Influence of Genotype and Environment on Fruit Phenolic Composition of Olive
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Locations
2.2. Analysis of Fruit Phenolic Compounds
2.3. Data Analysis
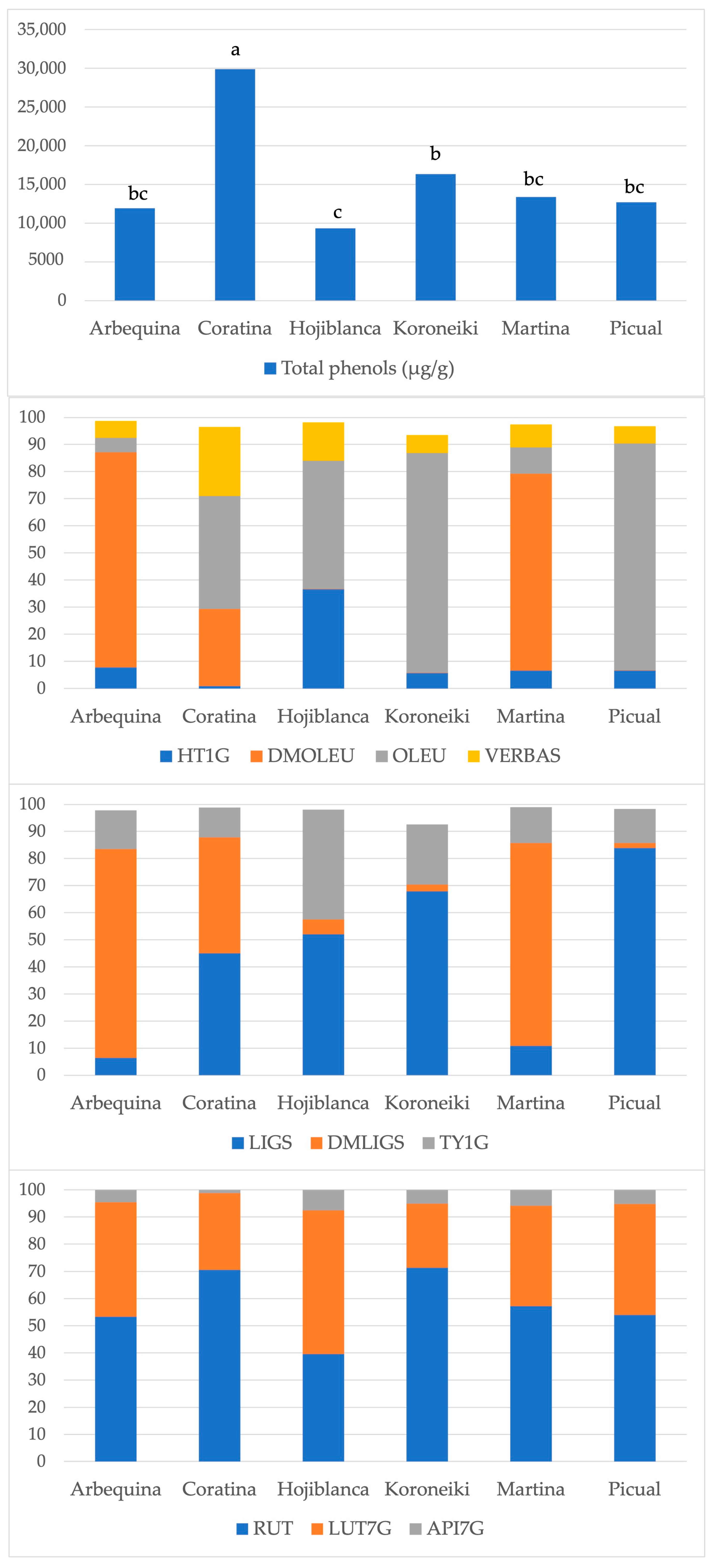
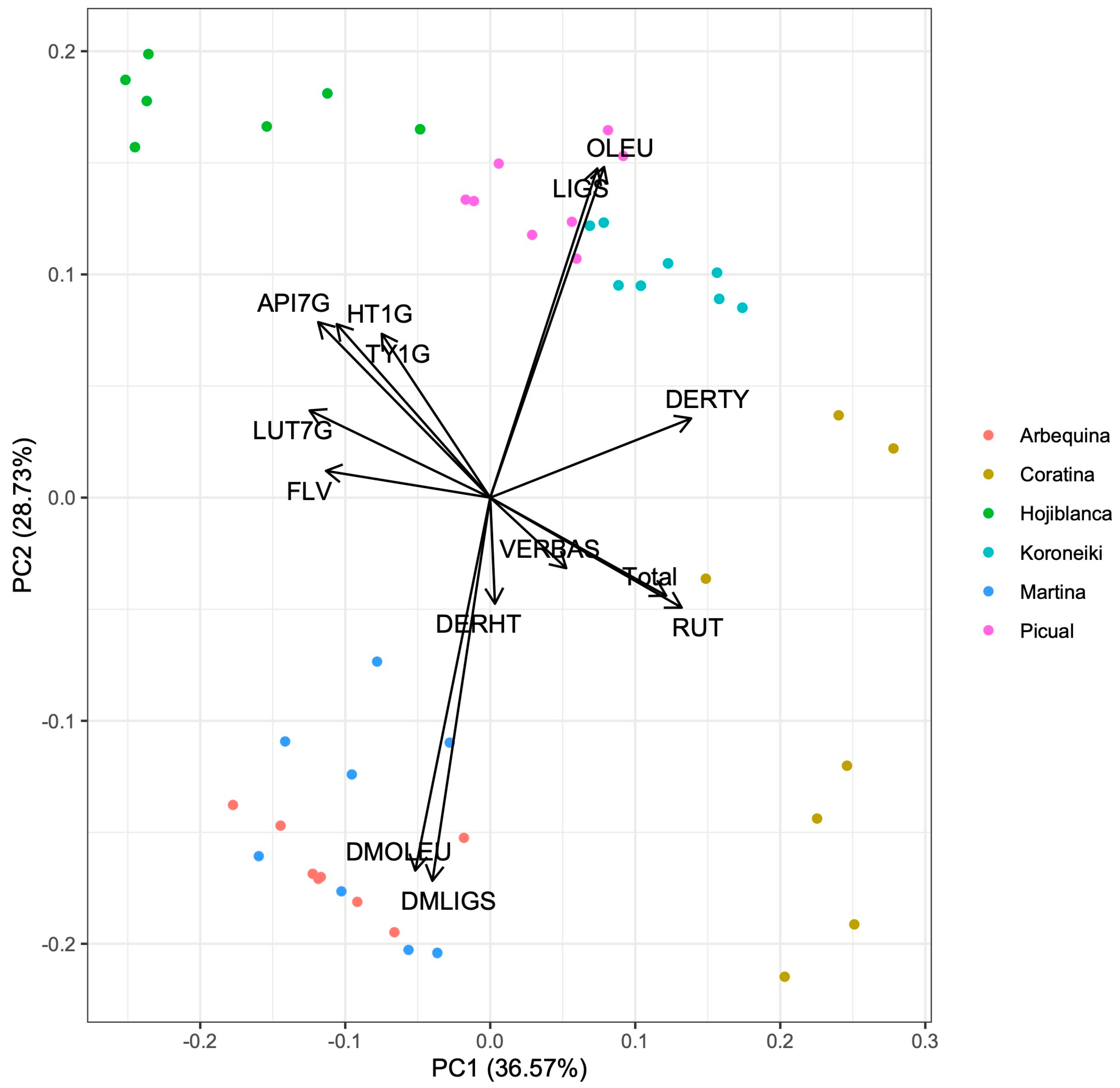
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lechhab, T.; Lechhab, W.; Cacciola, F.; Salmoun, F. Sets of Internal and External Factors Influencing Olive Oil (Olea europaea L.) Composition: A Review. Eur. Food Res. Technol. 2022, 248, 1069–1088. [Google Scholar] [CrossRef]
- Ray, N.B.; Hilsabeck, K.D.; Karagiannis, T.C.; McCord, D.E. Bioactive Olive Oil Polyphenols in the Promotion of Health. In The Role of Functional Food Security in Global Health; Elsevier: Amsterdam, The Netherlands, 2019; pp. 623–637. ISBN 978-0-12-813148-0. [Google Scholar]
- Granados-Principal, S.; Quiles, J.L.; Ramirez-Tortosa, C.L.; Sanchez-Rovira, P.; Ramirez-Tortosa, M.C. Hydroxytyrosol: From Laboratory Investigations to Future Clinical Trials. Nutr. Rev. 2010, 68, 191–206. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Rico, A.; Fregapane, G.; Salvador, M.D. Effect of Cultivar and Ripening on Minor Components in Spanish Olive Fruits and Their Corresponding Virgin Olive Oils. Food Res. Int. 2008, 41, 433–440. [Google Scholar] [CrossRef]
