Only 11 Simple Sequence Repeats Needed to Identify Chinese Cabbage (Brassica rapa L.) Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of Molecular Markers Showing Polymorphism among Chinese Cabbage Cultivars
2.1.1. Plant Materials and DNA Preparation
2.1.2. Amplification of Genome-Anchored Markers
2.2. Development of a Minimum Number of Markers for Identifying Cultivars
2.2.1. Plant Material, DNA Preparation, and Genotyping
2.2.2. Cluster Analysis
2.2.3. Procedure for Selecting the Minimum Number of Markers
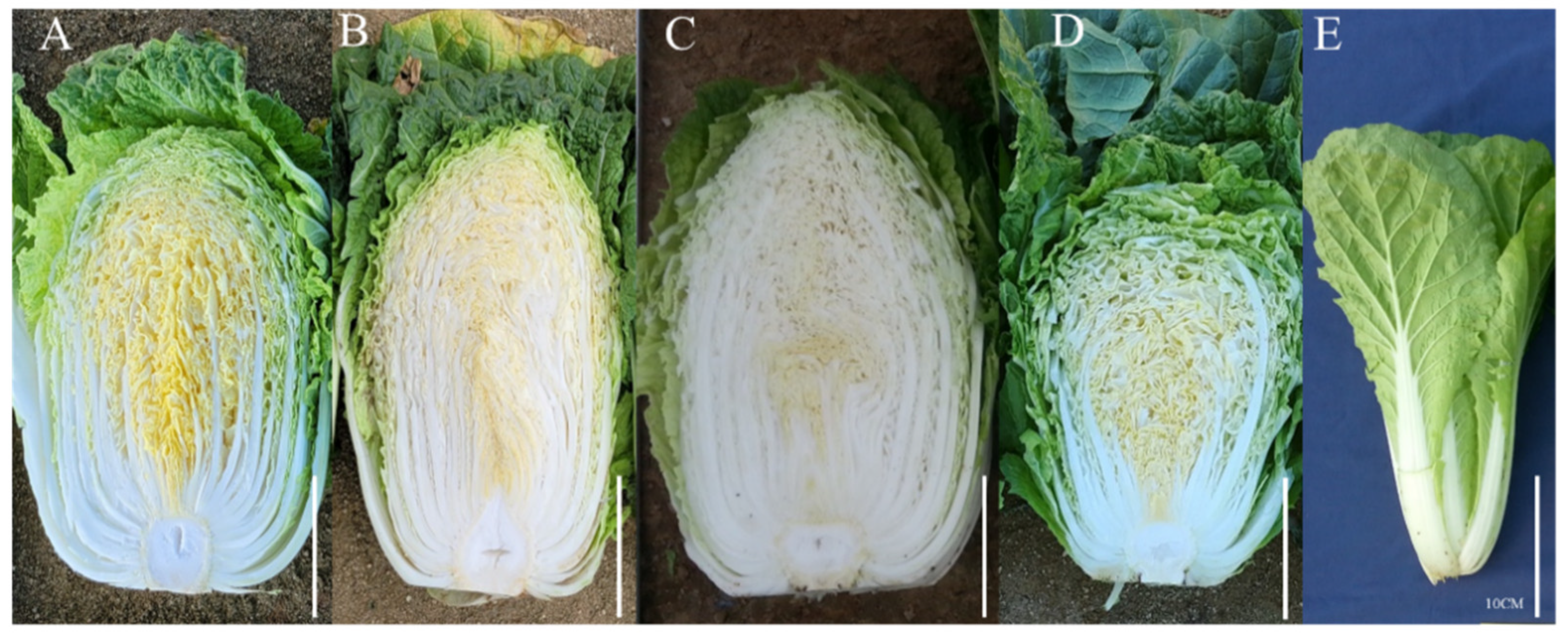
2.2.4. Phenotypic Observation of Chinese Cabbage Cultivars
3. Results
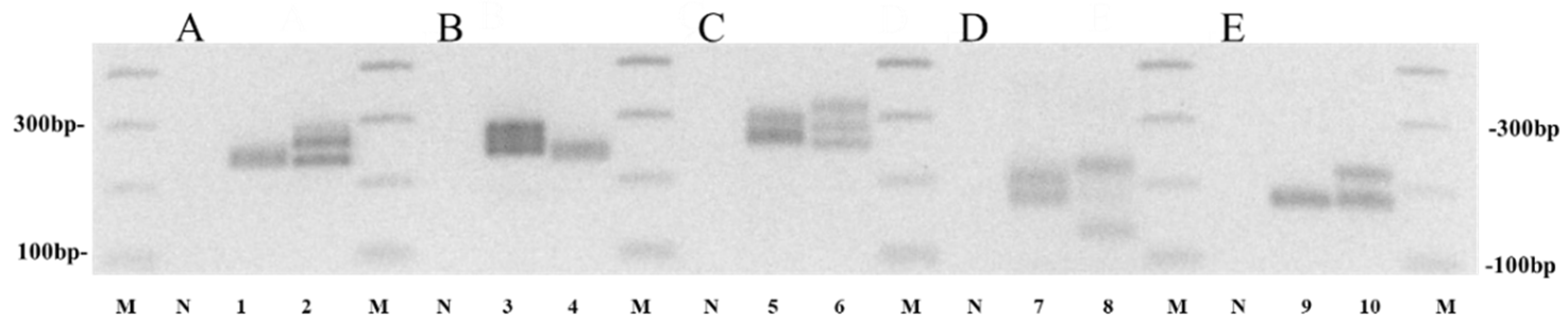
3.1. Selection of Markers Showing Polymorphism among Chinese Cabbage Cultivars
3.2. Development of the Minimum Number of Markers for Cultivar Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Islam, M.; Choi, H. Chinese cabbage (Brassica campestris L.) does not improve glucose tolerance, serum insulin, or blood lipid profiles in a rat model of type-2 diabetes. J. Food Sci. 2008, 73, 213–217. [Google Scholar] [CrossRef]
- Jang, D.-J.; Chung, K.R.; Yang, H.J.; Kim, K.-S.; Kwon, D.Y. Discussion on the origin of kimchi, representative of Korean unique fermented vegetables. J. Ethn. Foods 2015, 2, 126–136. [Google Scholar] [CrossRef]
