Genome-Wide Identification and Analysis of NF-Y Gene Family Reveal Its Potential Roles in Stress-Resistance in Chrysanthemum
Abstract
1. Introduction
2. Experimental Materials and Methods
2.1. Plant Material and Treatment
2.2. Identification and Analysis of CsNF-Y Family Genes in C. seticuspe
2.3. Phylogenetic Analysis of CsNF-Y Family Genes
2.4. Multiple Sequence Alignment, Gene Structure and Conserved Motif Analysis of CsNF-Y Protein
2.5. Genome Chromosomal Location and Gene Duplication Parameter Analysis
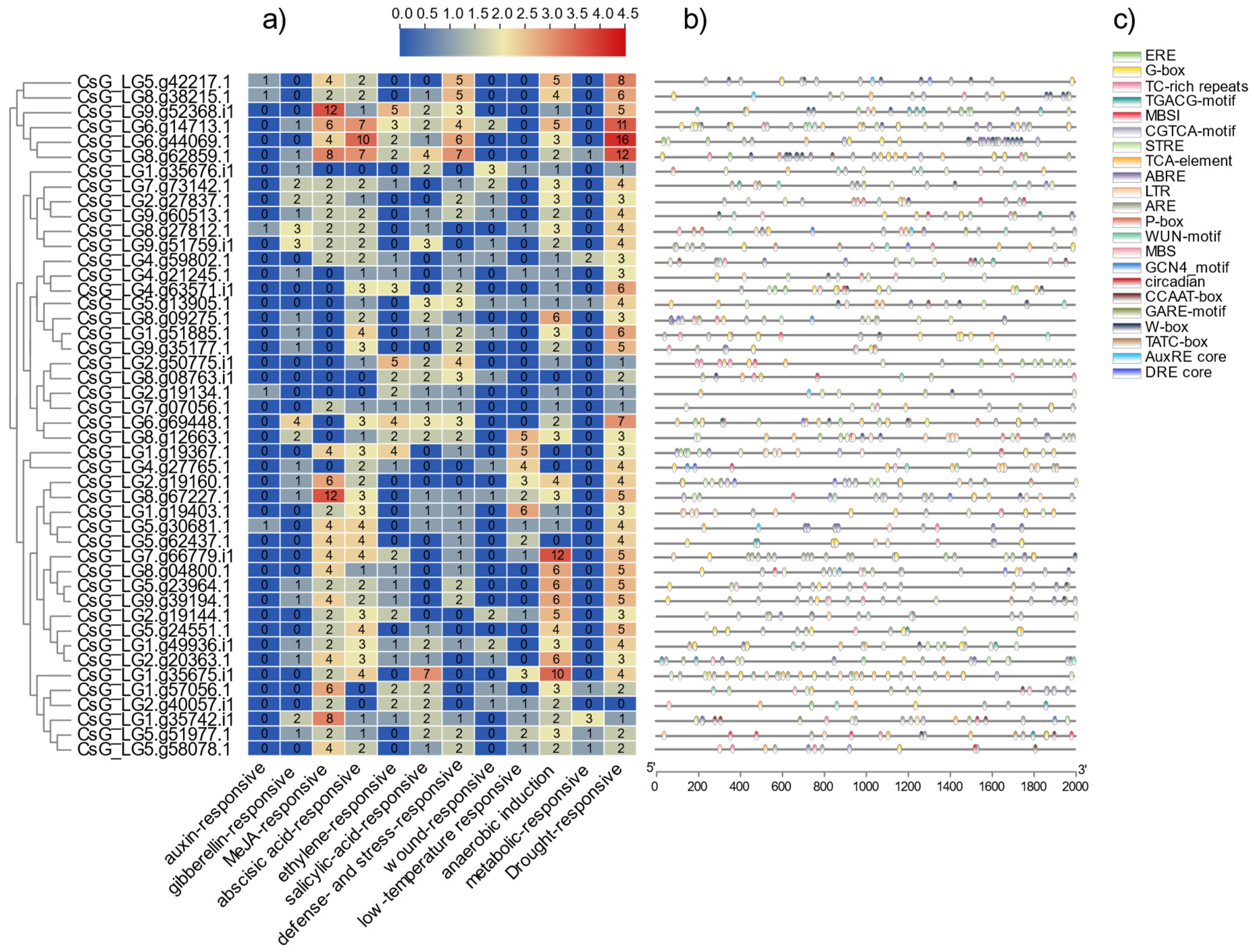
2.6. Analysis and Statistics of Stress Response Elements of CsNF-Y Promoters
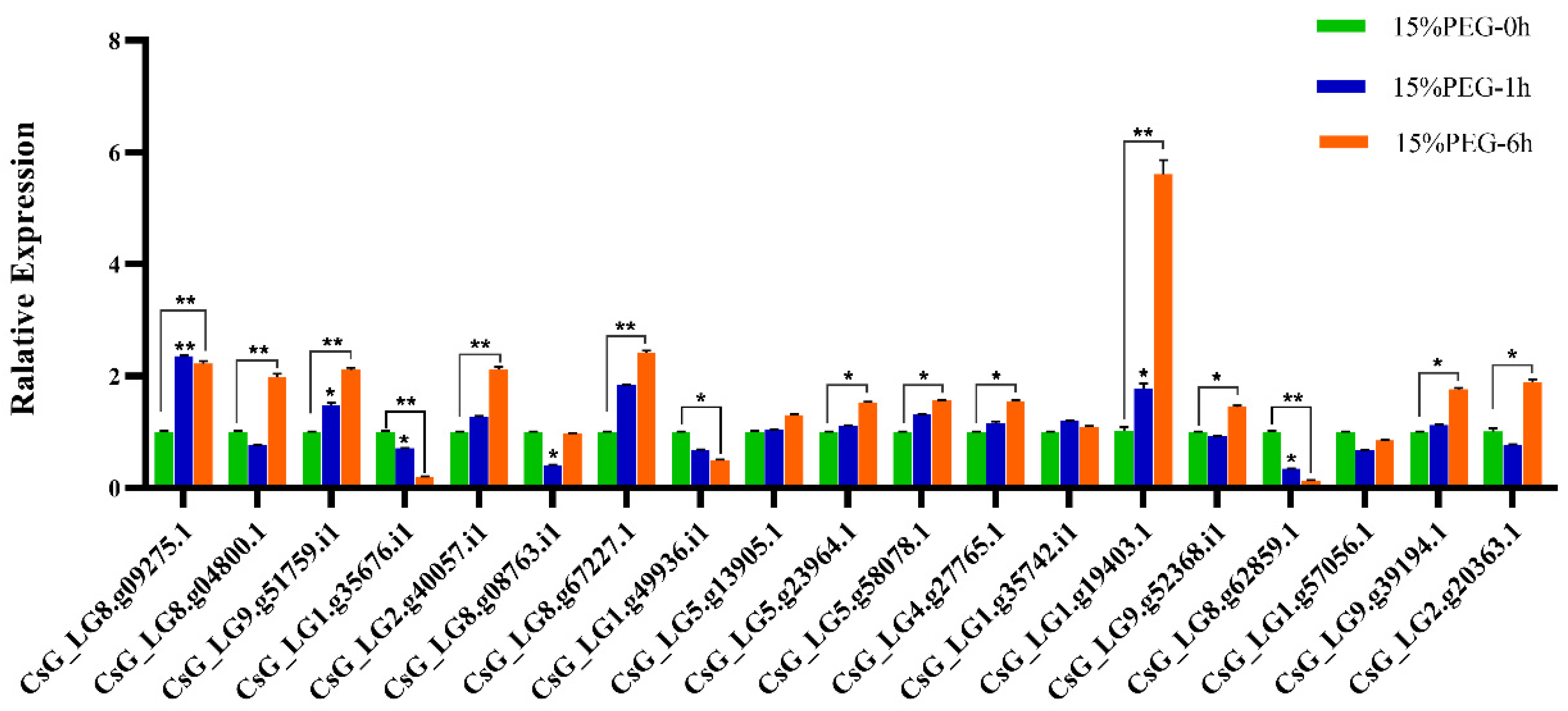
2.7. RNA Extraction and RT-PCR Analysis
3. Results
3.1. Identification of NF-Y Family Genes in Chrysanthemum
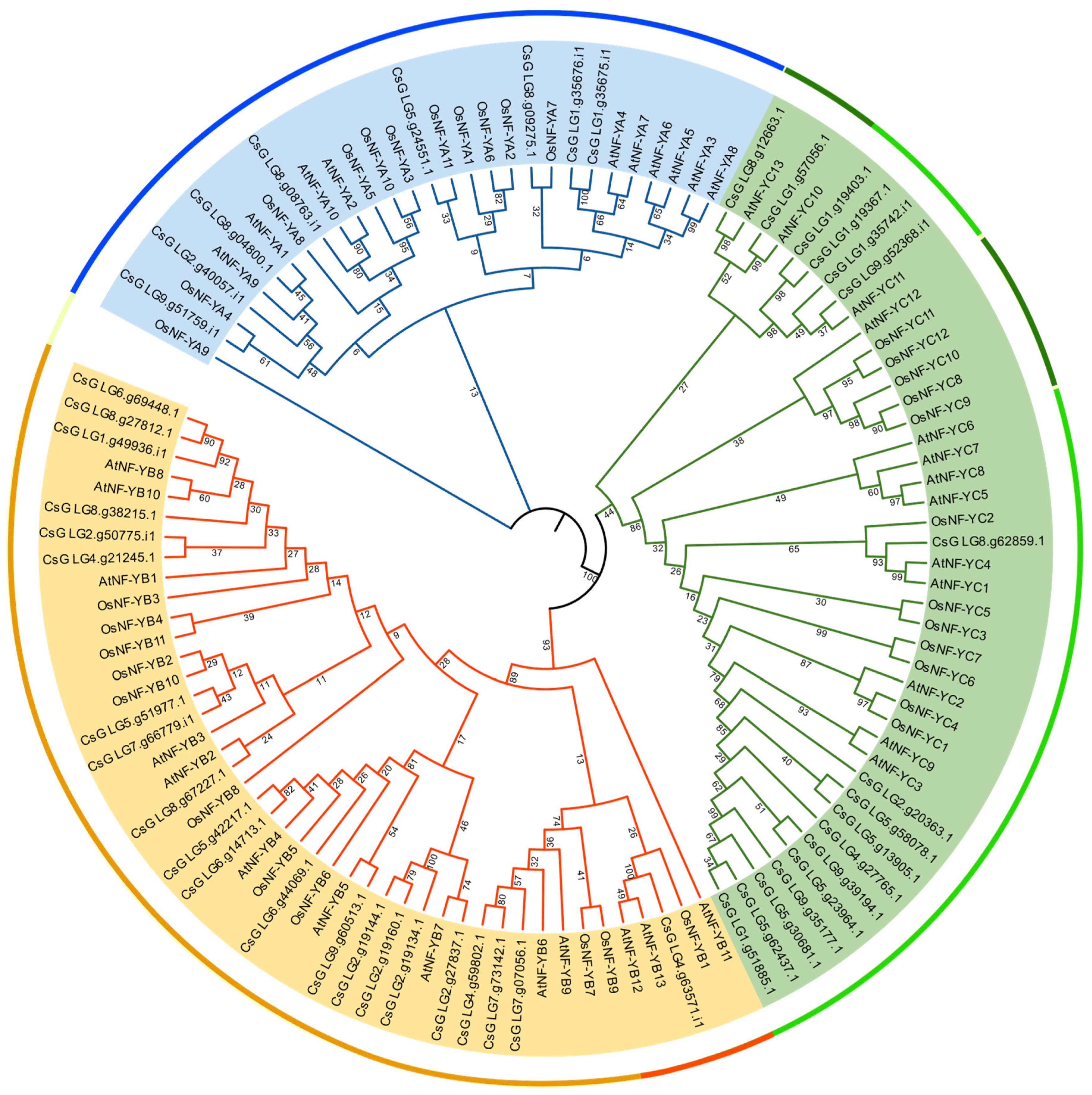
3.2. Phylogenetic Analysis of CsNF-Y Proteins
3.3. Multiple Sequence Alignment of CsNF-Y Genes in Chrysanthemum
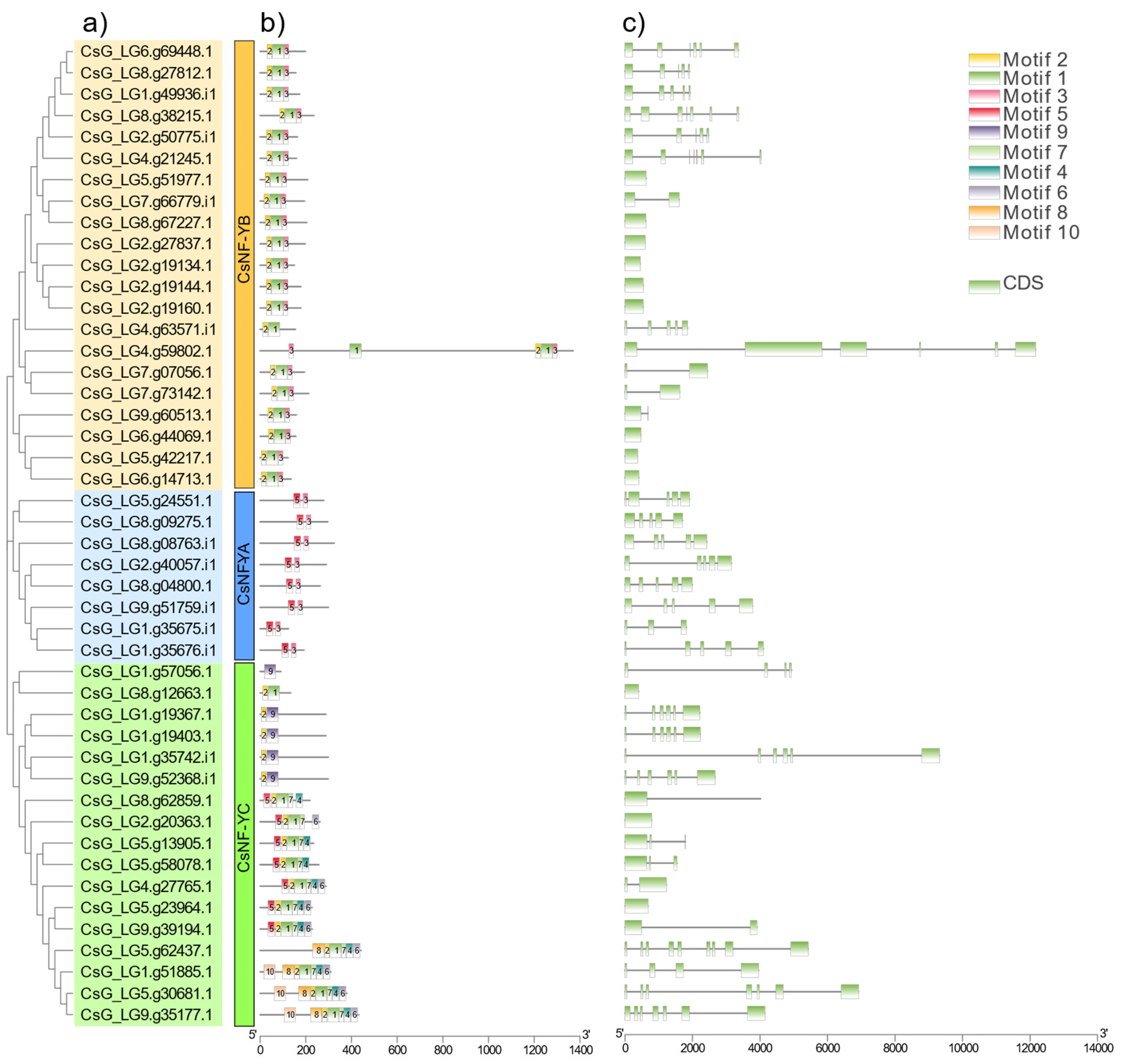
3.4. Analysis of Gene Structure and Conserved Motifs of CsNF-Y Genes in Chrysanthemum
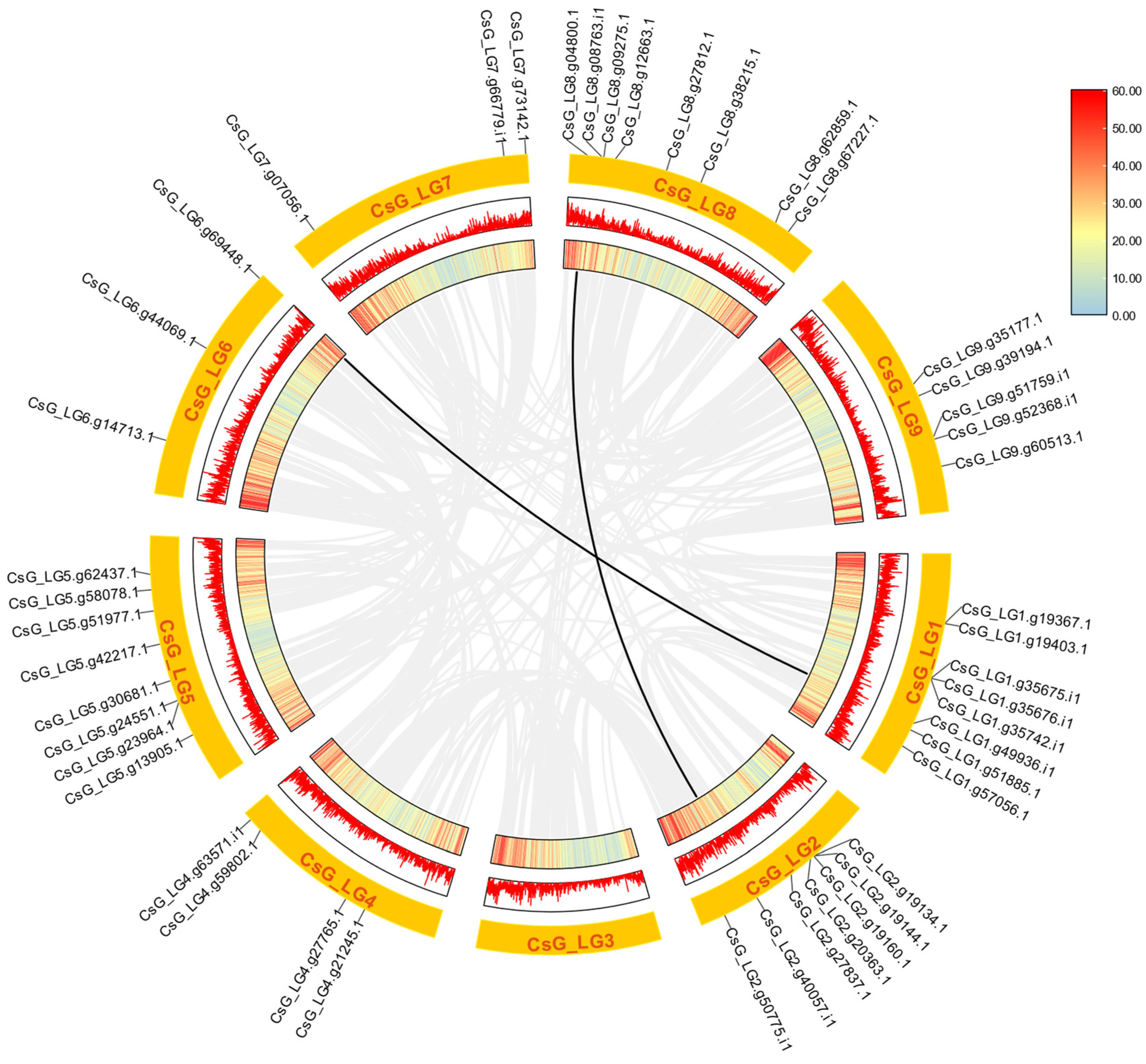
3.5. Analyses of Chromosomal Distribution and Synteny of CsNF-Y Genes in Chrysanthemum
3.6. Analysis of Cis-Elements in the Promoter Sequences of NF-Y Genes in Chrysanthemum
3.7. Expression Levels of CsNF-Y Genes in Chrysanthemum under Osmotic-Stress
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dolfini, D.; Gatta, R.; Mantovani, R. NF-Y and the transcriptional activation of CCAAT promoters. Crit. Rev. Biochem. Mol. Biol. 2012, 47, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Bucher, P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990, 212, 563–578. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Jan, A.; Todaka, D.; Maruyama, K.; Goto, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comparative functional analysis of six drought-responsive promoters in transgenic rice. Planta 2014, 239, 47–60. [Google Scholar] [CrossRef] [PubMed]
- Nardone, V.; Chaves-Sanjuan, A.; Nardini, M. Structural determinants for NF-Y/DNA interaction at the CCAAT box. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 571–580. [Google Scholar] [CrossRef]
- Benatti, P.; Basile, V.; Merico, D.; Fantoni, L.I.; Tagliafico, E.; Imbriano, C. A balance between NF-Y and p53 governs the pro- and anti-apoptotic transcriptional response. Nucleic Acids Res. 2008, 36, 1415–1428. [Google Scholar] [CrossRef]
- Coustry, F.; Sinha, S.; Maity, S.N.; Crombrugghe, B. The two activation domains of the CCAAT-binding factor CBF interact with the dTAFII110 component of the Drosophila TFIID complex. Biochem. J. 1998, 331 Pt 1, 291–297. [Google Scholar] [CrossRef]
- Wright, K.L.; Moore, T.L.; Vilen, B.J.; Brown, A.M.; Ting, J.P. Major histocompatibility complex class II-associated invariant chain gene expression is up-regulated by cooperative interactions of Sp1 and NF-Y. J. Biol. Chem. 1995, 270, 20978–20986. [Google Scholar] [CrossRef]
- Li, S.; Li, K.; Ju, Z.; Cao, D.; Fu, D.; Zhu, H.; Zhu, B.; Luo, Y. Genome-wide analysis of tomato NF-Y factors and their role in fruit ripening. BMC Genom. 2016, 17, 36. [Google Scholar] [CrossRef]
