Combined Metabolomic and Transcriptomic Analysis Reveals Candidate Genes for Anthocyanin Accumulation in Ginkgo biloba Seed Exocarp
Abstract
1. Introduction
2. Results
2.1. Observation of Growth and Development Phenotype of Ginkgo biloba Seed Exocarp and Determination of Color Parameters
2.2. Determination of Anthocyanin Content in Ginkgo biloba Seed Exocarp
2.3. Metabolome Analysis of Ginkgo biloba Seed Exocarp in Three Different Color Periods
2.4. Screening of Anthocyanin-Related Genes in the Transcriptome of Ginkgo biloba Seed Exocarp at Three Different Color Periods
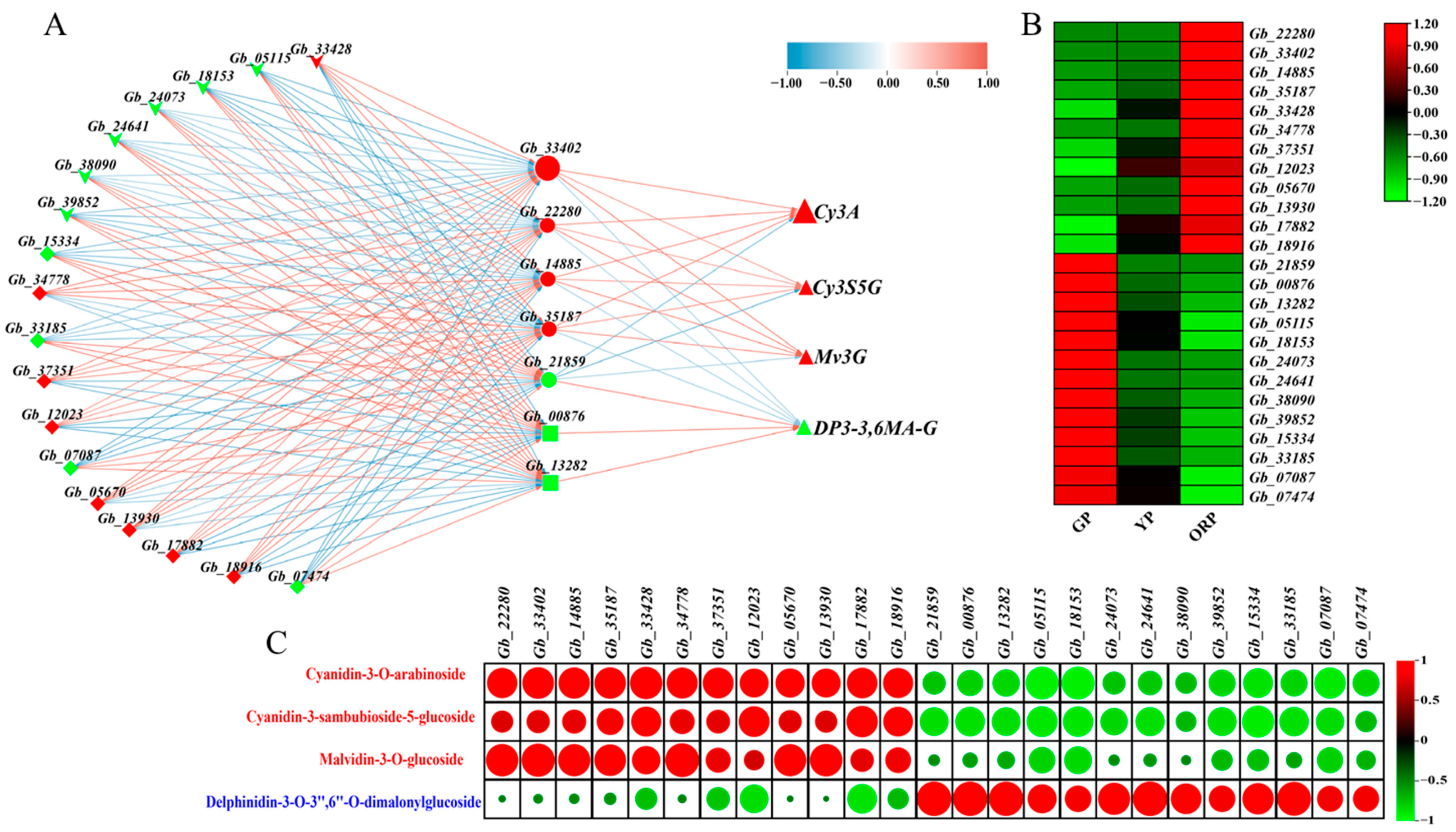
2.5. Candidate Genes for Color Change of Ginkgo biloba Seed Exocarp Were Obtained by Combining Transcriptome and Metabolome Analysis
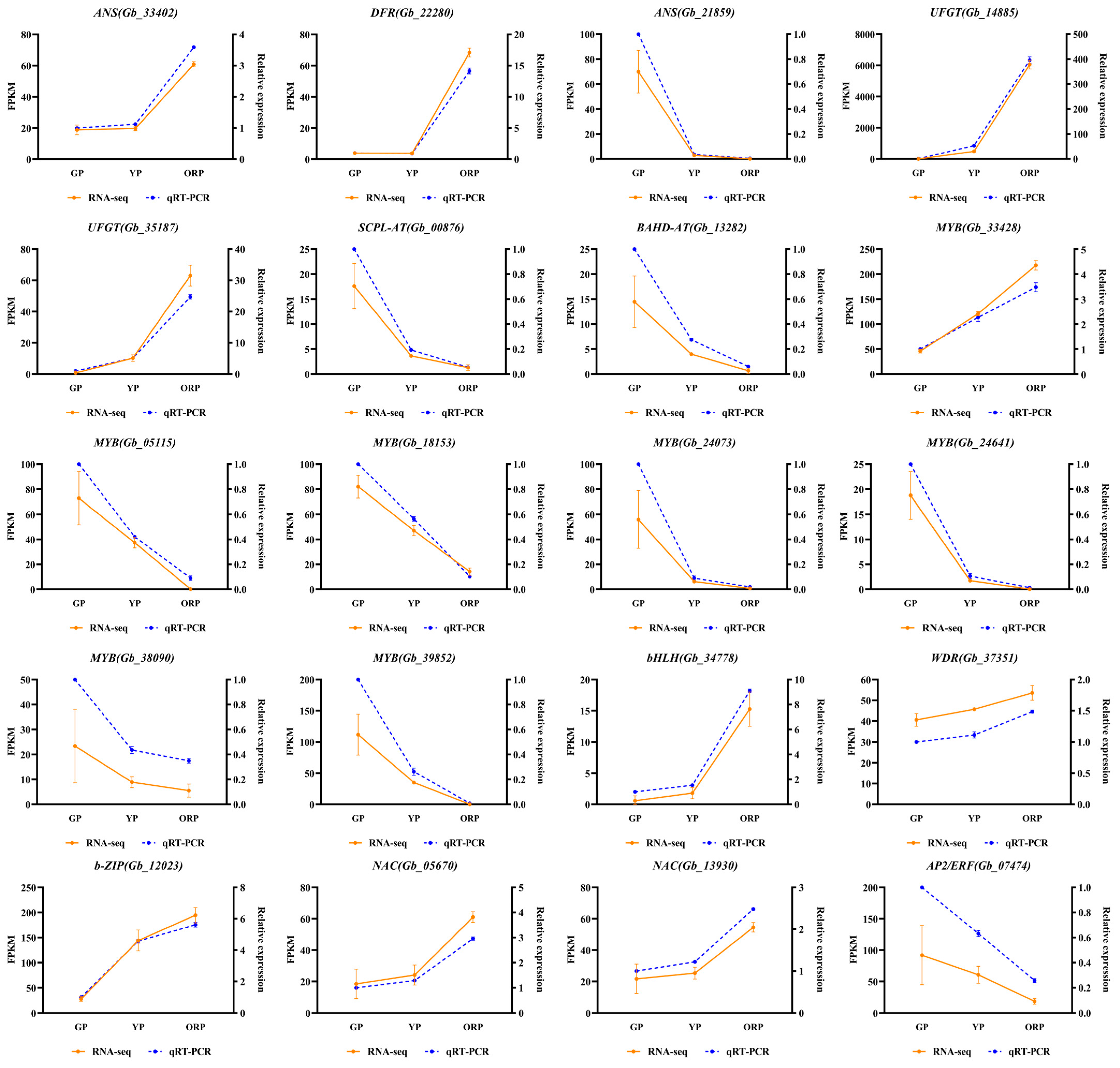
2.6. qRT-PCR Verification of Differentially Expressed Genes in Ginkgo biloba Seed Exocarp Transcriptome Data
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Determination of Color Parameters and Anthocyanin Content of Ginkgo biloba Seed Exocarp
4.3. Metabolome and Transcriptome Data Materials
4.4. Real-Time Fluorescence Quantitative PCR Verification
4.5. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Strømgaard, K.; Nakanishi, K. Chemistry and biology of terpene trilactones from Ginkgo biloba. Angew. Chem. (Int. Ed.) 2004, 43, 1640–1658. (In English) [Google Scholar] [CrossRef]
- Wang, H.; Shi, M.; Cao, F.; Su, E. Ginkgo biloba seed exocarp: A waste resource with abundant active substances and other components for potential applications. Food Res. Int. 2022, 160, 111637. [Google Scholar] [CrossRef]
- Zhu, M.; Ren, J.; Shen, N.; Xin, H.; Liu, Y.; Li, W.; Cui, Y. Research progress on chemical composition, biological activity and resource utilization of Ginkgo biloba seed exocarp. West. Chin. J. Pharm. 2022, 37, 587–593. (In Chinese) [Google Scholar]
- Cui, N.; Zhang, L.; Quan, M.; Xu, J. Profile of the main bioactive compounds and in vitro biological activity of different solvent extracts from Ginkgo biloba exocarp. RSC Adv. 2020, 10, 45105–45111. [Google Scholar] [CrossRef]
- Zhao, D. Extraction and Bioactivity of Active Components from Ginkgo biloba Seed Exocarp. Master’s Thesis, Nanjing Forestry University, Nanjing, China, 2013. [Google Scholar]
- Zhang, S.-L.; Lei, J.-H.; Du, X.; Du, Y.; Guo, L. Study on preparation of health beverage from fresh Ginkgo biloba seed exocarp. Preserv. Process. 2018, 18, 92–96. (In Chinese) [Google Scholar]
- Zhu, Z.; Lu, Y. Anthocyanin metabolism pathway and plant color variation. Chin. Bull. Bot. 2016, 51, 107–119. (In Chinese) [Google Scholar]
- Araceli, C.; Pacheco, M.; Páez-Hernández, M.; Rodríguez, J.; Galán-Vidal, C. Chemical studies of anthocyanins: A review. Food Chem. 2008, 113, 859–871. [Google Scholar]
