Abstract
As an important genetic improvement technique in current production practice, heterosis is widely used to enhance the productive traits of hybrid progeny from their parents. Alternative splicing (AS) analysis can be used as a method for exploring the molecular manifestations of heterosis. In our research, 16 hybrids and their parents were utilized to analyze the heterosis performance and AS events. Statistics of plant gross weight (PGW) showed that these hybrids had prominent heterosis, with the mid-parent heterosis values (MPV) ranging from 15.69% to 233.98%. Through pairwise comparison among the female parent, male parent, and hybrid, there were 2980–3205 AS events in each combination, with intron retention being the most common type followed by alternate 3’ splice site, alternative 5’ splice site, skipped exon, and mutually exclusive exon.There were 263–409 differential AS genes (DASGs) between the female parent and the hybrid, and 234–425 DASGs between the male parent and the hybrid in cross combinations. The DASGs were significantly enriched in 33 metabolic pathways in 16 cross combinations, and DASGs of different cross combinations were enriched in different metabolic pathways. Moreover, 76 DASGs in the strong heterosis combinations were identified and significantly enriched in the metabolic pathways related to amino acid metabolism. Further analysis revealed that most of these DASGs in amino acid metabolism were expressed differently in strong heterosis combinations. In addition, the expression levels of BraA06g014310.3C and BraA03g041700.3C in amino acid metabolism significantly correlated with PGW. These results could provide an index for future studies of the genetic and molecular mechanism of heterosis in hybrids.
1. Introduction
Heterosis typically refers to the phenomenon where a hybrid produced by mating two parents with genetic compositions that are comparatively different performs better than either parent alone or both parents in terms of traits such as fertility, growth, adaptability, stress resistance, biomass, yield, quality, and so on [1]. It benefits plant reproduction and environmental adaption, has been extensively used, and becomes one of the best ways to increase the grain output of many crops [2,3,4,5,6]. Since its discovery, heterosis has drawn the attention of numerous scientists, leading to the creation of many traditional experiments, hypotheses, and discussions [7,8,9,10]. There is no need for particular causes of this phenomenon, as there are many examples where heterosis has been explained at the genetic and molecular level [11,12,13,14,15].However, analyzing the molecular manifestations of this phenomenon remains relevant and useful for heterosis. Transcriptomics, metabolomics, proteomics, epigenetics, and biological regulatory network have all been used to elaborate on heterosis as a result of ongoing advancements in biological technology [16,17,18,19,20].
Alternative splicing(AS) is a frequent occurrence in eukaryotic gene expression [21,22,23]. During transcription, the preRNA requires a number of post transcriptional modifications to become mature mRNA, in which the exons are linked together and the introns are removed [24]. It occasionally only be cut in one way to produce one mRNA, but it can also occasionally be cut in several ways to produce various types of mRNA. This phenomenon is known as the AS of the mRNA precursor. Only one protein can be translated from the mRNA precursor, when AS occurs in the 5’or 3’ noncoding region. However, many proteins will be translated and even changed, when AS occurs in the coding area. The complexity of the transcriptome and proteome is considerably increased by AS, which can yield a wide range of transcriptome subtypes with comparable or entirely different functions [25]. AS can also lead to the introduction of stop codons in advance, thus affecting the stability of mRNA [26,27]. In addition, AS regulates gene expression by changing the efficiency of transcriptional extension and/or translation [26,28].
AS is an important post-transcriptional process that is crucial for crop development and environmental responsiveness in plants. More than 60% of the genes in tomato and barley, as do roughly 50% of the genes in soybeans, 46% of the genes in rice, 40% of the genes in maize, and 47% of the genes in rice undergo AS events [29,30,31,32]. There are now five primary categories of AS modes: skipped exon(SE), alternative 5’ splice site(A5SS), alternative 3’ splice site(A3SS), mutually exclusive exon(MXE), and intron retention (IR).The most common type of AS is IR in plants [33,34,35,36,37]. As a key regulation method of plant gene expression, AS involves in a great deal of crucial biological processes, such as response to biotic and abiotic stresses [38,39,40], photosynthesis [41], defense response [38], metabolic pathway [42], decomposition pathway [43], signal transduction, etc. In addition, AS also regulates key growth and development processes, such as flowering [44], the day and night cycle, and participates in establishing different organizational forms [40]. A growing number of genome sequences and transcriptome data are now available thanks to the advancement of high-throughput sequencing technology, which offers scientific support for identifying AS in various tissues, species, developmental stages, and environment-specific regulations. Additionally, it aids in understanding how AS dynamically varies in response to various developmental stages, environmental challenges, and pressures, as well as how the variation in AS patterns across various ecotypes and polyploidies affects the adaptability of plant species.