- Talhaoui, N.; Gómez-Caravaca, A.; León, L.; De La Rosa, R.; Fernández-Gutiérrez, A.; Segura-Carretero, A. From Olive Fruits to Olive Oil: Phenolic Compound Transfer in Six Different Olive Cultivars Grown under the Same Agronomical Conditions. Int. J. Mol. Sci. 2016, 17, 337. [Google Scholar] [CrossRef] [PubMed]
- Romero-Segura, C.; García-Rodríguez, R.; Sánchez-Ortiz, A.; Sanz, C.; Pérez, A.G. The Role of Olive β-Glucosidase in Shaping the Phenolic Profile of Virgin Olive Oil. Food Res. Int. 2012, 45, 191–196. [Google Scholar] [CrossRef]
- Hachicha Hbaieb, R.; Kotti, F.; García-Rodríguez, R.; Gargouri, M.; Sanz, C.; Pérez, A.G. Monitoring Endogenous Enzymes during Olive Fruit Ripening and Storage: Correlation with Virgin Olive Oil Phenolic Profiles. Food Chem. 2015, 174, 240–247. [Google Scholar] [CrossRef] [PubMed]
- Pérez, A.G.; León, L.; Sanz, C.; De La Rosa, R. Fruit Phenolic Profiling: A New Selection Criterion in Olive Breeding Programs. Front. Plant Sci. 2018, 9, 241. [Google Scholar] [CrossRef] [PubMed]
- Gucci, R.; Caruso, G.; Gennai, C.; Esposto, S.; Urbani, S.; Servili, M. Fruit Growth, Yield and Oil Quality Changes Induced by Deficit Irrigation at Different Stages of Olive Fruit Development. Agric. Water Manag. 2019, 212, 88–98. [Google Scholar] [CrossRef]