- Zhao, C.; Yu, Y.; Chen, Z.; Wen, G.; Wei, F.; Zheng, Q.; Wang, C.; Xiao, X. Stability-increasing effects of anthocyanin glycosyl acylation. Food Chem. 2017, 214, 119–128. [Google Scholar] [CrossRef]
- Fu, M.; Yang, X.; Zheng, J.; Wang, L.; Yang, X.; Tu, Y.; Ye, J.; Zhang, W.; Liao, Y.; Cheng, S.; et al. Unraveling the Regulatory Mechanism of Color Diversity in Camellia japonica Petals by Integrative Transcriptome and Metabolome Analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef]
- Wang, Y.; Li, S.; Zhu, Z.; Xu, Z.; Qi, S.; Xing, S.; Yu, Y.; Wu, Q. Transcriptome and chemical analyses revealed the mechanism of flower color formation in Rosa rugosa. Front. Plant Sci. 2022, 13, 1021521. [Google Scholar] [CrossRef]
- Lu, J.; Zhang, Q.; Lang, L.; Jiang, C.; Wang, X.; Sun, H. Integrated metabolome and transcriptome analysis of the anthocyanin biosynthetic pathway in relation to color mutation in miniature roses. BMC Plant Biol. 2021, 21, 257. [Google Scholar] [CrossRef]
- Sun, A.; Pei, X.; Zhang, S.; Han, Z.; Xie, Y.; Qu, G.; Hu, X.; Mulualem, T.; Zhao, X. Metabolome and Transcriptome Analyses Unravels Molecular Mechanisms of Leaf Color Variation by Anthocyanidin Biosynthesis in Acer triflorum. Horticulturae 2022, 8, 635. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Zhao, M.; Hu, Y.; Meng, F.; Song, X.; Mulualem, T.; Vincent, C.; Ronald, S.; Ma, W.; et al. Molecular and Metabolic Insights into Anthocyanin Biosynthesis for Leaf Color Change in Chokecherry (Padus virginiana). Int. J. Mol. Sci. 2021, 22, 10697. [Google Scholar] [CrossRef]
- Cai, J.; Lv, L.; Zeng, X.; Zhang, F.; Chen, Y.; Tian, W.; Li, J.; Li, X.; Li, Y. Integrative Analysis of Metabolomics and Transcriptomics Reveals Molecular Mechanisms of Anthocyanin Metabolism in the Zikui Tea Plant (Camellia sinensis cv. Zikui). Int. J. Mol. Sci. 2022, 23, 4780. [Google Scholar] [CrossRef]
- Zheng, T.; Zhang, Q.; Su, K.; Liu, S. Transcriptome and metabolome analyses reveal the regulation of peel coloration in green, red Chinese prickly ash (Zanthoxylum L.). Food Chem. Mol. Sci. 2020, 1, 100004. [Google Scholar] [CrossRef]
- Yi, D.; Zhang, H.; Lai, B.; Liu, L.; Pan, X.; Ma, Z.; Wang, Y.; Xie, J.; Shi, S.; Wei, Y. Integrative Analysis of the Coloring Mechanism of Red Longan Pericarp through Metabolome and Transcriptome Analyses. J. Agric. Food Chem. 2020, 69, 1806–1815. [Google Scholar] [CrossRef]
- Wang, Z.; Cui, Y.; Vainstein, A.; Chen, S.; Ma, H. Regulation of Fig (Ficus carica L.) Fruit Color: Metabolomic and Transcriptomic Analyses of the Flavonoid Biosynthetic Pathway. Front. Plant Sci. 2017, 8, 1990. [Google Scholar] [CrossRef]
- Xue, Q.; Zhang, X.; Yang, H.; Li, H.; Lv, Y.; Zhang, K.; Liu, Y.; Liu, F.; Wan, Y. Transcriptome and Metabolome Analysis Unveil Anthocyanin Metabolism in Pink and Red Testa of Peanut (Arachis hypogaea L.). Int. J. Genom. 2021, 2021, 5883901. [Google Scholar] [CrossRef]