With the in-depth study of the heterosis, researchers gradually pay attention to the AS events in hybrids. It has been reported that as an essential regulation mode of plant gene expression, AS often shows differences between hybrids and parents. Gao et al. found that 87.2% of the differential ASgenes in mule muscles conformed to over-dominant or dominant expression pattern. Further analysis showed that the differential ASgenes in mule muscle were significantly enriched in the “muscle contraction”pathway [45]. Mei et al. identified differential AS between Angus × Qinchuan cattle and Qinchuan, those results are helpful to understand further the complex metabolic regulation of cattle [46]. Relevant studies have identified many different AS events in hybrids, and provide a reference for subsequent research.
Chinese cabbage is one of the most extensively grown vegetables in Asia and is a major member of the cruciferous family. The nutritionally dense leaves can be stir-fried, added to raw foods, salted, or sauced, and the leaves that fall off the outer layer can still be fed to animals. Chinese cabbage also has the benefits of easy planting management, high yield, long supply times, easystorage and transportation, and low input costs, which contribute to its significant role in Chinese vegetable production and trade. Heterosis is frequently utilized in Chinese cabbage breeding, but the research on the subject is lagging. Research on heterosis primarily focuses on related traits, the molecular level is relatively few, and the AS is even less. In our study, 16 hybrids and their parents were used as materials to explore the AS events in hybrids.
2. Materials and Methods
2.1. Plant Materials
The inbred line parents of Chinese cabbage were used for artificial cross-pollination to obtain the hybrids (Table 1). All the hybrids and their parents were selected to analyze phenotype and AS events.

Table 1.
The code of inbred lines and hybrids of Chinese cabbage.
The experiment was conducted at Yangling Wuquan test field in Shaanxi, China. At the middle stage of heading (about 70 days), the first outer leaf of Chinese cabbage from top to bottom was collected in three biological replicates with the help of sterile scissors. The samples were wrapped in tin foil, quickly frozen with liquid nitrogen, stored at −80 °C, and used for RNA-Seq. At maturity (about 100 days), parents’ and hybrids’plant gross weight (PGW) was investigated.
2.2. RNA Isolation, cDNA Library Construction, and RNA-Seq
Following the manufacturer’s instructions, RNA was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA) before the decontamination of genomic DNA using DNaseI (TaKaRa, Otsu, Japan). A NanoDrop 8000 spectrophotometer (Thermo Scientific, Waltham, MA, USA), an Agilent 2100 Bioanalyzer(Agilent Technologies, Santa Clara, CA, USA), and 1.0% agarose gels were used to evaluate the quality, purity, and integrity of the RNA.
Total RNA was isolated from the sample, followed by the enrichment of mRNA using Oligo (dT) magnetic beads, shortening of the acquired mRNA by adding a fragmentation buffer, and using the short fragmented mRNA as a template. The first strand of cDNA was created using six-base random primers (random hexamers), while the second strand was created by adding buffer, dNTPs, RNase H, and DNA polymerase I. Then the cDNA fragments were purified with a QiaQuick PCR extraction kit, end-repaired, poly (A) added, and ligated to Illumina sequencing adapters. The ligation products were size selected by agarose gel electrophoresis, PCR-amplified, and sequenced using Illumina HiSeqTM by Genedenovo Biotechnology Co., Ltd. (Guangzhou, China). The obtained raw data from constructed cDNA libraries were deposited in NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/Traces/sra/ (accessed on 1 June 2022)) under the accession number: BioProject PRJNA876066.