- Frontini, M.; Imbriano, C.; Manni, I.; Mantovani, R. Cell cycle regulation of NF-YC nuclear localization. Cell Cycle 2004, 3, 217–222. [Google Scholar] [CrossRef]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Sinha, S.; Maity, S.N.; Lu, J.; de Crombrugghe, B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. USA 1995, 92, 1624–1628. [Google Scholar] [CrossRef] [PubMed]
- Nardini, M.; Gnesutta, N.; Donati, G.; Gatta, R.; Forni, C.; Fossati, A.; Vonrhein, C.; Moras, D.; Romier, C.; Bolognesi, M.; et al. Sequence-specific transcription factor NF-Y displays histone-like DNA binding and H2B-like ubiquitination. Cell 2013, 152, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Soltis, P.S.; Soltis, D.E. Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 2016, 30, 159–165. [Google Scholar] [CrossRef]
- Yang, J.; Xie, Z.; Glover, B.J. Asymmetric evolution of duplicate genes encoding the CCAAT-binding factor NF-Y in plant genomes. New Phytol. 2005, 165, 623–631. [Google Scholar] [CrossRef] [PubMed]
- Zanetti, M.E.; Rípodas, C.; Niebel, A. Plant NF-Y transcription factors: Key players in plant-microbe interactions, root development and adaptation to stress. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 645–654. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Sun, J.; Ren, L.; Zhou, M.; Han, X.; Ding, L.; Zhang, F.; Guan, Z.; Fang, W.; Chen, S.; et al. CmBBX8 accelerates flowering by targeting CmFTL1 directly in summer chrysanthemum. Plant Biotechnol. J. 2020, 18, 1562–1572. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, S.B.; Shen, Y.; Chang, H.C.; Hou, Y.; Harris, A.; Ma, S.F.; McPartland, M.; Hymus, G.J.; Adam, L.; Marion, C.; et al. The flowering time regulator CONSTANS is recruited to the FLOWERING LOCUS T promoter via a unique cis-element. New Phytol. 2010, 187, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.; Kumimoto, R.W.; Gnesutta, N.; Calogero, A.M.; Mantovani, R.; Holt, B.F., III. A distal CCAAT/NUCLEAR FACTOR Y complex promotes chromatin looping at the FLOWERING LOCUS T promoter and regulates the timing of flowering in Arabidopsis. Plant Cell 2014, 26, 1009–1017. [Google Scholar] [CrossRef]
- Siriwardana, C.L.; Gnesutta, N.; Kumimoto, R.W.; Jones, D.S.; Myers, Z.A.; Mantovani, R.; Holt, B.F., III. NUCLEAR FACTOR Y, Subunit A (NF-YA) Proteins Positively Regulate Flowering and Act Through FLOWERING LOCUS T. PLoS Genet. 2016, 12, e1006496. [Google Scholar] [CrossRef]
- Gnesutta, N.; Kumimoto, R.W.; Swain, S.; Chiara, M.; Siriwardana, C.; Horner, D.S.; Holt, B.F., III; Mantovani, R. CONSTANS Imparts DNA Sequence Specificity to the Histone Fold NF-YB/NF-YC Dimer. Plant Cell 2017, 29, 1516–1532. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Liu, H.; Guan, Z.; Yan, J.; Zheng, T.; Yan, W.; Wu, C.; Zhang, Q.; Yin, P.; Xing, Y. Structural Insight into DNA Recognition by CCT/NF-YB/YC Complexes in Plant Photoperiodic Flowering. Plant Cell 2020, 32, 3469–3484. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Zhou, J.; Liu, C.; Liu, L.; Shen, L.; Yu, H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat. Commun. 2014, 5, 4601. [Google Scholar] [CrossRef] [PubMed]