- Yao, X.; Yao, Y.; An, L.; Li, X.; Bai, Y.; Cui, Y.; Wu, K. Accumulation and regulation of anthocyanins in white and purple Tibetan Hulless Barley (Hordeum vulgare L. var. nudum Hook. f.) revealed by combined de novo transcriptomics and metabolomics. BMC Plant Biol. 2022, 22, 391. [Google Scholar] [CrossRef]
- Sivankalyani, V.; Feygenberg, O.; Diskin, S.; Wright, B.; Alkan, N. Increased anthocyanin and flavonoids in mango fruit peel are associated with cold and pathogen resistance. Postharvest Biol. Technol. 2016, 111, 132–139. [Google Scholar] [CrossRef]
- Kim, J.; Lee, W.; Thanh, V.; Jeong, C.; Hong, S.; Lee, H. High accumulation of anthocyanins via the ectopic expression of AtDFR confers significant salt stress tolerance in Brassica napus L. Plant Cell Rep. 2017, 36, 1215–1224. [Google Scholar] [CrossRef]
- Landi, M.; Tattini, M.; Gould, K.S. Multiple functional roles of anthocyanins in plant-environment interactions. Environ. Exp. Bot. 2015, 119, 4–17. [Google Scholar] [CrossRef]
- Fernandes, I.; Faria, A.; Calhau, C.; Freitas, V.; Mateus, N. Bioavailability of anthocyanins and derivatives. J. Funct. Foods 2014, 7, 54–66. [Google Scholar] [CrossRef]
- Li, X.; Qian, X.; Lǚ, X.; Wang, X.; Ji, N.; Zhang, M.; Ren, M. Upregulated structural and regulatory genes involved in anthocyanin biosynthesis for coloration of purple grains during the middle and late grain-filling stages. Plant Physiol. Biochem. 2018, 130, 235–247. [Google Scholar] [CrossRef]
- Li, M.; Cao, Y.; Ye, S.; Irshad, M.; Pan, T.; Qiu, D. Isolation of CHS Gene from Brunfelsia acuminata Flowers and Its Regulation in Anthocyanin Biosysthesis. Molecules 2016, 22, 44. [Google Scholar] [CrossRef]
- Peng, Y.; Kui, L.; Cooney, J.; Wang, T.; Espley, R.; Allan, A. Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic. Res. 2019, 6, 1–16. [Google Scholar] [CrossRef]
- Yang, Y.; Yao, G.; Zheng, D.; Zhang, S.; Wang, C.; Zhang, M.; Wu, J. Expression differences of anthocyanin biosynthesis genes reveal regulation patterns for red pear coloration. Plant Cell Rep. 2015, 34, 189–198. [Google Scholar] [CrossRef]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Vincent, C.; Ronald, S.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Lloyd, A.; Brockman, A.; Aguirre, L.; Campbell, A.; Bean, A.; Cantero, A.; Gonzalez, A. Advances in the MYB-bHLH-WD Repeat (MBW) Pigment Regulatory Model: Addition of a WRKY Factor and Co-option of an Anthocyanin MYB for Betalain Regulation. Plant Cell Physiol. 2017, 58, 1431–1441. [Google Scholar] [CrossRef]