2.3. AS Event Statistics
The rMATS software (http://rnaseq-mats.sourceforge.net/index.html (accessed on 1 June 2022)) was used to analyze AS events. AS events were divided into five categories: SE, A5SS, A3SS, MXE, and IR. As a unit, each comparison group was used for differential AS analysis. We counted the types and numbers of AS events that occur, calculated the expression amount of each type of AS event, and calculated each type of AS event. In the quantitative process, the quantitative method adopted by rMATs is read on target and junction counts.
2.4. Functional Enrichment Analysis
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) annotations were performed for AS genes (ASGs) and differential AS genes (DASGs). A GO term was considered significantly enriched if the false discovery rate (FDR) cut-off was <0.05. The KEGG pathways were assigned using the KEGG software package (http://www.kegg.jp/ (accessed on 1 June 2022)), and considered significant if p < 0.05.
2.5. Statistical Analysis
The data were presented as the mean and statistical analyses were conducted using the SPSS software version 16.0. Duncan’s multiple-range test was determined for all the data at the 0.05 confidence level.
3. Result
3.1. Phenotypes of 16 Hybrids and 8 Parents in Chinese Cabbage
In this study, sixteen hybrids and eight parents of Chinese cabbage were used to investigate the PGW at the heading maturity stage.The results showed that the PGW of sixteen hybrids ranged from2.32 kg to 6.50 kg, which was significantly higher than the PGW of their female parent, male parent, and mid-parent PGW values, except for BH and DH hybrid (Figure 1). Statistics of mid-parent heterosis value(MPV) showed that these hybrids have obvious heterosis, with the MPV ranging from15.69 to 233.98% (Table 2). These hybrids were separated into three groups according to the MPV of PGW. When the MPV of PGW is higher than 140, the hybrid belongs to a strong heterosis combination, including AF, CE, DE, and CF hybrid. When the MPV of PGW is lower than 40, the hybrid belongs to a weak heterosis combination, including AH, BG, BH, and DH hybrid. When the MPV of PGW is between 40 and 140, the hybrid was in the moderate heterosis group.
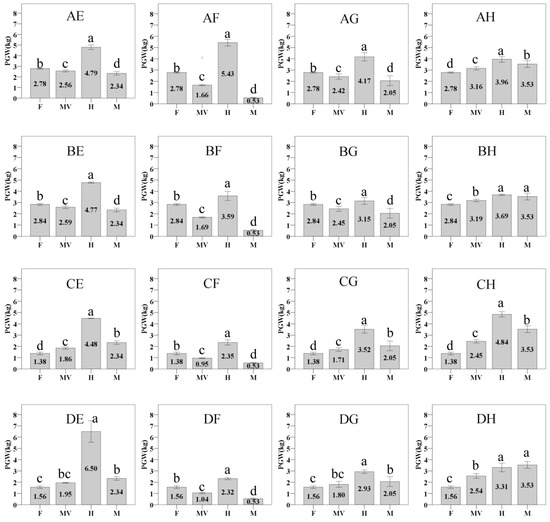
Figure 1.
The plant’s gross weight of sixteen hybrids and eight parents. F: female parent; M: male parent; H: hybrid; MV:mid-parent value. The values are means ± SD. The different letters above each column are significantly different at p < 0.05 by Duncan’s test.

Table 2.
Mid-parent heterosis value analysis of plant gross weight in hybrids.
3.2. Statistics of ASEvent
Through pairwise comparison among female parent, male parent, and hybrid, 2980–3205 AS events were found in each combination. Among them, there were 846–912 A3SS events, accounting for 28.22–29.01% of all AS events; there were 510–591 A5SS events, accounting for 16.89–18.44% of all AS events; there were 9–12 MXE events, accounting for 0.28–0.40% of all AS events; there were 1483–1573 IR events, accounting for 48.72–50.33% of all AS events; there are 113–144 SE events, accounting for 3.70–4.52% of all AS events (Figure 2, Table S1). Among all AS events, IR was the most abundant AS event, followed by A3SS, A5SS, SE, and MXE.