- Medina, G.; Sanz, C.; León, L.; Pérez, A.G.; De La Rosa, R. Phenolic Variability in Fruit from the ‘Arbequina’ Olive Cultivar under Mediterranean and Subtropical Climatic Conditions. Grasas Aceites 2021, 72, e438. [Google Scholar] [CrossRef]
- Pérez, A.G.; León, L.; Pascual, M.; Romero-Segura, C.; Sánchez-Ortiz, A.; de la Rosa, R.; Sanz, C. Variability of Virgin Olive Oil Phenolic Compounds in a Segregating Progeny from a Single Cross in Olea europaea L. and Sensory and Nutritional Quality Implications. PLoS ONE 2014, 9, e92898. [Google Scholar] [CrossRef] [PubMed]
- Navas-López, J.F.; Cano, J.; de la Rosa, R.; Velasco, L.; León, L. Genotype by Environment Interaction for Oil Quality Components in Olive Tree. Eur. J. Agron. 2020, 119, 126115. [Google Scholar] [CrossRef]
- Belaj, A.; Ninot, A.; Gómez-Gálvez, F.J.; El Riachy, M.; Gurbuz-Veral, M.; Torres, M.; Lazaj, A.; Klepo, T.; Paz, S.; Ugarte, J.; et al. Utility of EST-SNP Markers for Improving Management and Use of Olive Genetic Resources: A Case Study at the Worldwide Olive Germplasm Bank of Córdoba. Plants 2022, 11, 921. [Google Scholar] [CrossRef] [PubMed]
- García-Rodríguez, R.; Romero-Segura, C.; Sanz, C.; Sánchez-Ortiz, A.; Pérez, A.G. Role of Polyphenol Oxidase and Peroxidase in Shaping the Phenolic Profile of Virgin Olive Oil. Food Res. Int. 2011, 44, 629–635. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2023; Available online: https://www.R-project.org/ (accessed on 1 July 2023).