- Wang, L.; Tang, W.; Hu, Y.; Zhang, Y.; Sun, J.; Guo, X.; Lu, H.; Yang, Y.; Fang, C.; Niu, X.; et al. A MYB/bHLH complex regulates tissue-specific anthocyanin biosynthesis in the inner pericarp of red-centered kiwifruit Actinidia chinensis cv. Hongyang. Plant J. Cell Mol. Biol. 2019, 99, 359–378. [Google Scholar] [CrossRef]
- Yao, P.; Zhao, H.; Luo, X.; Gao, F.; Li, C.; Yao, H.; Chen, H.; Sang-Un, P.; Wu, Q. Fagopyrum tataricum FtWD40 Functions as a Positive Regulator of Anthocyanin Biosynthesis in Transgenic Tobacco. J. Plant Growth Regul. 2017, 36, 755–765. [Google Scholar] [CrossRef]
- Duan, S.; Wang, J.; Gao, C.; Jin, C.; Li, D.; Peng, D.; Du, G.; Li, Y.; Chen, M. Functional characterization of a heterologously expressed Brassica napus WRKY41-1 transcription factor in regulating anthocyanin biosynthesis in Arabidopsis thaliana. Plant Sci. 2018, 268, 47–53. [Google Scholar] [CrossRef]
- Zhou, H.; Kui, L.; Wang, H.; Gu, C.; Andrew, P.; Richard, V.; He, H.; Andrew, C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. Cell Mol. Biol. 2015, 82, 105–121. [Google Scholar] [CrossRef]
- Jiang, Y.; Liu, C.; Yan, D.; Wen, X.; Liu, Y.-L.; Wang, H.; Dai, J.; Zhang, Y.; Liu, Y.-F.; Zhou, B.; et al. MdHB1 down-regulation activates anthocyanin biosynthesis in the white-fleshed apple cultivar ‘Granny Smith’. J. Exp. Bot. 2017, 68, 1055–1069. [Google Scholar] [CrossRef]
- Xu, Z.; Yang, X.; Wang, Y.; Du, S. Leaf pigment change and color mechanism of Chinese Pistacia chinensis. J. Nanjing For. Univ. 2024, 48, 97–104. [Google Scholar]
- Li, Q. Analysis of Anthocyanin Components and Color Factors of Three Species of Bauhinia purpurea L. Master’s Thesis, South China Agricultural University, Guangzhou, China, 2018. [Google Scholar]
- Zhang, Q.; Wang, L.; Liu, Z.; Zhao, Z.; Zhao, J.; Wang, Z.; Zhou, G.; Liu, P.; Liu, M. Transcriptome and metabolome profiling unveil the mechanisms of Ziziphus jujuba Mill. peel coloration. Food Chem. 2020, 312, 125903. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Han, Y.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef]
- Jaakola, L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013, 18, 477–483. [Google Scholar] [CrossRef]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- An, J.-P.; Qu, F.-J.; Yao, J.-F.; Wang, X.-N.; You, C.-X.; Wang, X.-F.; Hao, Y.-J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023. [Google Scholar] [CrossRef]
- Yu, P. Verification and Influencing factors of colorimeter. Technol. Mark. 2017, 24, 146. [Google Scholar]
- Cheng, L.; Xu, Y.; Grotewold, E.; Jin, Z.; Wu, F.; Fu, C.; Zhao, D. Characterization of Anthocyanidin Synthase (ANS) Gene and anthocyanidin in rare medicinal plant-Emphasis Type = ItalicSaussurea medusa/Emphasis. Plant Cell Tissue Organ Cult. 2007, 89, 63–73. [Google Scholar] [CrossRef]