- Hwang, K.; Susila, H.; Nasim, Z.; Jung, J.Y.; Ahn, J.H. Arabidopsis ABF3 and ABF4 Transcription Factors Act with the NF-YC Complex to Regulate SOC1 Expression and Mediate Drought-Accelerated Flowering. Mol. Plant 2019, 12, 489–505. [Google Scholar] [CrossRef]
- Kim, S.K.; Park, H.Y.; Jang, Y.H.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.K. OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 2016, 243, 563–576. [Google Scholar] [CrossRef]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z.; et al. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef]
- Zhou, H.; Zeng, R.F.; Liu, T.J.; Ai, X.Y.; Ren, M.K.; Zhou, J.J.; Hu, C.G.; Zhang, J.Z. Drought and low temperature-induced NF-YA1 activates FT expression to promote citrus flowering. Plant Cell Environ. 2022, 45, 3505–3522. [Google Scholar] [CrossRef]
- Chen, C.; Hussain, N.; Wang, Y.; Li, M.; Liu, L.; Qin, M.; Ma, N.; Gao, J.; Sun, X. An Ethylene-inhibited NF-YC Transcription Factor RhNF-YC9 Regulates Petal Expansion in Rose. Hortic. Plant J. 2020, 6, 419–427. [Google Scholar] [CrossRef]
- Wang, J.; Li, G.; Li, C.; Zhang, C.; Cui, L.; Ai, G.; Wang, X.; Zheng, F.; Zhang, D.; Larkin, R.M.; et al. NF-Y plays essential roles in flavonoid biosynthesis by modulating histone modifications in tomato. New Phytol. 2021, 229, 3237–3252. [Google Scholar] [CrossRef]
- Zhang, W.; Tang, Y.; Hu, Y.; Yang, Y.; Cai, J.; Liu, H.; Zhang, C.; Liu, X.; Hou, X. Arabidopsis NF-YCs play dual roles in repressing brassinosteroid biosynthesis and signaling during light-regulated hypocotyl elongation. Plant Cell 2021, 33, 2360–2374. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef]
- Jo, L.; Pelletier, J.M.; Harada, J.J. Central role of the LEAFY COTYLEDON1 transcription factor in seed development. J. Integr. Plant Biol. 2019, 61, 564–580. [Google Scholar] [CrossRef]
- Kwong, R.W.; Bui, A.Q.; Lee, H.; Kwong, L.W.; Fischer, R.L.; Goldberg, R.B.; Harada, J.J. LEAFY COTYLEDON1-LIKE defines a class of regulators essential for embryo development. Plant Cell 2003, 15, 5–18. [Google Scholar] [CrossRef]
- Pelletier, J.M.; Kwong, R.W.; Park, S.; Le, B.H.; Baden, R.; Cagliari, A.; Hashimoto, M.; Munoz, M.D.; Fischer, R.L.; Goldberg, R.B.; et al. LEC1 sequentially regulates the transcription of genes involved in diverse developmental processes during seed development. Proc. Natl. Acad. Sci. USA 2017, 114, E6710–E6719. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Toyoshima, R.; Kagaya, M.; Takeda, S.; Hattori, T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. Cell Mol. Biol. 2009, 58, 843–856. [Google Scholar] [CrossRef]
- Miyoshi, K.; Ito, Y.; Serizawa, A.; Kurata, N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. Cell Mol. Biol. 2003, 36, 532–540. [Google Scholar] [CrossRef]
- Leyva-González, M.A.; Ibarra-Laclette, E.; Cruz-Ramírez, A.; Herrera-Estrella, L. Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS ONE 2012, 7, e48138. [Google Scholar] [CrossRef]
- Lee, D.K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Choi, Y.D.; Kim, J.K. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. Int. J. Exp. Plant Biol. 2015, 241, 199–210. [Google Scholar] [CrossRef]