- Bodoira, R.; Torres, M.; Pierantozzi, P.; Taticchi, A.; Servili, M.; Maestri, D. Oil Biogenesis and Antioxidant Compounds from “Arauco” Olive (Olea europaea L.) Cultivar during Fruit Development and Ripening. Eur. J. Lipid Sci. Technol. 2015, 117, 377–388. [Google Scholar] [CrossRef]
- Servili, M.; Montedoro, G. Contribution of phenolic compounds to virgin olive oil quality. Eur. J. Lipid Sci. Technol. 2002, 104, 602–613. [Google Scholar] [CrossRef]
- Uylaşer, V. Changes in Phenolic Compounds during Ripening in Gemlik Variety Olive Fruits Obtained from Different Locations. CyTA—J. Food 2015, 13, 167–173. [Google Scholar] [CrossRef]
- Romero, M.-P.; Motilva, M.-J. Effect of Climatic Conditions on Quality of Virgin Olive Oil. In Olives and Olive Oil in Health and Disease Prevention; Elsevier: Amsterdam, The Netherlands, 2010; pp. 43–50. ISBN 978-0-12-374420-3. [Google Scholar]
- Arslan, D.; Özcan, M.M. Phenolic Profile and Antioxidant Activity of Olive Fruits of the Turkish Variety “Sarıulak” from Different Locations. Grasas Aceites 2011, 62, 453–461. [Google Scholar] [CrossRef]
- Navas-Lopez, J.F.; León, L.; Trentacoste, E.R.; de la Rosa, R. Multi-Environment Evaluation of Oil Accumulation Pattern Parameters in Olive. Plant Physiol. Biochem. 2019, 139, 485–494. [Google Scholar] [CrossRef] [PubMed]
- Navas-Lopez, J.F.; León, L.; Rapoport, H.F.; Moreno-Alías, I.; Lorite, I.J.; De La Rosa, R. Genotype, Environment and Their Interaction Effects on Olive Tree Flowering Phenology and Flower Quality. Euphytica 2019, 215, 184. [Google Scholar] [CrossRef]
- García-González, D.L.; Tena, N.; Aparicio, R. Quality Characterization of the New Virgin Olive Oil Var. Sikitita by Phenols and Volatile Compounds. J. Agric. Food Chem. 2010, 58, 8357–8364. [Google Scholar] [CrossRef] [PubMed]
| Average Temp (°C) | Max Temp (°C) | Min Temp (°C) | Rainfall (mm) | |||||
|---|---|---|---|---|---|---|---|---|
| Tenerife | Cordoba | Tenerife | Cordoba | Tenerife | Cordoba | Tenerife | Cordoba | |
| January | 14.69 | 7.91 | 19.29 | 14.10 | 11.46 | 2.95 | 135.30 | 70.00 |
| February | 14.61 | 12.10 | 19.21 | 18.24 | 11.10 | 7.56 | 85.55 | 45.80 |
| March | 15.32 | 13.52 | 20.54 | 21.83 | 11.54 | 6.36 | 5.20 | 9.40 |
| April | 16.38 | 16.54 | 21.10 | 23.33 | 12.96 | 10.97 | 18.30 | 83.80 |