- He, C.; Cai, Y.; Li, M.; Dong, J.; Liu, W.; Wang, J.; Wang, Y. Screening and expression analysis of related genes based on transcriptome sequencing at three differentiation stages of Ginkgo biloba flower buds. Chin. J. Hortic. 2018, 45, 1479–1490. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔC(T) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
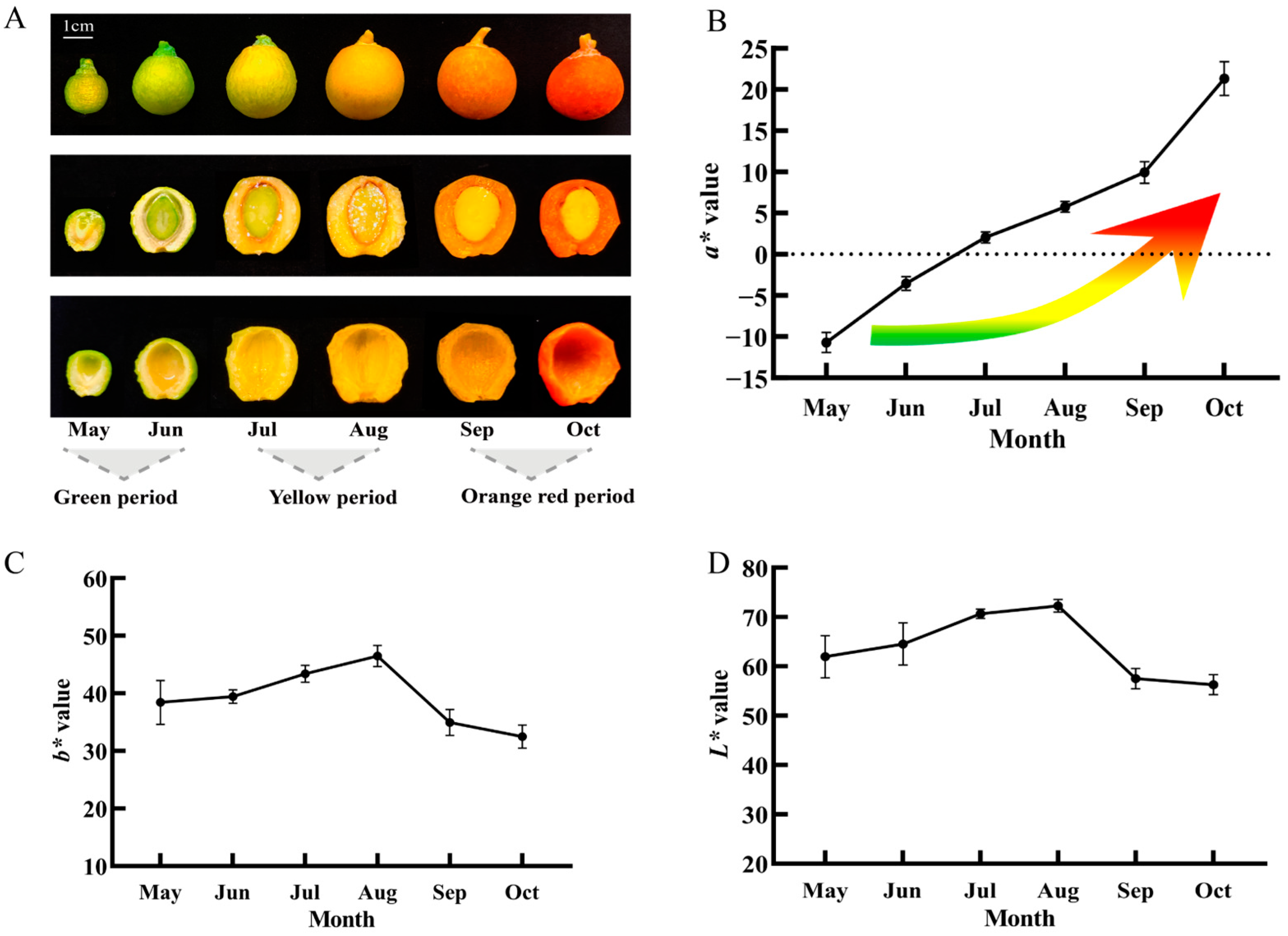

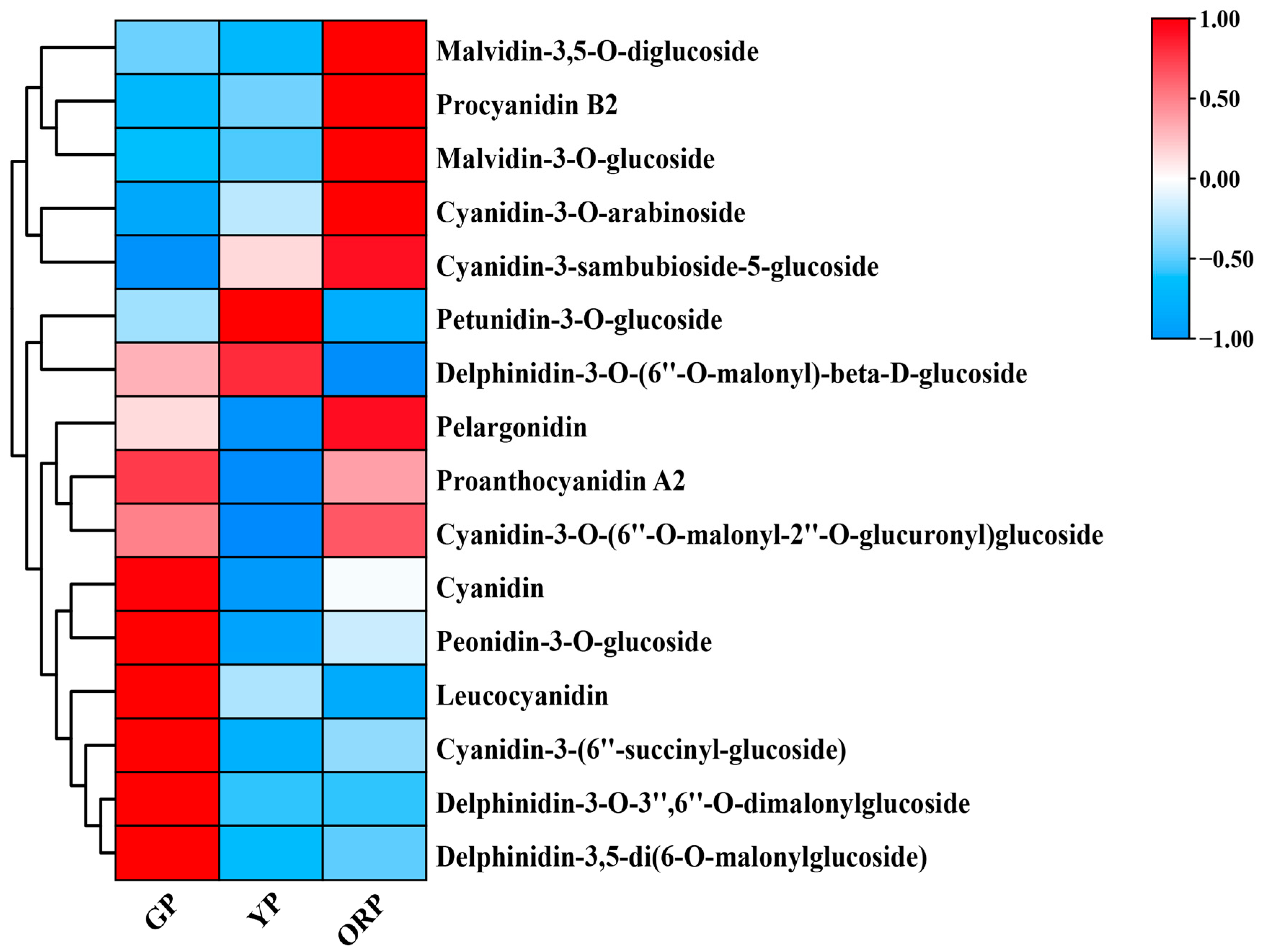
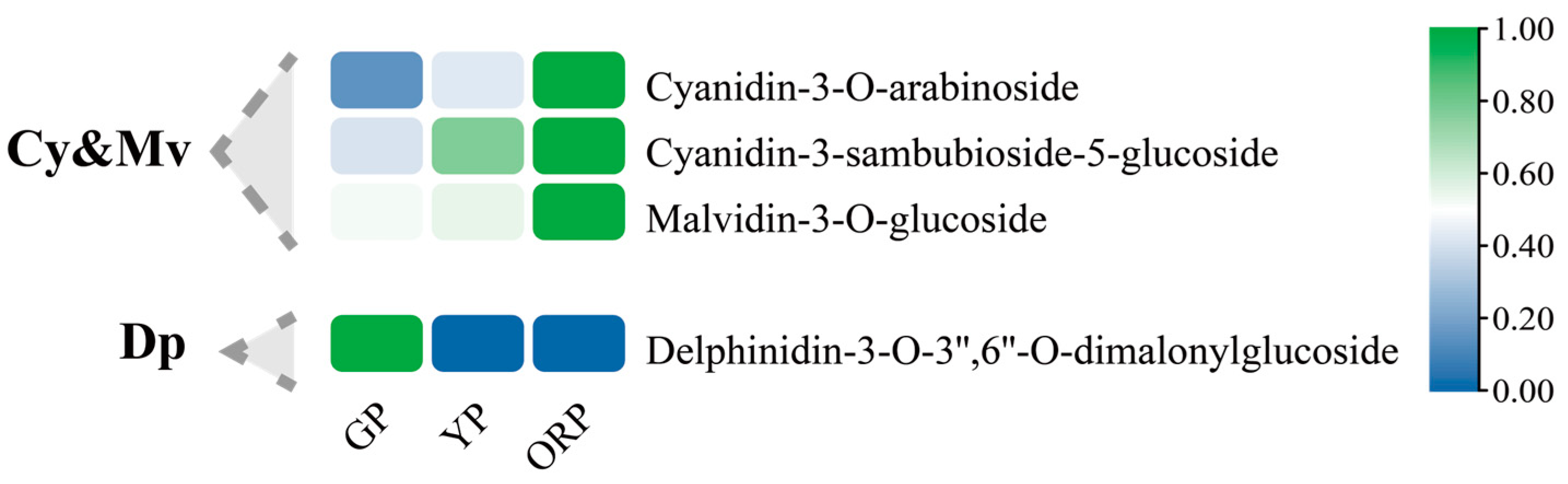

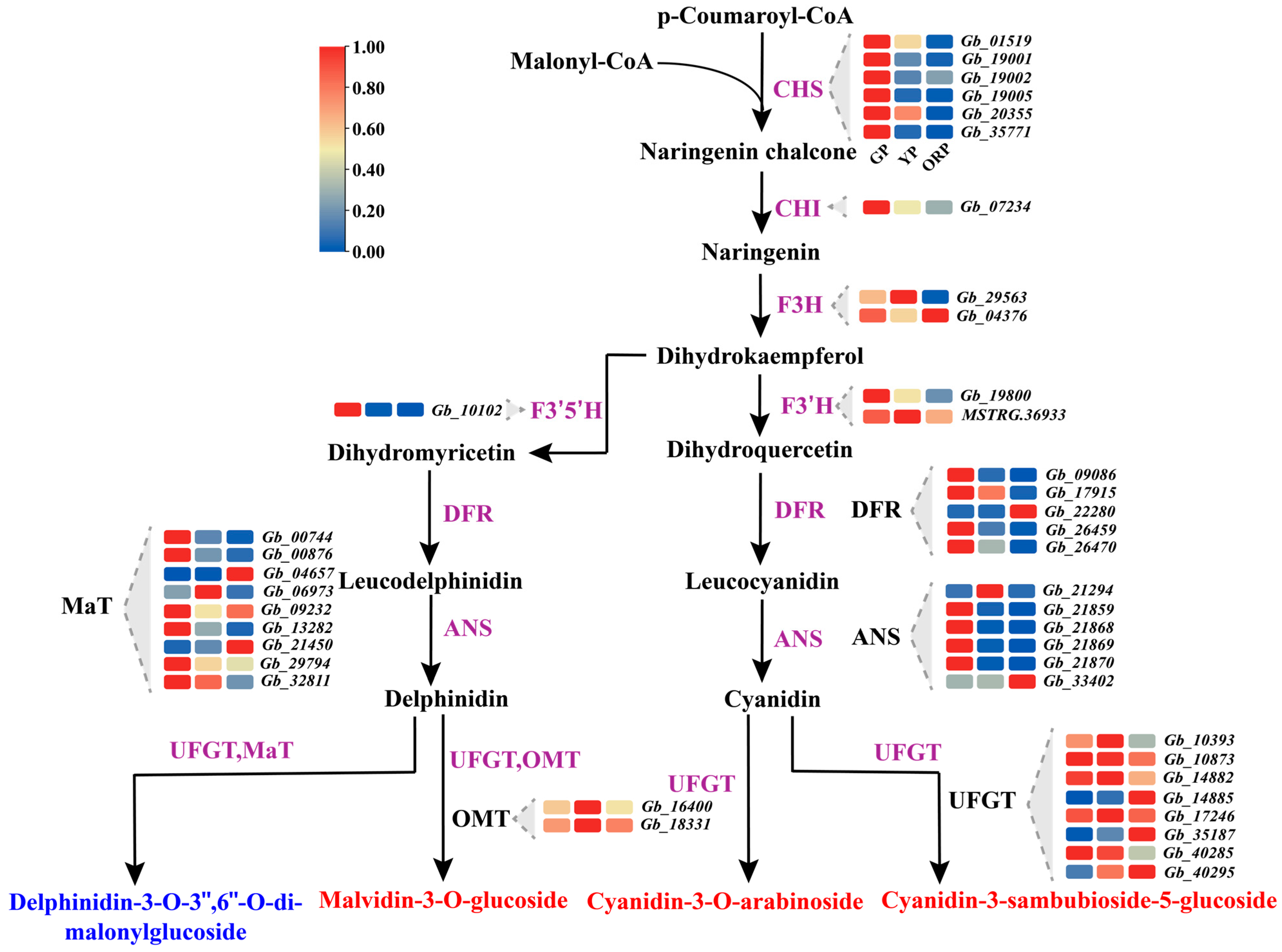
| Developmental Stage | Picking Period | CIEL*a*b* | Anthocyanin Content (mg/g) | ||
|---|---|---|---|---|---|
| Brightness (L*) | Redness (a*) | Yellowness (b*) | |||
| Green period | May | 61.96 ± 4.29 bc | −10.70 ± 1.22 f | 38.42 ± 3.83 cd | 0.1896 ± 0.0304 d |
| Jun | 64.55 ± 4.27 b | −3.55 ± 0.83 e | 39.44 ± 1.16 bc | 0.2540 ± 0.0188 bc | |
| Yellow period | Jul | 70.69 ± 0.94 a | 2.05 ± 0.67 d | 43.38 ± 1.46 ab | 0.2373 ± 0.0140 c |
| Aug | 72.29 ± 1.29 a | 5.76 ± 0.65 c | 46.48 ± 1.83 a | 0.2620 ± 0.0129 bc | |
| Orange-red period | Sep | 57.53 ± 2.03 cd | 9.94 ± 1.32 b | 34.94 ± 2.23 de | 0.2867 ± 0.0225 b |
| Oct | 56.29 ± 2.01 d | 21.33 ± 2.04 a | 32.48 ± 1.99 e | 0.3569 ± 0.0275 a | |
| Index | L* | a* | b* | AN |
|---|---|---|---|---|
| L* | 1 | |||
| a* | −0.379 | 1 | ||
| b* | 0.862 ** | −0.402 | 1 | |
| AN | −0.428 | 0.883 ** | −0.509 * | 1 |
| a* | |
|---|---|
| Proanthocyanidin A2 | −0.026 |
| Leucocyanidin | −0.163 |
| Procyanidin B2 | 0.572 * |
| Cyanidin | −0.040 |
| Pelargonidin | 0.118 |
| Cyanidin 3-arabinoside | 0.914 ** |
| Cyanidin 3-(6″-succinyl-glucoside) | −0.556 * |
| Peonidin-3-glucoside | −0.554 * |
| Cyanidin 3-sambubioside 5-glucoside | 0.736 ** |
| Cyanidin-3-O-(6″-O-malonyl-2″-O-glucuronyl) glucoside | 0.102 |
| Petunidin 3-O-glucoside | −0.162 |
| Malvidin 3-glucoside | 0.783 ** |
| Malvidin 3,5-diglucoside | 0.509 * |
| Delphinidin 3-O-(6″-O-malonyl)-beta-D-glucoside | −0.635 ** |
| Delphinidin 3-O-3″,6″-O-dimalonylglucoside | −0.743 ** |
| Delphinidin 3,5-di(6-O-malonylglucoside) | −0.488 * |
| Gene Category | Anthocyanin | Cyanidin 3-Arabinoside | Cyanidin 3-Sambubioside 5-Glucoside | Malvidin 3-O-Glucoside | Delphinidin 3-O-3″,6″-O-Dimalonylglucoside | |
|---|---|---|---|---|---|---|
| Gene_ID | ||||||
| CHS | Gb_01519 | −0.9454 ** | −0.8984 ** | −0.8650 ** | 0.7533 * | |
| Gb_19001 | −0.7933 * | −0.8832 ** | −0.6161 | 0.8927 ** | ||
| Gb_19005 | −0.7489 * | −0.8642 ** | −0.5462 | 0.9376 ** | ||
| Gb_20355 | −0.9414 ** | −0.8303 ** | −0.8707 ** | 0.6744 * | ||
| Gb_35771 | −0.7504 * | −0.8693 ** | −0.5488 | 0.9314 ** | ||
| CHI | Gb_07234 | −0.8739 ** | −0.8606 ** | −0.7040 * | 0.9352 ** | |
| DFR | Gb_22280 | 0.8948 ** | 0.7334 * | 0.9368 ** | −0.4432 | |
| Gb_09086 | −0.7420 * | −0.8550 ** | −0.5661 | 0.9060 ** | ||
| Gb_17915 | −0.9338 ** | −0.8243 ** | −0.9139 ** | 0.6163 | ||
| Gb_26459 | −0.7812 * | −0.8777 ** | −0.5852 | 0.9231 ** | ||