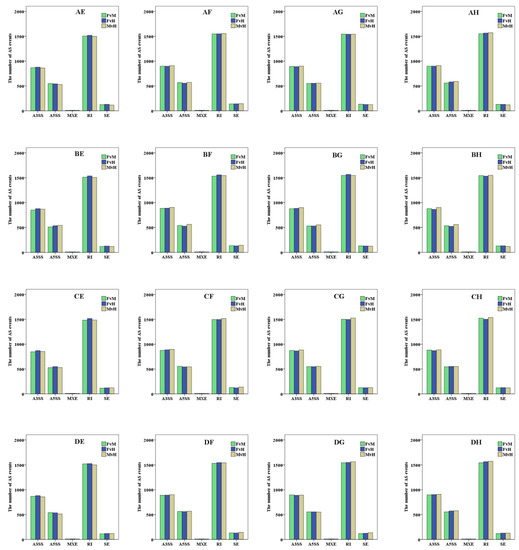
Figure 2.
Statistics aboutthe number of alternative splicing (AS) events including skipped exon (SE), alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), mutually exclusive exon (MXE), and intron retention (IR).
3.3. Analysis of Structure in Gene with AS Events
The exons of genes with AS or without AS were analyzed and compared to understand the relationship between gene structure and AS events. The results demonstrated that the genes with A3SS, A5SS, IR, SE, and MXE had considerably more exons than those without AS (Figure 3a). The genes with A3SS, A5SS, IR, and SE had longer exon length and higher exon GC content than that without AS, while the genes with MXE had no similar performance (Figure 3b,c). There were few genes with MXE, so the greater number, the longer length, and the lower GC content of exons, the more genes that undergo AS.
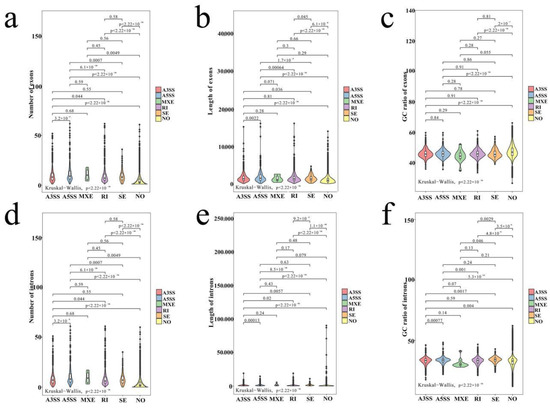
Figure 3.
The structure of genes with A3SS, A5SS, MXE, IR, SE, and non-AS events. (a) The exon number of genes with A3SS, A5SS, IR, SE, and non-AS events. (b) The exon length of genes with A3SS, A5SS, IR, SE, and non-AS events. (c) The exon GC content of genes with A3SS, A5SS, IR, SE, and non-AS events. (d) The GC content of genes with A3SS, A5SS, IR, SE, and non-AS events. (e) The intron number of genes with A3SS, A5SS, RI, SE, and non-AS events. (f) The intron length of genes with A3SS, A5SS, RI, SE, and non-AS events.
Moreover, analysis of intron structures showed that the genes with A3SS, A5SS, IR, SE, and MXE had significantly more introns than those without AS (Figure 3d). Genes with A3SS, A5SS, IR, and SE had considerably longer intron lengths than genes without AS (Figure 3e). The genes with A3SS, A5SS, IR, and SE had considerably greater intron GC contents than those without AS (Figure 3f). The more introns, the longer intron length, and the higher the GC content of introns, the gene was more prone to conduct AS events.
3.4. ASGs Functional Enrichment Analysis
In all crossing combinations, 2151 genes were commonly identified to have taken place AS event (Figure 4a). A GO enrichment analysis was carried out to better understand the ASGs. The significantly enriched GO terms were chosen based on the threshold, adjusted p values < 0.05 after all ASGs were mapped to various functional GO terms. As a result, 46 GO terms were significantly enriched, including 15 GO terms in the cellular component group, 10 GO terms in the molecular function group, and 21 GO terms in the biological process group (Figure 4b).The two most significantly enriched GO terms in the cellular component ontology were cell and cell part. The binding was the most significantly enriched in the molecular function ontology. As for the biological process ontology, the cellular process was the most significantly enriched.