| May | 19.01 | 20.65 | 24.52 | 29.35 | 14.47 | 11.80 | 0.10 | 9.40 |
| June | 19.69 | 24.61 | 25.19 | 33.32 | 15.45 | 15.83 | 1.00 | 12.80 |
| July | 24.04 | 28.57 | 29.42 | 37.60 | 19.33 | 18.87 | 0.00 | 0.00 |
| August | 23.45 | 28.75 | 28.84 | 37.60 | 19.27 | 19.83 | 1.10 | 0.00 |
| September | 21.28 | 23.94 | 26.25 | 31.72 | 17.86 | 17.20 | 0.20 | 40.60 |
| November | 16.45 | 10.94 | 21.15 | 18.43 | 13.79 | 4.96 | 0.00 | 27.20 |
| October | 20.67 | 19.34 | 26.25 | 27.86 | 16.55 | 12.46 | 54.20 | 19.40 |
| December | 15.66 | 10.45 | 20.31 | 17.16 | 12.81 | 4.76 | 13.90 | 122.80 |
| Average | 18.47 | 18.17 | 23.54 | 25.96 | 14.74 | 11.16 | 314.85 * | 441.20 * |
| Average | Coefficient of Variation (%) | Min | Max | |
|---|---|---|---|---|
| Total phenols (μg/g) | 16,577 | 48.53 | 6019 | 38,380 |
| DERHT (%) | 88.72 | 3.19 | 80.80 | 94.06 |
| HT1G | 8.82 | 145.42 | 0.09 | 60.08 |
| DMOLEU | 29.59 | 119.11 | 0.01 | 86.61 |
| OLEU | 48.52 | 70.87 | 0.31 | 97.73 |
| VERBAS | 10.14 | 78.89 | 0.69 | 33.13 |
| DERTY (%) | 5.55 | 49.85 | 2.02 | 14.34 |
| LIGS | 48.83 | 69.20 | 0.73 | 96.67 |
| DMLIGS | 32.21 | 106.66 | 0.22 | 97.65 |
| TY1G | 16.23 | 89.41 | 0.03 | 70.16 |
| FLV (%) | 5.73 | 50.07 | 1.15 | 15.79 |
| RUT | 57.65 | 19.29 | 36.99 | 78.04 |
| LUT7G | 37.44 | 26.56 | 18.74 | 56.32 |
| API7G | 4.91 | 41.55 | 0.23 | 8.57 |
| Sum of Squares (%) | |||||||
|---|---|---|---|---|---|---|---|
| G | HD | G × HD | Error | ||||
| Total phenols | 90 | *** | 7 | *** | 1 | ns | 3 |
| DERHT | 20 | ns | 2 | ns | 15 | ns | 63 |
| HT1G | 72 | *** | 8 | *** | 19 | *** | 1 |
| DMOLEU | 99 | *** | 0 | ** | 1 | *** | 0 |
| OLEU | 94 | *** | 2 | *** | 4 | *** | 0 |
| VERBAS | 93 | *** | 0 | ns | 2 | ns | 4 |
| DERTY | 89 | *** | 5 | *** | 4 | * | 2 |
| LIGS | 88 | *** | 3 | *** | 7 | *** | 1 |
| DMLIGS | 98 | *** | 0 | ns | 1 | ns | 1 |
| TY1G | 42 | *** | 10 | ** | 40 | *** | 7 |
| FLV | 52 | * | 6 | ns | 6 | ns | 36 |
| RUT | 94 | *** | 0 | ns | 2 | ns | 3 |
| LUT7G | 94 | *** | 0 | ns | 2 | ns | 4 |
| API7G | 90 | *** | 2 | * | 2 | ns | 6 |
| Sum of Squares (%) | |||||||
|---|---|---|---|---|---|---|---|
| G | L | G × L | Error | ||||
| Total phenols | 76 | *** | 14 | *** | 6 | ns | 4 |
| DERHT | 8 | ns | 5 | ns | 29 | ns | 57 |
| HT1G | 79 | *** | 5 | *** | 14 | *** | 1 |