| Gb_26470 | −0.8823 ** | −0.9206 ** | −0.7377 * | 0.8827 ** | ||
| ANS | Gb_21859 | −0.7498 * | −0.8596 ** | −0.5288 | 0.9764 ** | |
| Gb_21868 | −0.7308 * | −0.8559 ** | −0.5271 | 0.9408 ** | ||
| Gb_21869 | −0.7421 * | −0.8644 ** | −0.5401 | 0.9268 ** | ||
| Gb_21870 | −0.7367 * | −0.8581 ** | −0.5281 | 0.9510 ** | ||
| Gb_33402 | 0.9217 ** | 0.7555 * | 0.9519 ** | −0.4899 | ||
| UFGT | Gb_14885 | 0.9144 ** | 0.7710 * | 0.9321 ** | −0.5035 | |
| Gb_35187 | 0.9174 ** | 0.8260 ** | 0.9217 ** | −0.5417 | ||
| Gb_40295 | 0.8296 ** | 0.8995 ** | 0.6740 * | −0.8719 ** | ||
| Gene Category | Anthocyanin | Cyanidin 3-Arabinoside | Cyanidin 3-Sambubioside 5-Glucoside | |
|---|---|---|---|---|
| Gene_ID | ||||
| F3′H | Gb_19800 | −0.9343 ** | −0.9054 ** | |
| Gene Category | Anthocyanin | Malvidin 3-O-Glucoside | Delphinidin 3-O-3″,6″-O-Dimalonylglucoside | |
|---|---|---|---|---|
| Gene_ID | ||||
| F3′5′H | Gb_10102 | −0.5153 | 0.8876 ** | |
| Gene Category | Anthocyanin | Delphinidin 3-O-3″,6″-O-Dimalonylglucoside | |
|---|---|---|---|
| Gene_ID | |||
| SCPL-AT | Gb_00744 | 0.7883 * | |
| Gb_00876 | 0.9746 ** | ||
| Gb_29794 | 0.9563 ** | ||
| Gb_32811 | 0.5962 | ||
| BAHD-AT | Gb_04657 | −0.4095 | |
| Gb_21450 | −0.5352 | ||
| Gb_13282 | 0.9730 ** | ||
| Gene Category | Anthocyanin | Cyanidin 3-Arabinoside | Cyanidin 3-Sambubioside 5-Glucoside | Malvidin 3-O-Glucoside | Delphinidin 3-O-3″,6″-O-Dimalonylglucoside | |
|---|---|---|---|---|---|---|
| Gene_ID | ||||||
| R2R3MYB | Gb_33428 | 0.9344 ** | 0.8886 ** | 0.8570 ** | −0.7317 * | |
| Gb_05115 | −0.9410 ** | −0.9000 ** | −0.8146 ** | 0.8692 ** | ||
| Gb_18153 | −0.9411 ** | −0.8928 ** | −0.8418 ** | 0.8202 ** | ||
| Gb_19348 | −0.8389 ** | −0.7203 * | −0.7460 * | 0.6809 * | ||
| Gb_24073 | −0.7473 * | −0.8339 ** | −0.5216 | 0.9309 ** | ||
| Gb_24641 | −0.7752 * | −0.8657 ** | −0.5611 | 0.9777 ** | ||
| Gb_38090 | −0.7171 * | −0.7002 * | −0.4960 | 0.9016 ** | ||
| Gb_39484 | −0.9234 ** | −0.7769 * | −0.9480 ** | 0.4516 | ||
| Gb_39852 | −0.8228 ** | −0.8621 ** | −0.7127 * | 0.8204 ** | ||
| R3MYB | Gb_15334 | −0.8798 ** | −0.9232 ** | −0.7282 * | 0.9159 ** | |
| bHLH | Gb_34778 | 0.9096 ** | 0.7898 * | 0.9647 ** | −0.4727 | |
| Gb_14057 | −0.9162 ** | −0.8361 ** | −0.9029 ** | 0.5916 | ||
| Gb_21995 | −0.8852 ** | −0.7791 * | −0.7970 * | 0.7345 * | ||
| Gb_33185 | −0.8234 ** | −0.8723 ** | −0.6158 | 0.9653 ** | ||
| WDR | Gb_37351 | 0.9052 ** | 0.7658 * | 0.7915 * | −0.7554 * | |
| Gb_40146 | 0.8264 ** | 0.7905 * | 0.8317 ** | −0.5701 | ||
| Gb_30748 | −0.8180 ** | −0.7298 * | −0.9117 ** | 0.3287 | ||
| b-ZIP | Gb_12023 | 0.8683 ** | 0.9037 ** | 0.7016 * | −0.8585 ** | |
| Gb_07087 | −0.9076 ** | −0.8475 ** | −0.8071 ** | 0.8107 ** | ||