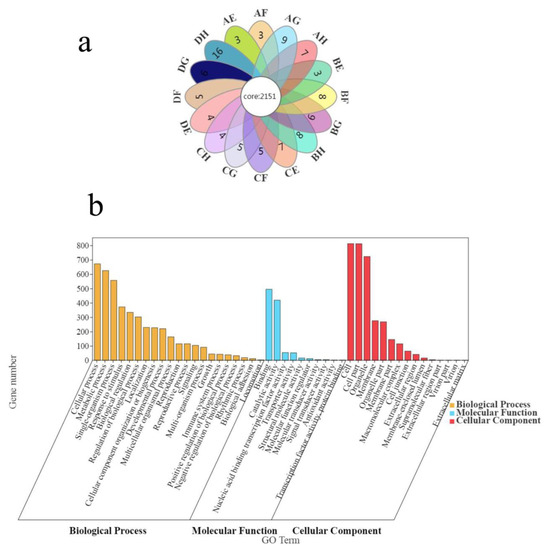
Figure 4.
Analysis of AS genes. (a) Venn map of AS genes in different crosscombinations. (b) GO classification of AS genes in different crosscombinations.
3.5. DASGs between Hybrid and Parents
Based on the restrictive threshold, 353–600DASGs between parents, and411–621DASGs between parents and hybrid in different combinations were identified. (Figure 5a,b). In all DASGs between parent and hybrid, 56.5–75.22% of genes occurred differential AS events between parents, so most of the difference AS between parent and hybrid is generated by the diversity between parents (Figure 5c).
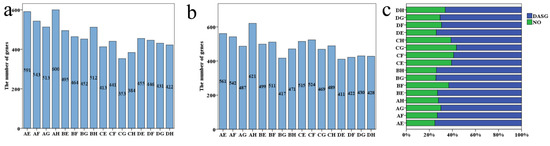
Figure 5.
Different AS events. (a) Different AS events between parents. (b) Different AS events between parents and hybrid. (c) Percentage of different AS genes between parents in different AS genes between hybrid and parents.
3.6. Functional Enrichment Analysis of DASGs
According to GO classification, DASGswere divided into 38–44 functional groups, including 20 biological processes, 10 molecular functions, and 15 cellular components in 16 cross combinations (Table S2). In the classification of biological processes, DASGswere mainly concentrated in the processes of the metabolic process (GO:0008152), cellular process (GO:0009987), single organization process (GO:0044699), and so on. In molecular functional classification, DASGswere mainly concentrated in catalytic activity (GO:0003824), binding (GO:0005488), and so on. In the classification of cell components, DASGswere mainly concentrated in the cell (GO:0005623), cell part (GO:0044464), and organelle (GO:0043226). Those results show that DASGs generally exist in some functional groups in cross combinations. Furthermore, KEGG pathway enrichment analysis was conducted for these DASGs. A total of 33 metabolic pathways were significantly enriched in 16 cross combinations, 3–13 metabolic pathways were significantly enriched in each combination, and DASGs of different cross combinations were enriched in different metabolic pathway (Table S3). There were six pathways in which DASGssignificantly enriched in eight or more cross combinations, and those metabolic pathways could be significantly enriched in most cross combinations. Those significantly enriched pathways included ubiquitin-mediated protein (ko04120), circadian rhythms-plant (ko04712), spliceosome (ko03040), microbial metabolism in diverse environments (ko01120), 2-oxocarboxylic acid metabolism (ko01210),and flavonol biosynthesis (ko00944).
3.7. Metabolic Pathways Influencing the Degree of Heterosis
By further analysis,76 DASGs inthe strong heterosis combinations and 71 DASGsin weak heterosis combinations were identified (Figure 6a,b). A GO enrichment analysis was conducted to further investigate the function of DASGs in strong heterosis combinations. The identified DASGs in strong heterosis combination were significantly enriched in carboxy-lyaseactivity (GO:0016831), carbon-carbon lyase activity (GO:0016830), positive regulation of the development process (GO:0051094), and so on (Figure 6c). For weak heterosis combination, DASGs were significantly enriched in metal ion binding (GO:0046872), transition metal ion binding (GO:0046914), hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides (GO:0016812) (Figure 6d). The KEGG pathway analysis results show that DASGsin strong heterosis combinations were significantly enriched in beta alanine metabolism (ko00410), alanine, aspartate, and glutamate metabolism (ko00250), arginine and proline metabolism (ko00330), glycine, serine, and threonine metabolism (ko00260) (Figure 6e). Coincidentally, these metabolic pathways belonged to amino acid metabolism. The DASGsin weak heterosis combinations were significantly enriched in flavor and flavor biosynthesis (ko00944) and glycosaminoglycan degradation (ko00531) (Figure 6f).