- Ji, F.D.; Ji, B.P.; Li, B.; Lu, F. Effect of fermentation on nitrate, nitrite and organic acid contents in traditional pickled Chinese cabbage. J. Food Process. Preserv. 2009, 33, 175–186. [Google Scholar] [CrossRef]
- Iacovou, M.; Tan, V.; Muir, J.G.; Gibson, P.R. The low FODMAP diet and its application in East and Southeast Asia. J. Neurogastroenterol. Motil. 2015, 21, 459. [Google Scholar] [CrossRef] [PubMed]
- Saeki, N.; Kawanabe, T.; Ying, H.; Shimizu, M.; Kojima, M.; Abe, H.; Okazaki, K.; Kaji, M.; Taylor, J.M.; Sakakibara, H. Molecular and cellular characteristics of hybrid vigour in a commercial hybrid of Chinese cabbage. BMC Plant Biol. 2016, 16, 45. [Google Scholar] [CrossRef]
- Cho, M.; Kwak, J.; Jeong, H.; Jang, S.; Park, S.; Kwon, Y.; Kim, C.; Choi, M.; Han, J.; Moon, J. Overview of Korean vegetable breeding: Past, present and future. Korean Soc. Breed. Sci. 2020, 52, 112–143. [Google Scholar] [CrossRef]
- Korea Seed and Variety Service. Status of Cultivar Protection Applications and Registrations. 2023. Available online: http://www.seed.go.kr (accessed on 4 September 2023).