- Nakano, M.; Taniguchi, K.; Masuda, Y.; Kozuka, T.; Aruga, Y.; Han, J.; Motohara, K.; Nakata, M.; Sumitomo, K.; Hisamatsu, T.; et al. A pure line derived from a self-compatible Chrysanthemum seticuspe mutant as a model strain in the genus Chrysanthemum. Plant Sci. 2019, 287, 110174. [Google Scholar] [CrossRef]
- Yu, T.F.; Liu, Y.; Fu, J.D.; Ma, J.; Fang, Z.W.; Chen, J.; Zheng, L.; Lu, Z.W.; Zhou, Y.B.; Chen, M.; et al. The NF-Y-PYR module integrates the abscisic acid signal pathway to regulate plant stress tolerance. Plant Biotechnol. J. 2021, 19, 2589–2605. [Google Scholar] [CrossRef]
- Li, S.; Zhang, N.; Zhu, X.; Ma, R.; Liu, S.; Wang, X.; Yang, J.; Si, H. Genome-Wide Analysis of NF-Y Genes in Potato and Functional Identification of StNF-YC9 in Drought Tolerance. Front. Plant Sci. 2021, 12, 749688. [Google Scholar] [CrossRef]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Zhang, D.; Liu, Y.; Luo, C.; Zhou, Y.; Zhang, L. Overexpression of a NF-YB3 transcription factor from Picea wilsonii confers tolerance to salinity and drought stress in transformed Arabidopsis thaliana. Plant Physiol. Biochem. PPB 2015, 94, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant nuclear factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef]
- Wang, B.; Li, Z.; Ran, Q.; Li, P.; Peng, Z.; Zhang, J. ZmNF-YB16 Overexpression Improves Drought Resistance and Yield by Enhancing Photosynthesis and the Antioxidant Capacity of Maize Plants. Front. Plant Sci. 2018, 9, 709. [Google Scholar] [CrossRef]
- Wang, T.; Wei, Q.; Wang, Z.; Liu, W.; Zhao, X.; Ma, C.; Gao, J.; Xu, Y.; Hong, B. CmNF-YB8 affects drought resistance in chrysanthemum by altering stomatal status and leaf cuticle thickness. J. Integr. Plant Biol. 2022, 64, 741–755. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y.; Wang, X.; Han, X.; An, Y.; Lin, S.; Shen, C.; Wen, J.; Liu, C.; Yin, W.; et al. Root-specific NF-Y family transcription factor, PdNF-YB21, positively regulates root growth and drought resistance by abscisic acid-mediated indoylacetic acid transport in Populus. New Phytol. 2020, 227, 407–426. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, J.; He, J.; Geng, Z.; Li, S.; Zhang, J.; Li, P.; Zhang, L.; Wang, Z.; Wang, L.; et al. Whole-transcriptome profiles of Chrysanthemum seticuspe improve genome annotation and shed new light on mRNA-miRNA-lncRNA networks in ray florets and disc florets. BMC Plant Biol. 2022, 22, 515. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Chen, S.; Jiang, J.; Zhang, F.; Chen, F. Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci. Rep. 2015, 5, 8094. [Google Scholar] [CrossRef]
- Gu, C.; Chen, S.; Liu, Z.; Shan, H.; Luo, H.; Guan, Z.; Chen, F. Reference gene selection for quantitative real-time PCR in Chrysanthemum subjected to biotic and abiotic stress. Mol. Biotechnol. 2011, 49, 192–197. [Google Scholar] [CrossRef]
- Li, M.; Li, G.; Liu, W.; Dong, X.; Zhang, A. Genome-wide analysis of the NF-Y gene family in peach (Prunus persica L.). BMC Genom. 2019, 20, 612. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Yin, X.; Lin, Z.; Zheng, Q.; Liu, G.; Zhao, G. Identification and characterization of NF-Y transcription factor families in Canola (Brassica napus L.). Planta 2014, 239, 107–126. [Google Scholar] [CrossRef] [PubMed]