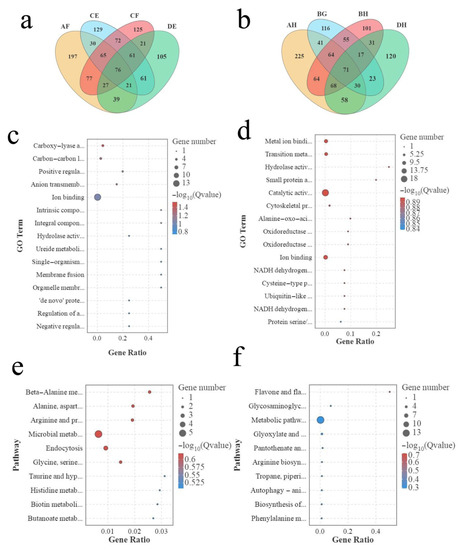
Figure 6.
Analysis of different AS genes in strong heterosis combinations and weak heterosis combinations. (a) Venn map of different AS genes in strong heterosis combinations. (b) Venn map of different AS genes in weak heterosis combinations. (c) GO enrichment of different AS genes in strong heterosis combinations. (d) GO enrichment analysis of different AS genes in weak heterosis combinations. (e) KEGG enrichment analysis of different AS genes in strong heterosis combinations. (f) KEGG enrichment analysis of different AS genes in weak heterosis combinations.
3.8. Amino Acid Metabolic Pathways Influencing the Heterosis
In strong heterosis combinations, DASGs were mainly enriched in some metabolic pathways related to aminoacid metabolism. In these pathways, six related genes were found. Of them, four genes conformed to IR, one gene conformed to A3SS, and two genes conformed to A5SS (Table 3).

Table 3.
Ddifferent AS genes in amino acid metabolism pathways.
To further understand those genes, gene expression levels from RNA-seq data were used for analysis. Among those genes, four genes (BraA06g024290.3C, BraA07g031850.3C, BraA07g035330.3C, BraA10g021330.3C) were differentially expressed in strong heterosis combinations between hybrid and parents, and two genes (BraA06g014310.3C, BraA03g041700.3C) were differentially expressed in most strong heterosis combinations between hybrid and parents (Figure 7a).
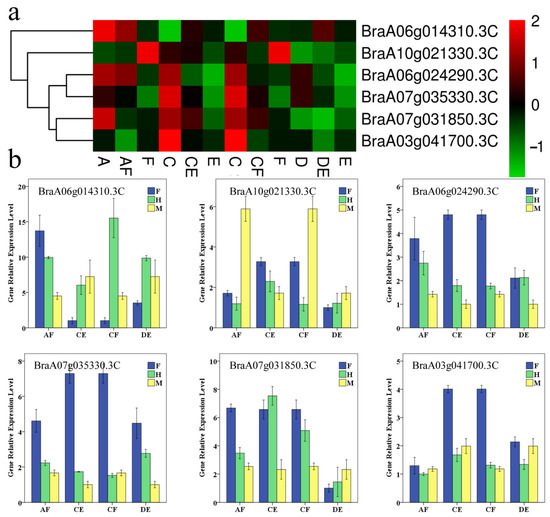
Figure 7.
Gene expression of different AS genes involved in amino acid metabolism. (a) using RNA-seq. data. (b) using qPCR data. Bars represent mean ± SE of triplicate tests. F: female parent; M: male parent; H: hybrid.
The results of qPCR further confirmed transcriptome data.For those genes, the expression levels in hybrid were different from that in either parent alone or both parents among strong heterosis combinations. Although the relative expression levels in a few of the selected genes differed from the data determined by RNA-seq. It is important that the most expression trends between hybrid and parents were consistent with those obtained from RNA-seq in those genes (Figure 7b).