- Kwon, Y.; Prak, D.; Song, I.; Yi, S.; Yoon, W.; Moon, J. AFLP analysis for cultivar discrimination in radish and Chinese cabbage. Korean J. Breed. 2003, 35, 319–328. (In Korean) [Google Scholar]
- Choe, Y.; Park, D.; Shin, H.; Kwon, Y.; Yoon, W.; Moon, J. Studies for similarity evaluation of radish and Chinese cabbage cultivars. Hortic. Sci. Technol. 2002, 20, 160–167. (In Korean) [Google Scholar]
- Kumar, P.; Gupta, V.; Misra, A.; Modi, D.; Pandey, B. Potential of molecular markers in plant biotechnology. Plant Omics 2009, 2, 141–162. [Google Scholar]
- Choi, S.; Sim, S.; Hong, J.; Choi, K.; Jin, M.; Park, B.; Kim, D.; Kwon, Y. Genetic characterisation of commercial Chinese cabbage varieties using SSR markers. Seed Sci. Technol. 2016, 44, 595–608. [Google Scholar] [CrossRef]
- Li, P.; Su, T.; Yu, S.; Wang, H.; Wang, W.; Yu, Y.; Zhang, D.; Zhao, X.; Wen, C.; Zhang, F. Identification and development of a core set of informative genic SNP markers for assaying genetic diversity in Chinese cabbage. Hortic. Environ. Biotechnol. 2019, 60, 411–425. [Google Scholar] [CrossRef]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Ghislain, M.; Spooner, D.; Rodríguez, F.; Villamón, F.; Nunez, J.; Vásquez, C.; Waugh, R.; Bonierbale, M. Selection of highly informative and user-friendly microsatellites (SSRs) for genotyping of cultivated potato. Theor. Appl. Genet. 2004, 108, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Hong, H.; Lee, J.; Chae, W. An economic method to identify cultivars and elite lines in radish (Raphanus sativus L.) for small seed companies and independent breeders. Horticulturae 2023, 9, 140. [Google Scholar] [CrossRef]
- Kim, H.; Choi, S.R.; Bae, J.; Hong, C.P.; Lee, S.Y.; Hossain, M.J.; Van Nguyen, D.; Jin, M.; Park, B.-S.; Bang, J.-W. Sequenced BAC anchored reference genetic map that reconciles the ten individual chromosomes of Brassica rapa. BMC Genomics 2009, 10, 432. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. Isolation of plant DNA from fresh tissue. Focus 1990, 12, 13–15. [Google Scholar]
- Jaccard, P. Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 1908, 44, 223–270. [Google Scholar]
- Rohlf, F.J. NTSYS–Pc: Numerical Taxonomy and Multivariate Analysis System, Version 2.1; Applied Biostatistics Inc.: New York, NY, USA, 2000. [Google Scholar]
- Anderson, J.A.; Churchill, G.; Autrique, J.; Tanksley, S.; Sorrells, M. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Earl, D.A.; VonHoldt, B.M. STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv. Genet. Gesources 2012, 4, 359–361. [Google Scholar] [CrossRef]
- Suwabe, K.; Iketani, H.; Nunome, T.; Kage, T.; Hirai, M. Isolation and characterization of microsatellites in Brassica rapa L. Theor. Appl. Genet. 2002, 104, 1092–1098. [Google Scholar] [CrossRef]
- Lowe, A.; Moule, C.; Trick, M.; Edwards, K. Efficient large-scale development of microsatellites for marker and mapping applications in Brassica crop species. Theor. Appl. Genet. 2004, 108, 1103–1112. [Google Scholar] [CrossRef] [PubMed]