- Xing, Y.; Fikes, J.D.; Guarente, L. Mutations in yeast HAP2/HAP3 define a hybrid CCAAT box binding domain. EMBO J. 1993, 12, 4647–4655. [Google Scholar] [CrossRef] [PubMed]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Xu, G.; Guo, C.; Shan, H.; Kong, H. Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1187–1192. [Google Scholar] [CrossRef]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.Q.; Wang, J.; Wong, G.K.; Yu, J. KaKs_Calculator: Calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Zhao, T.J.; Sun, S.; Liu, Y.; Liu, J.M.; Liu, Q.; Yan, Y.B.; Zhou, H.M. Regulating the drought-responsive element (DRE)-mediated signaling pathway by synergic functions of trans-active and trans-inactive DRE binding factors in Brassica napus. J. Biol. Chem. 2006, 281, 10752–10759. [Google Scholar] [CrossRef]
- Sato, H.; Suzuki, T.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. NF-YB2 and NF-YB3 Have Functionally Diverged and Differentially Induce Drought and Heat Stress-Specific Genes. Plant Physiol. 2019, 180, 1677–1690. [Google Scholar] [CrossRef]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F., III; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed]
- Kumimoto, R.W.; Zhang, Y.; Siefers, N.; Holt, B.F., III. NF-YC3, NF-YC4 and NF-YC9 are required for CONSTANS-mediated, photoperiod-dependent flowering in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 2010, 63, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Mu, J.; Tan, H.; Hong, S.; Liang, Y.; Zuo, J. Arabidopsis Transcription Factor Genes NF-YA1, 5, 6, and 9 Play Redundant Roles in Male Gametogenesis, Embryogenesis, and Seed Development. Mol. Plant 2013, 6, 188–201. [Google Scholar] [CrossRef] [PubMed]
- Siriwardana, C.L.; Kumimoto, R.W.; Jones, D.S.; Holt, B.F., III. Gene Family Analysis of the Arabidopsis NF-YA Transcription Factors Reveals Opposing Abscisic Acid Responses During Seed Germination. Plant Mol. Biol. Report. 2014, 32, 971–986. [Google Scholar] [CrossRef]
- Sorin, C.; Declerck, M.; Christ, A.; Blein, T.; Ma, L.; Lelandais-Brière, C.; Njo, M.F.; Beeckman, T.; Crespi, M.; Hartmann, C. A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol. 2014, 202, 1197–1211. [Google Scholar] [CrossRef]
- Zhang, M.; Hu, X.; Zhu, M.; Xu, M.; Wang, L. Transcription factors NF-YA2 and NF-YA10 regulate leaf growth via auxin signaling in Arabidopsis. Sci. Rep. 2017, 7, 1395. [Google Scholar] [CrossRef]
- Niu, B.; Zhang, Z.; Zhang, J.; Zhou, Y.; Chen, C. The rice LEC1-like transcription factor OsNF-YB9 interacts with SPK, an endosperm-specific sucrose synthase protein kinase, and functions in seed development. Plant J. Cell Mol. Biol. 2021, 106, 1233–1246. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, P.; Wu, Y.; Lian, G.; Yang, Z.; Liu, W.; Buerte, B.; Zhou, C.; Zhang, W.; Li, D.; et al. Rice LEAFY COTYLEDON1 Hinders Embryo Greening During the Seed Development. Front. Plant Sci. 2022, 13, 887980. [Google Scholar] [CrossRef]
- Kumimoto, R.W.; Adam, L.; Hymus, G.J.; Repetti, P.P.; Reuber, T.L.; Marion, C.M.; Hempel, F.D.; Ratcliffe, O.J. The Nuclear Factor Y subunits NF-YB2 and NF-YB3 play additive roles in the promotion of flowering by inductive long-day photoperiods in Arabidopsis. Planta 2008, 228, 709–723. [Google Scholar] [CrossRef]