| Gb_41342 | −0.7924 * | −0.7049 * | −0.8346 ** | 0.4522 | ||
| NAC | Gb_05670 | 0.8730 ** | 0.7530 * | 0.9317 ** | −0.4294 | |
| Gb_13930 | 0.8682 ** | 0.7361 * | 0.9382 ** | −0.4134 | ||
| Gb_17882 | 0.9013 ** | 0.9164 ** | 0.7601 * | −0.8778 ** | ||
| Gb_18916 | 0.8913 ** | 0.8834 ** | 0.8043 ** | −0.7130 * | ||
| Gb_21446 | −0.9266 ** | −0.7874 * | −0.8803 ** | 0.7077 * | ||
| AP2/ERF | Gb_07474 | −0.8226 ** | −0.7134 * | −0.6785 * | 0.8140 ** | |
| Gb_26662 | −0.8952 ** | −0.7758 * | −0.7770 * | 0.7500 * | ||
| MADS-box | Gb_41549 | −0.9357 ** | −0.8845 ** | −0.8765 ** | 0.7646 * | |
| MSTRG.2320 | −0.9227 ** | −0.8616 ** | −0.8772 ** | 0.7407 * | ||
| Gene Category | Structure Gene | Gb_33402 (ANS) | Gb_22280 (DFR) | Gb_21859 (ANS) | Gb_14885 (UFGT) | Gb_35187 (UFGT) | Gb_00876 (SCPL-AT) | Gb_13282 (BAHD-AT) | |
|---|---|---|---|---|---|---|---|---|---|
| TFs | |||||||||
| R2R3MYB | Gb_33428 | 0.9006 ** | 0.8951 ** | −0.8135 ** | 0.9265 ** | 0.9434 ** | −0.8481 ** | −0.8521 ** | |
| Gb_05115 | −0.8540 ** | −0.8199 ** | 0.8795 ** | −0.8552 ** | −0.8715 ** | 0.9197 ** | 0.9539 ** | ||
| Gb_18153 | −0.8624 ** | −0.8401 ** | 0.8730 ** | −0.8759 ** | −0.8934 ** | 0.9020 ** | 0.9236 ** | ||
| Gb_24073 | −0.5431 | −0.5242 | 0.9581 ** | −0.5800 | −0.6157 | 0.9656 ** | 0.9145 ** | ||
| Gb_24641 | −0.5793 | −0.5462 | 0.9983 ** | −0.6058 | −0.6465 | 0.9959 ** | 0.9786 ** | ||
| Gb_38090 | −0.5076 | −0.4672 | 0.8544 ** | −0.5107 | −0.5204 | 0.8848 ** | 0.8789 ** | ||
| Gb_39852 | −0.7271 * | −0.7008 * | 0.8811 ** | −0.7464 * | −0.7825 * | 0.8818 ** | 0.8996 ** | ||
| R3MYB | Gb_15334 | −0.7411 * | −0.7112 * | 0.9565 ** | −0.7594 * | −0.7950 * | 0.9678 ** | 0.9713 ** | |
| bHLH | Gb_34778 | 0.9765 ** | 0.9789 ** | −0.5496 | 0.9755 ** | 0.9765 ** | −0.6107 | −0.6491 | |
| Gb_33185 | −0.6451 | −0.6155 | 0.9786 ** | −0.6687 * | −0.6995 * | 0.9923 ** | 0.9773 ** | ||
| WDR | Gb_37351 | 0.8520 ** | 0.8367 ** | −0.7892 * | 0.8683 ** | 0.8591 ** | −0.8435 ** | −0.8580 ** | |
| b-ZIP | Gb_12023 | 0.7350 * | 0.7176 * | −0.9292 ** | 0.7699 * | 0.8037 ** | −0.9385 ** | −0.9164 ** | |
| Gb_07087 | −0.8532 ** | −0.8426 ** | 0.8440 ** | −0.8706 ** | −0.8783 ** | 0.8887 ** | 0.9006 ** | ||
| NAC | Gb_05670 | 0.9414 ** | 0.9482 ** | −0.5347 | 0.9510 ** | 0.9524 ** | −0.5888 | −0.6123 | |
| Gb_13930 | 0.9394 ** | 0.9396 ** | −0.5114 | 0.9406 ** | 0.9410 ** | −0.5632 | −0.6031 | ||
| Gb_17882 | 0.7941 * | 0.7724 * | −0.9309 ** | 0.8182 ** | 0.8472 ** | −0.9501 ** | −0.9507 ** | ||
| Gb_18916 | 0.8635 ** | 0.8599 ** | −0.8040 ** | 0.8960 ** | 0.9208 ** | −0.8297 ** | −0.8270 ** | ||
| AP2/ERF | Gb_07474 | −0.7397 * | −0.7125 * | 0.7788 * | −0.7387 * | −0.7299 * | 0.8331 ** | 0.8669 ** | |
| Primer Name | Sequence (5′→3′) |
|---|---|
| GAPDH-F | CAAGGACTCCAACACCTTACTC |
| GAPDH-R | CCGTGGATTCAACCACATACT |