| DMOLEU | 94 | *** | 2 | *** | 3 | ** | 1 |
| OLEU | 84 | *** | 7 | *** | 8 | *** | 1 |
| VERBAS | 69 | *** | 1 | ns | 29 | *** | 2 |
| DERTY | 80 | *** | 1 | ns | 17 | *** | 2 |
| LIGS | 82 | *** | 8 | *** | 8 | ** | 2 |
| DMLIGS | 90 | *** | 4 | *** | 5 | ** | 1 |
| TY1G | 77 | *** | 4 | ns | 9 | ns | 9 |
| FLV | 47 | ns | 6 | ns | 11 | ns | 36 |
| RUT | 89 | *** | 1 | ns | 6 | ns | 4 |
| LUT7G | 90 | *** | 1 | ns | 5 | ns | 4 |
| API7G | 74 | *** | 2 | ns | 18 | ** | 6 |
| Sum of Squares (%) | |||||||
|---|---|---|---|---|---|---|---|
| G | Y | G × Y | Error | ||||
| Total phenols | 88 | *** | 1 | ns | 8 | ** | 3 |
| DERHT | 16 | ns | 2 | ns | 17 | ns | 65 |
| HT1G | 96 | *** | 2 | ** | 1 | ns | 1 |
| DMOLEU | 93 | *** | 2 | ** | 3 | * | 2 |
| OLEU | 90 | *** | 2 | * | 6 | * | 3 |
| VERBAS | 82 | *** | 7 | *** | 7 | * | 4 |
| DERTY | 92 | *** | 1 | ns | 3 | ns | 4 |
| LIGS | 83 | *** | 1 | ns | 11 | * | 5 |
| DMLIGS | 87 | *** | 4 | *** | 7 | ** | 2 |
| TY1G | 81 | *** | 2 | ns | 6 | ns | 12 |
| FLV | 45 | ns | 4 | ns | 7 | ns | 45 |
| RUT | 93 | *** | 1 | ns | 1 | ns | 4 |
| LUT7G | 93 | *** | 1 | ns | 1 | ns | 5 |
| API7G | 82 | *** | 0 | ns | 4 | ns | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yılmaz-Düzyaman, H.; Medina-Alonso, M.G.; Sanz, C.; Pérez, A.G.; de la Rosa, R.; León, L. Influence of Genotype and Environment on Fruit Phenolic Composition of Olive. Horticulturae 2023, 9, 1087. https://doi.org/10.3390/horticulturae9101087
Yılmaz-Düzyaman H, Medina-Alonso MG, Sanz C, Pérez AG, de la Rosa R, León L. Influence of Genotype and Environment on Fruit Phenolic Composition of Olive. Horticulturae. 2023; 9(10):1087. https://doi.org/10.3390/horticulturae9101087
Chicago/Turabian StyleYılmaz-Düzyaman, Hande, María G. Medina-Alonso, Carlos Sanz, Ana G. Pérez, Raúl de la Rosa, and Lorenzo León. 2023. "Influence of Genotype and Environment on Fruit Phenolic Composition of Olive" Horticulturae 9, no. 10: 1087. https://doi.org/10.3390/horticulturae9101087
APA StyleYılmaz-Düzyaman, H., Medina-Alonso, M. G., Sanz, C., Pérez, A. G., de la Rosa, R., & León, L. (2023). Influence of Genotype and Environment on Fruit Phenolic Composition of Olive. Horticulturae, 9(10), 1087. https://doi.org/10.3390/horticulturae9101087