- Hwang, Y.H.; Kim, S.K.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.K. Functional conservation of rice OsNF-YB/YC and Arabidopsis AtNF-YB/YC proteins in the regulation of flowering time. Plant Cell Rep. 2016, 35, 857–865. [Google Scholar] [CrossRef]
- Jo, B.S.; Choi, S.S. Introns: The Functional Benefits of Introns in Genomes. Genom. Inform. 2015, 13, 112–118. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Kang, J.Y.; Cho, D.I.; Park, J.H.; Kim, S.Y. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. Cell Mol. Biol. 2004, 40, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Itzhaki, H.; Maxson, J.M.; Woodson, W.R. An ethylene-responsive enhancer element is involved in the senescence-related expression of the carnation glutathione-S-transferase (GST1) gene. Proc. Natl. Acad. Sci. USA 1994, 91, 8925–8929. [Google Scholar] [CrossRef] [PubMed]
- An, Y.; Suo, X.; Niu, Q.; Yin, S.; Chen, L. Genome-Wide Identification and Analysis of the NF-Y Transcription Factor Family Reveal Its Potential Roles in Salt Stress in Alfalfa (Medicago sativa L.). Int. J. Mol. Sci. 2022, 23, 6426. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wu, M.; Liu, H.L.; Gao, Y.M.; Chen, J.; Yan, H.W.; Xiang, Y. Genome-wide identification and expression analysis of the NF-Y transcription factor family in Populus. Physiol. Plant. 2021, 171, 309–327. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Zhu, J.; Ma, W.; Li, Z.; Bi, Z.; Sun, C.; Bai, J.; Zhang, J.; Liu, Y. Genome-Wide Identification and Analysis of the NF-Y Gene Family in Potato (Solanum tuberosum L.). Front. Genet. 2021, 12, 739989. [Google Scholar] [CrossRef]
- Mai, Y.; Shui, L.; Huo, K.; Niu, J. Genome-wide characterization of the NUCLEAR FACTOR-Y (NF-Y) family in Citrus grandis identified CgNF-YB9 involved in the fructose and glucose accumulation. Genes Genom. 2019, 41, 1341–1355. [Google Scholar] [CrossRef]
- Panahi, B.; Mohammadi, S.A.; Ruzicka, K.; Abbasi Holaso, H.; Zare Mehrjerdi, M. Genome-wide identification and co-expression network analysis of nuclear factor-Y in barley revealed potential functions in salt stress. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2019, 25, 485–495. [Google Scholar] [CrossRef]
- Pereira, S.L.S.; Martins, C.P.S.; Sousa, A.O.; Camillo, L.R.; Araújo, C.P.; Alcantara, G.M.; Camargo, D.S.; Cidade, L.C.; de Almeida, A.F.; Costa, M.G.C. Genome-wide characterization and expression analysis of citrus NUCLEAR FACTOR-Y (NF-Y) transcription factors identified a novel NF-YA gene involved in drought-stress response and tolerance. PLoS ONE 2018, 13, e0199187. [Google Scholar] [CrossRef]
- Quach, T.N.; Nguyen, H.T.; Valliyodan, B.; Joshi, T.; Xu, D.; Nguyen, H.T. Genome-wide expression analysis of soybean NF-Y genes reveals potential function in development and drought response. Mol. Genet. Genom. MGG 2015, 290, 1095–1115. [Google Scholar] [CrossRef]