The expression level of related genes and the PGWwere subjected to correlation analysis. PGW was highly correlated with the expression level of BraA06g014310.3C and significantly correlated with BraA03g041700.3C. Inaddition, there was a significant correlation between BraA06g014310.3C and BraA03g041700.3C (Table S4).
4. Discussion
Heterosis is a crucial breeding technology that is constantly used and researched.The study of heterosis has an opportunity to uncover fresh perspectives due to the rapid advancement of science and technology. The molecular mechanism of heterosis has also been explored in terms of genome, transcriptome, epigenetics, andprotein, soit is hoped that a deeper understanding of heterosis will be carried out.
AS plays an important role in the growth and development of plants, such as inducing flowering [26,47], responding to abiotic stress, etc. [48,49,50]. With the in-depth study of AS, researchers have gradually focused on the AS events in hybrids. The number of AS events in wheat decreased after domestication in both diploids and tetraploids. Moreover, AS occurrence frequency tended to decrease after polyploidization [51]. In maize, 19 of 33 nonadditive genes in response to heat stress had differential ASevents under heat stress, so ASevent was the result of heat stress response in maize hybrids [52]. These results indicate that DAS genes can indicate the divergence between hybrids and either of their parents.However, there are very few studies on the heterosis mechanism from the aspect of AS, especially in plants. In our research, 16 hybrid combinations were used to explore the AS events in hybrids, and the relevant results provide the basis and reference for subsequent research.
In Chinese cabbage, 2980–3205 AS events were found in each combination through pairwise comparison among female parent, male parent, and hybrid. Those AS greatly increased the complexity of transcriptome. The five main AS patterns known are SE, A5SS, A3SS, MXE, and IR. These AS patterns occur at different frequencies in different species. In plants, IR is the most common AS pattern [25,31,33,34,35,36,37,53]. In Chinese cabbage, whether between parents or between hybrids and parents, the IR is also the main type of AS, and the frequency is much higher than other types. In order, the other types are A3SS, A5SS, SE, and MXE. The frequency of ASevents was related to gene structure. In Wheat, the GC content of genesmight be correlated with the occurrence of SE and IR.The genes that conformed to SE exhibited a significantly lower GC content than that of other exons, where as the genes that conformed to IR had a significantly higher GC content [54]. In Chinese cabbage, the number, length, and GC content of exons and introns have a significant relationship with the frequency of AS.
DASGs between hybrid and parents were screened out. Functional enrichment analysis showed the metabolic pathways enriched by DASGs were different in different combinations. There were six pathways in which DASGs were significantly enriched in eight or more hybrids, and those metabolic pathways were believed to be significantly enriched in most hybrid combinations. Those significantly enriched pathways included ubiquitin-mediated protein (ko04120), circadian rhythms-plant (ko04712), spliceosome (ko03040), microbial metabolism in diverse environments (ko01120), 2-oxocarboxylic acid metabolism (ko01210) and flavor and flavor biosynthesis (ko00944). The reasons for the formation of heterosis are complex and diverse, and these results provide a basis and reference for subsequent research.
By comparing transcriptomic data collected from hybrid individuals and their parents, 76 DASGs in strong heterosis combinations were identified. Those DASGs were mainly enriched in some metabolic pathways related to amino acid metabolism. In these pathways, six related genes were found. Gene expression level analysis showed that the six related genes were frequently differentially expressed between hybrid and parents in the strong heterosis combinations. Moreover, among those genes, the expression level of BraA06g014310.3C and BraA03g041700.3C highly correlated with PGW. In brief, our results indicated that AS events in amino acid metabolism were linked to heterosis in hybrids. Relevant studies also provide support for our results. In super hybrid rice, allele-specific expression genes were most significantly enriched in the ionotropic glutamate receptor signaling pathway, which was hypothesized to be potential amino acid sensors in plants [55]. In maize hybrids, most amino acids display negative mid-parent heterosis and have lower values than parents’ average [56]. These results indicated that AS events in amino acid metabolism could be related to heterosis. Further experimental verification will be required to reveal the regulatory mechanism in hybrids and their parents.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae9010017/s1, Table S1: Proportion of various types of AS events, Table S2: Go term classification table of DASGs in different cross combinations, Table S3: KEGG enrichment analysis of DASGs in different cross combinations, Table S4: Correlation analysis between PGW and gene expression levels in amino acid metabolism.