- Ramchiary, N.; Nguyen, V.D.; Li, X.; Hong, C.P.; Dhandapani, V.; Choi, S.R.; Yu, G.; Piao, Z.Y.; Lim, Y.P. Genic microsatellite markers in Brassica rapa: Development, characterization, mapping, and their utility in other cultivated and wild Brassica relatives. DNA Res. 2011, 18, 305–320. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Ge, T.; Li, Y.; Hou, X. Genome-wide identification of SSR and SNP markers from the non-heading Chinese cabbage for comparative genomic analyses. BMC Genomics 2015, 16, 328. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Li, J.; Wang, F.; Zhang, Y.; Li, H.; Zhang, J.; Gao, J. Characterization and development of EST-SSRs by deep transcriptome sequencing in Chinese cabbage (Brassica rapa L. ssp. pekinensis). Int. J. Genomics 2015, 2015, 473028. [Google Scholar] [CrossRef] [PubMed]



| Cultivars | Genetic Similarity z | |
|---|---|---|
| Goranengjiyeoreum | Yeoreimsingwan | 0.97 |
| Victory | CR-chunhailpoom | 0.97 |
| CR-saesinrokutgari | Chamsin | 0.97 |
| Chunhanoran | Norangbom | 0.95 |
| CR-ok | Lipoomyeoreum | 0.93 |
| Primers | Linkage Group | NA z | Allele Frequency | PIC y | |||
|---|---|---|---|---|---|---|---|
| Allele 1 | Allele 2 | Allele 3 | Allele 4 | ||||
| Cnu_m474a | 1 | 3 | 0.439 | 0.140 | 0.037 | – | 0.786 |
| Cnu_m046a | 2 | 4 | 0.523 | 0.355 | 0.224 | 0.019 | 0.549 |
| Cnu_m241a | 3 | 4 | 0.551 | 0.280 | 0.150 | 0.140 | 0.575 |
| Cnu_m256a | 4 | 3 | 0.486 | 0.262 | 0.178 | – | 0.664 |
| Cnu_m471a | 5 | 3 | 0.617 | 0.308 | 0.187 | – | 0.489 |
| Cnu_m257a | 5 | 3 | 0.869 | 0.664 | 0.299 | – | – |
| Cnu_m049a | 6 | 3 | 0.411 | 0.383 | 0.112 | – | 0.671 |
| Cnu_m308a | 7 | 3 | 0.402 | 0.243 | 0.121 | – | 0.757 |
| Nia_m095a | 8 | 4 | 0.570 | 0.159 | 0.140 | 0.019 | 0.630 |
| Cnu_m008a | 9 | 3 | 0.542 | 0.449 | 0.075 | – | 0.499 |
| Nia_m034a | 10 | 3 | 0.729 | 0.355 | 0.103 | – | 0.332 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Lim, J.; Jeong, Y.; Yu, H.; Park, Y.; Lim, C.; Chae, W. Only 11 Simple Sequence Repeats Needed to Identify Chinese Cabbage (Brassica rapa L.) Cultivars. Horticulturae 2023, 9, 1123. https://doi.org/10.3390/horticulturae9101123
Kim J, Lim J, Jeong Y, Yu H, Park Y, Lim C, Chae W. Only 11 Simple Sequence Repeats Needed to Identify Chinese Cabbage (Brassica rapa L.) Cultivars. Horticulturae. 2023; 9(10):1123. https://doi.org/10.3390/horticulturae9101123
Chicago/Turabian StyleKim, Jiwon, Jihyeon Lim, Yunjeong Jeong, Hyewon Yu, Yong Park, Chaewan Lim, and Wonbyoung Chae. 2023. "Only 11 Simple Sequence Repeats Needed to Identify Chinese Cabbage (Brassica rapa L.) Cultivars" Horticulturae 9, no. 10: 1123. https://doi.org/10.3390/horticulturae9101123
APA StyleKim, J., Lim, J., Jeong, Y., Yu, H., Park, Y., Lim, C., & Chae, W. (2023). Only 11 Simple Sequence Repeats Needed to Identify Chinese Cabbage (Brassica rapa L.) Cultivars. Horticulturae, 9(10), 1123. https://doi.org/10.3390/horticulturae9101123