| Gb_33402-F | CCTGGTGCTCTGATTGTGAATATCG |
| Gb_33402-R | TGACATCCTTGATTGGTCCTTATGC |
| Gb_22280-F | CGAATAGCGGCACAGAGCAGAA |
| Gb_22280-R | TGACAGTAGCATGGACGTTGTAACC |
| Gb_21859-F | TGGTGCCTGGTCTCCAACTCTT |
| Gb_21859-R | AGCCCACTCTTGTATTTGCCATTGC |
| Gb_14885-F | GGCGTTCCAATGCTCAGTGTTC |
| Gb_14885-R | CGCTTATTCAGTCGCAATCCAGTCT |
| Gb_35187-F | CCATTAGAGACGCAGAAGGAGAGT |
| Gb_35187-R | AATGTGAACGGTCGCAGCATAA |
| Gb_00876-F | TCTAAGCCTCTGGTTCTGTGGTTGA |
| Gb_00876-R | GAGCGACTTTCCATCCGAGTTGAC |
| Gb_13282-F | TCCGCCCTTATCTCCGTCTTTCTTT |
| Gb_13282-R | TGCTCCATACCACATTCCGTCACT |
| Gb_33428-F | GCAGCAATCTGGAGCCGAAGG |
| Gb_33428-R | CCGTCGCCCTTGTACTCTCATTTC |
| Gb_05115-F | CCGTGCTGCGAGAAGGTTGG |
| Gb_05115-R | GCTCTTTCCACACCGTAACAGACC |
| Gb_18153-F | TAGCATAAGCAGAGCCACCACAG |
| Gb_18153-R | ACAGCAGCCTCCTCCATTGATT |
| Gb_24073-F | ACAGCGGTTAAGATGGCGGTTG |
| Gb_24073-R | TGCGATTGGAATCCTGCGGTTG |
| Gb_24641-F | ATCCATTTGCGTGTTCCCGACTAG |
| Gb_24641-R | GCTTCCGCTCGCTATCTCATCAG |
| Gb_38090-F | CCGTCCCCATTAACTCCACCAAC |
| Gb_38090-R | GCTGTGCCTCCTTCATGCTGTC |
| Gb_39852-F | GCTGCTTGGATACAGATCAGGATG |
| Gb_39852-R | CCGCCGAGAATTTGGGAGAG |
| Gb_34778-F | TCTGGGTGTCGGAAGCGGATAA |
| Gb_34778-R | CTGAGCCTCCACGAAATTCCACTT |
| Gb_37351-F | GCTTCAACCTTGGCAGTGTCATC |
| Gb_37351-R | GCTTCCTCTTCTTCATCACGCTCTA |
| Gb_12023-F | ATCATTTGACGGCTTTGGCGGTAG |
| Gb_12023-R | GATTGGTGGGCAGGATAGGCGATA |
| Gb_05670-F | AAGAAACTGTTACTCCAGCCGAAGA |
| Gb_05670-R | AGCAGCCTGTAGCACCAATTCATT |
| Gb_13930-F | TCCACCAGGTCAAGAGATCCAATCC |
| Gb_13930-R | GTATGGCGGCATCCAATGTCTGT |
| Gb_07474-F | CGCCCATCACTCAGCATTTCC |
| Gb_07474-R | AGCCTCCTTCAGAACGCCATAA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, J.; Feng, Z.; Xiang, X.; Wang, Y.; Li, M. Combined Metabolomic and Transcriptomic Analysis Reveals Candidate Genes for Anthocyanin Accumulation in Ginkgo biloba Seed Exocarp. Horticulturae 2024, 10, 540. https://doi.org/10.3390/horticulturae10060540
Tang J, Feng Z, Xiang X, Wang Y, Li M. Combined Metabolomic and Transcriptomic Analysis Reveals Candidate Genes for Anthocyanin Accumulation in Ginkgo biloba Seed Exocarp. Horticulturae. 2024; 10(6):540. https://doi.org/10.3390/horticulturae10060540
Chicago/Turabian StyleTang, Jianlu, Zhi Feng, Xiangyue Xiang, Yiqiang Wang, and Meng Li. 2024. "Combined Metabolomic and Transcriptomic Analysis Reveals Candidate Genes for Anthocyanin Accumulation in Ginkgo biloba Seed Exocarp" Horticulturae 10, no. 6: 540. https://doi.org/10.3390/horticulturae10060540
APA StyleTang, J., Feng, Z., Xiang, X., Wang, Y., & Li, M. (2024). Combined Metabolomic and Transcriptomic Analysis Reveals Candidate Genes for Anthocyanin Accumulation in Ginkgo biloba Seed Exocarp. Horticulturae, 10(6), 540. https://doi.org/10.3390/horticulturae10060540