- Ren, C.; Zhang, Z.; Wang, Y.; Li, S.; Liang, Z. Genome-wide identification and characterization of the NF-Y gene family in grape (vitis vinifera L.). BMC Genom. 2016, 17, 605. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Liu, C.; Zhao, J.; Ye, X.; Wu, Q.; Yao, T.; Peng, L.; Zou, L.; Zhao, G. Genome-wide analysis of the NF-Y gene family and their roles in relation to fruit development in Tartary buckwheat (Fagopyrum tataricum). Int. J. Biol. Macromol. 2021, 190, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, X.; Zhang, C.; Zou, H.; Wu, Z. Isolation, structural analysis, and expression characteristics of the maize nuclear factor Y gene families. Biochem. Biophys. Res. Commun. 2016, 478, 752–758. [Google Scholar] [CrossRef]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana Nuclear Factor Y Transcription Factors. Front. Plant Sci. 2016, 7, 2045. [Google Scholar] [CrossRef] [PubMed]
- Rao, S.; Gupta, A.; Bansal, C.; Sorin, C.; Crespi, M.; Mathur, S. A conserved HSF:miR169:NF-YA loop involved in tomato and Arabidopsis heat stress tolerance. Plant J. Cell Mol. Biol. 2022, 112, 7–26. [Google Scholar] [CrossRef]
- Bello, B.K.; Hou, Y.; Zhao, J.; Jiao, G.; Wu, Y.; Li, Z.; Wang, Y.; Tong, X.; Wang, W.; Yuan, W.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef]
- Manimaran, P.; Venkata Reddy, S.; Moin, M.; Raghurami Reddy, M.; Yugandhar, P.; Mohanraj, S.S.; Balachandran, S.M.; Kirti, P.B. Activation-tagging in indica rice identifies a novel transcription factor subunit, NF-YC13 associated with salt tolerance. Sci. Rep. 2017, 7, 9341. [Google Scholar] [CrossRef]
- Xiong, Y.; Ren, Y.; Li, W.; Wu, F.; Yang, W.; Huang, X.; Yao, J. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [CrossRef]
- Das, S.; Parida, S.K.; Agarwal, P.; Tyagi, A.K. Transcription factor OsNF-YB9 regulates reproductive growth and development in rice. Planta 2019, 250, 1849–1865. [Google Scholar] [CrossRef]


| Sequence ID | PL (aa) | MW (kDa) | pI | Subcellular Localization | Arabidopsis |
|---|---|---|---|---|---|
| CsG_LG1.g19367.1 | 288 | 32.17 | 5.19 | chlo: 4, mito: 4, golg: 2 | AtNF-YC |
| CsG_LG1.g19403.1 | 288 | 32.24 | 5.2 | chlo: 4, mito: 4, golg: 2 | AtNF-YC |
| CsG_LG1.g35675.i1 | 125 | 14.39 | 9.78 | nucl: 11, extr: 2, cysk: 1 | AtNF-YA |
| CsG_LG1.g35676.i1 | 193 | 21.80 | 8.82 | nucl: 13, chlo: 1 | AtNF-YA |
| CsG_LG1.g35742.i1 | 299 | 32.57 | 5.23 | chlo: 4, nucl: 3.5, mito: 3 | AtNF-YC |
| CsG_LG1.g49936.i1 | 174 | 19.24 | 6.21 | nucl: 14 | AtNF-YB |
| CsG_LG1.g51885.1 | 310 | 35.12 | 8.47 | cyto: 7.5, cyto_nucl: 5 | AtNF-YC |
| CsG_LG1.g57056.1 | 91 | 10.52 | 9.26 | cyto: 7, chlo: 3, plas: 3 | AtNF-YC |
| CsG_LG2.g19134.1 | 151 | 16.59 | 7.66 | nucl: 4, cyto: 4, chlo: 2 | AtNF-YB |
| CsG_LG2.g19144.1 | 179 | 19.94 | 6.17 | chlo: 9, nucl: 2, cyto: 1 | AtNF-YB |
| CsG_LG2.g19160.1 | 179 | 19.92 | 5.8 | chlo: 10, nucl: 1, cyto: 1 | AtNF-YB |
| CsG_LG2.g20363.1 | 263 | 29.42 | 5.34 | nucl: 9, chlo: 2, cyto: 2 | AtNF-YC |
| CsG_LG2.g27837.1 | 198 | 22.53 | 5.76 | nucl: 14 | AtNF-YB |
| CsG_LG2.g40057.i1 | 290 | 32.16 | 6.72 | nucl: 14 | AtNF-YA |
| CsG_LG2.g50775.i1 | 164 | 17.71 | 6.08 | nucl: 12, pero: 2 | AtNF-YB |
| CsG_LG4.g21245.1 | 160 | 17.80 | 7.54 | nucl: 13, pero: 1 | AtNF-YB |
| CsG_LG4.g27765.1 | 289 | 32.54 | 6.25 | cyto: 8, nucl: 4, chlo: 1 | AtNF-YC |
| CsG_LG4.g59802.1 | 1370 | 154.75 | 8.03 | chlo: 4, vacu: 3, plas: 2 | AtNF-YB |
| CsG_LG4.g63571.i1 | 155 | 17.20 | 4.67 | cyto: 6, mito: 4, chlo: 3 | AtNF-YB |
