12114. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene duplication and evolution in recurring polyploidization–diploidization cycles in plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef]
- Wu, Y.; Li, D.; Hu, Y.; Li, H.; Ramstein, G.P.; Zhou, S.; Zhang, X.; Bao, Z.; Zhang, Y.; Song, B.; et al. Phylogenomic discovery of deleterious mutations facilitates hybrid potato breeding. Cell 2023, 186, 2313–2328. [Google Scholar] [CrossRef]
- Chen, J.; Wang, Z.; Tan, K.; Huang, W.; Shi, J.; Li, T.; Hu, J.; Wang, K.; Wang, C.; Xin, B. A complete telomere-to-telomere assembly of the maize genome. Nat. Genet. 2023, 55, 1221–1231. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, Y.; Yan, L.; Zhang, G.; Wang, X.; Zeng, Y.; Zhang, J.; Ma, X.; Tan, Y.; Long, N. Genome of plant maca (Lepidium meyenii) illuminates genomic basis for high-altitude adaptation in the central Andes. Mol. Plant 2016, 9, 1066–1077. [Google Scholar] [CrossRef]
- Del Pozo, J.C.; Ramirez-Parra, E. Whole genome duplications in plants: An overview from Arabidopsis. J. Exp. Bot. 2015, 66, 6991–7003. [Google Scholar] [CrossRef]
- Yu, T.; Bai, Y.; Liu, Z.; Wang, Z.; Yang, Q.; Wu, T.; Feng, S.; Zhang, Y.; Shen, S.; Li, Q. Large-scale analyses of heat shock transcription factors and database construction based on whole-genome genes in horticultural and representative plants. Hortic. Res. 2022, 9, uhac035. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.-H.; Bancroft, I. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 2011, 43, 1035–1039. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Liu, D.; Wang, X.; Ji, C.; Cheng, F.; Liu, B.; Hu, Z.; Chen, S.; Pental, D.; Ju, Y. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 2016, 48, 1225–1232. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xu, Y.; Zhang, X.; Ma, X.; Zhang, L.; Liao, Z.; Zhang, Q.; Wan, X.; Cheng, Y.; Zhang, J. The genome of kenaf (Hibiscus cannabinus L.) provides insights into bast fibre and leaf shape biogenesis. Plant Biotechnol. J. 2020, 18, 1796–1809. [Google Scholar] [CrossRef]
- Schmutz, J.; Cannon, S.B.; Schlueter, J.; Ma, J.; Mitros, T.; Nelson, W.; Hyten, D.L.; Song, Q.; Thelen, J.J.; Cheng, J. Genome sequence of the palaeopolyploid soybean. Nature 2010, 463, 178–183. [Google Scholar] [CrossRef]
- Tuskan, G.A.; Difazio, S.; Jansson, S.; Bohlmann, J.; Grigoriev, I.; Hellsten, U.; Putnam, N.; Ralph, S.; Rombauts, S.; Salamov, A.; et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 2006, 313, 1596–1604. [Google Scholar]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef]
- Hu, R.; Qi, G.; Kong, Y.; Kong, D.; Gao, Q.; Zhou, G. Comprehensive Analysis of NAC Domain Transcription Factor Gene Family in Populus trichocarpa. BMC Plant Biol. 2010, 10, 145. [Google Scholar] [CrossRef]
- Wang, J.L.; Wang, H.W.; Cao, Y.N.; Kan, S.L.; Liu, Y.Y. Comprehensive evolutionary analysis of the TCP gene family: Further insights for its origin, expansion, and diversification. Front. Plant Sci. 2022, 13, 994567. [Google Scholar] [CrossRef]
- Rouhier, N.; Couturier, J.; Jacquot, J.P. Genome-wide analysis of plant glutaredoxin systems. J. Exp. Bot. 2006, 57, 1685–1696. [Google Scholar] [CrossRef]
- Long, J.A.; Barton, M.K. The development of apical embryonic pattern in Arabidopsis. Development 1998, 125, 3027–3035. [Google Scholar] [CrossRef]
- Li, G.; Yang, J.; Chen, Y.; Zhao, X.; Chen, Y.; Kimura, S.; Hu, S.; Hou, H. SHOOT MERISTEMLESS participates in the heterophylly of Hygrophila difformis (Acanthaceae). Plant Physiol. 2022, 190, 1777–1791. [Google Scholar] [CrossRef]
- Ragni, L.; Belles-Boix, E.; Gunl, M.; Pautot, V. Interaction of KNAT6 and KNAT2 with BREVIPEDICELLUS and PENNYWISE in Arabidopsis inflorescences. Plant Cell 2008, 20, 888–900. [Google Scholar] [CrossRef]
- Butenko, M.A.; Shi, C.-L.; Aalen, R.B. KNAT1, KNAT2 and KNAT6 act downstream in the IDA-HAE/HSL2 signaling pathway to regulate floral organ abscission. Plant Signal. Behav. 2012, 7, 135–138. [Google Scholar] [CrossRef] [PubMed]
- Belles-Boix, E.; Hamant, O.; Witiak, S.M.; Morin, H.; Traas, J.; Pautot, V. KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 2006, 18, 1900–1907. [Google Scholar] [CrossRef]
- Reiser, L.; Sánchez-Baracaldo, P.; Hake, S. Knots in the family tree: Evolutionary relationships and functions of knox homeobox genes. Plant Mol. Evol. 2000, 151–166. [Google Scholar] [CrossRef]
- Richardson, A.; Rebocho, A.B.; Coen, E. Ectopic KNOX expression affects plant development by altering tissue cell polarity and identity. Plant Cell 2016, 28, 2079–2096. [Google Scholar] [CrossRef]
- Yan, F.; Deng, W.; Pang, X.; Gao, Y.; Chan, H.; Zhang, Q.; Hu, N.; Chen, J.; Li, Z. Overexpression of the KNOX gene Tkn4 affects pollen development and confers sensitivity to gibberellin and auxin in tomato. Plant Sci. 2019, 281, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Bolduc, N.; Yilmaz, A.; Mejia, M.K.; Morohashi, K.; O’Connor, D.; Grotewold, E.; Hake, S. Unraveling the KNOTTED1 regulatory network in maize meristems. Genes Dev. 2012, 26, 1685–1690. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the origin and evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef]
- Da, X.; Yu, K.; Shen, S.; Zhang, Y.; Wu, J.; Yi, H. Identification of differentially expressed genes in a spontaneous altered leaf shape mutant of the navel orange [Citrus sinensis (L.) Osbeck]. Plant Physiol. Biochem. 2012, 56, 97–103. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, Z.-M.; Ai, X.-Y.; Hu, C.-G.; Zhang, J.-Z. Genome-Wide Classification and Evolutionary Analysis of the KNOX Gene Family in Plants. Horticulturae 2023, 9, 1174. https://doi.org/10.3390/horticulturae9111174
Gan Z-M, Ai X-Y, Hu C-G, Zhang J-Z. Genome-Wide Classification and Evolutionary Analysis of the KNOX Gene Family in Plants. Horticulturae. 2023; 9(11):1174. https://doi.org/10.3390/horticulturae9111174
Chicago/Turabian StyleGan, Zhi-Meng, Xiao-Yan Ai, Chun-Gen Hu, and Jin-Zhi Zhang. 2023. "Genome-Wide Classification and Evolutionary Analysis of the KNOX Gene Family in Plants" Horticulturae 9, no. 11: 1174. https://doi.org/10.3390/horticulturae9111174
APA StyleGan, Z.-M., Ai, X.-Y., Hu, C.-G., & Zhang, J.-Z. (2023). Genome-Wide Classification and Evolutionary Analysis of the KNOX Gene Family in Plants. Horticulturae, 9(11), 1174. https://doi.org/10.3390/horticulturae9111174