| CsG_LG5.g13905.1 | 235 | 26.50 | 7.02 | nucl: 11, cyto: 1, plas: 1 | AtNF-YC |
| CsG_LG5.g23964.1 | 228 | 25.86 | 6.15 | nucl: 6, extr: 3, cyto: 2 | AtNF-YC |
| CsG_LG5.g24551.1 | 280 | 31.32 | 9.12 | nucl: 9.5, cyto_nucl: 6 | AtNF-YA |
| CsG_LG5.g30681.1 | 380 | 42.64 | 6.52 | cyto: 14 | AtNF-YC |
| CsG_LG5.g42217.1 | 123 | 13.82 | 5.4 | cyto: 6, nucl: 2.5, chlo: 2 | AtNF-YB |
| CsG_LG5.g51977.1 | 209 | 21.95 | 6.31 | nucl: 14 | AtNF-YB |
| CsG_LG5.g58078.1 | 258 | 28.87 | 6.96 | nucl: 6, cyto: 5, chlo: 1 | AtNF-YC |
| CsG_LG5.g62437.1 | 442 | 49.77 | 5.91 | cyto: 10, chlo: 4 | AtNF-YC |
| CsG_LG6.g14713.1 | 136 | 15.09 | 7.74 | mito: 9, chlo: 3, nucl: 2 | AtNF-YB |
| CsG_LG6.g44069.1 | 157 | 17.69 | 4.93 | chlo: 6, nucl: 5, extr: 2 | AtNF-YB |
| CsG_LG6.g69448.1 | 199 | 21.67 | 8.36 | nucl: 14 | AtNF-YB |
| CsG_LG7.g07056.1 | 195 | 21.99 | 5.64 | nucl: 6, mito: 4, cyto: 3 | AtNF-YB |
| CsG_LG7.g66779.i1 | 195 | 20.73 | 5.95 | nucl: 14 | AtNF-YB |
| CsG_LG7.g73142.1 | 213 | 23.85 | 6.17 | cyto: 7, nucl: 2, plas: 2 | AtNF-YB |
| CsG_LG8.g04800.1 | 264 | 29.60 | 6.56 | nucl: 13, plas: 1 | AtNF-YA |
| CsG_LG8.g08763.i1 | 325 | 35.09 | 9.26 | nucl: 14 | AtNF-YA |
| CsG_LG8.g09275.1 | 297 | 32.99 | 9.2 | nucl: 14 | AtNF-YA |
| CsG_LG8.g12663.1 | 134 | 15.36 | 9.45 | nucl: 13, chlo: 1 | AtNF-YC |
| CsG_LG8.g27812.1 | 157 | 17.15 | 5.76 | nucl: 8, mito: 6 | AtNF-YB |
| CsG_LG8.g38215.1 | 236 | 25.67 | 9.02 | nucl: 14 | AtNF-YB |
| CsG_LG8.g62859.1 | 219 | 23.91 | 5.14 | nucl: 9, chlo: 2, cyto: 2 | AtNF-YC |
| CsG_LG8.g67227.1 | 205 | 21.70 | 6.3 | nucl: 14 | AtNF-YB |
| CsG_LG9.g35177.1 | 432 | 48.67 | 8.94 | vacu: 5, golg: 3, chlo: 2 | AtNF-YC |
| CsG_LG9.g39194.1 | 229 | 25.97 | 5.88 | nucl: 7, cyto: 2, plas: 1.5 | AtNF-YC |
| CsG_LG9.g51759.i1 | 300 | 33.32 | 9.19 | nucl: 13, chlo: 1 | AtNF-YA |
| CsG_LG9.g52368.i1 | 299 | 32.83 | 5.25 | nucl: 3.5, cyto_nucl: 3.5 | AtNF-YC |
| CsG_LG9.g60513.1 | 160 | 18.15 | 5.76 | nucl: 8, cyto: 3, chlo: 1 | AtNF-YB |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, R.; Yin, M.; Song, A.; Guan, Z.; Fang, W.; Chen, F.; Jiang, J. Genome-Wide Identification and Analysis of NF-Y Gene Family Reveal Its Potential Roles in Stress-Resistance in Chrysanthemum. Horticulturae 2023, 9, 70. https://doi.org/10.3390/horticulturae9010070
Hu R, Yin M, Song A, Guan Z, Fang W, Chen F, Jiang J. Genome-Wide Identification and Analysis of NF-Y Gene Family Reveal Its Potential Roles in Stress-Resistance in Chrysanthemum. Horticulturae. 2023; 9(1):70. https://doi.org/10.3390/horticulturae9010070
Chicago/Turabian StyleHu, Rongqian, Mengru Yin, Aiping Song, Zhiyong Guan, Weimin Fang, Fadi Chen, and Jiafu Jiang. 2023. "Genome-Wide Identification and Analysis of NF-Y Gene Family Reveal Its Potential Roles in Stress-Resistance in Chrysanthemum" Horticulturae 9, no. 1: 70. https://doi.org/10.3390/horticulturae9010070
APA StyleHu, R., Yin, M., Song, A., Guan, Z., Fang, W., Chen, F., & Jiang, J. (2023). Genome-Wide Identification and Analysis of NF-Y Gene Family Reveal Its Potential Roles in Stress-Resistance in Chrysanthemum. Horticulturae, 9(1), 70. https://doi.org/10.3390/horticulturae9010070