Author Contributions
Conceptualization, L.Z. and R.L.; methodology, L.Z. and R.L.; data curation, R.L. and M.T.; validation, R.L. and M.T.; writing—original draft preparation, L.Z., S.N. and R.L.; writing—review and editing, L.Z., S.N., R.L. and M.T.; funding acquisition, L.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Key Research and Development Program of China (2017YFD0101802) and the Key Research and Development Program of Yangling Seed Innovative Center (Ylzy-sc-04).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The RNA-seq data have been deposited with the NCBI with the dataset identifier PRJNA876066.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hochholdinger, F.; Baldauf, J.A. Heterosis in plants. Curr. Biol. 2018, 28, R1089–R1092. [Google Scholar] [CrossRef] [PubMed]
- Schnable, P.S.; Springer, N.M. Progress Toward Understanding Heterosis in Crop Plants. Annu. Rev. Plant Biol. 2013, 64, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, R.; Uezono, K.; Ishikura, S.; Osabe, K.; Peacock, W.J.; Dennis, E.S. Recent research on the mechanism of heterosis is important for crop and vegetable breeding systems. Breed. Sci. 2018, 68, 145–158. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, M.; Zhang, Q.; Wei, X.; Huang, X. Exploring the molecular basis of heterosis for plant breeding. J. Integr. Plant Biol. 2020, 62, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.U.; Dang, T.; Qamar, S.; Ilyas, A.; Fatema, R.; Kafle, M.; Hussain, Z.; Masood, S.; Iqbal, S.; Shahzad, K. Revisiting Plant Heterosis—From Field Scale to Molecules. Genes 2021, 12, 1688. [Google Scholar] [CrossRef]
- Yu, D.; Gu, X.; Zhang, S.; Dong, S.; Miao, H.; Gebretsadik, K.; Bo, K. Molecular basis of heterosis and related breeding strategies reveal its importance in vegetable breeding. Hortic. Res. 2021, 8, 120. [Google Scholar] [CrossRef]
- Bruce, A.B. The mendelian theory of heredity and the augmentation of vigor. Science 1910, 32, 627–628. [Google Scholar] [CrossRef]
- Crow, J.F. 90 Years Ago: The Beginning of Hybrid Maize. Genetics 1998, 148, 923–928. [Google Scholar] [CrossRef]
- Davenport, C.B. Degeneration, albinism and inbreeding. Science 1908, 28, 454–455. [Google Scholar] [CrossRef]
- Jones, D.F. Dominance of Linked Factors as a Means of Accounting for Heterosis. Proc. Natl. Acad. Sci. USA 1917, 3, 310–312. [Google Scholar] [CrossRef]
- Fiévet, J.B.; Nidelet, T.; Dillmann, C.; de Vienne, D. Heterosis Is a Systemic Property Emerging From Non-linear Genotype-Phenotype Relationships: Evidence From in Vitro Genetics and Computer Simulations. Front. Genet. 2018, 9, 159. [Google Scholar] [CrossRef] [PubMed]
- Kaeppler, S. Heterosis: One boat at a time, or a rising tide? New Phytol. 2011, 189, 900–902. [Google Scholar] [CrossRef] [PubMed]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef] [PubMed]
- Steinmetz, L.M.; Sinha, H.; Richards, D.R.; Spiegelman, J.I.; Oefner, P.J.; McCusker, J.H.; Davis, R.W. Dissecting the architecture of a quantitative trait locus in yeast. Nature 2002, 416, 326–330. [Google Scholar] [CrossRef]
- Sinha, H.; Nicholson, B.P.; Steinmetz, L.M.; McCusker, J.H. Complex Genetic Interactions in a Quantitative Trait Locus. PLoS Genet. 2006, 2, e13. [Google Scholar] [CrossRef]
- Fu, D.; Xiao, M.; Hayward, A.; Jiang, G.; Zhu, L.; Zhou, Q.; Li, J.; Zhang, M. What is crop heterosis: New insights into an old topic. J. Appl. Genet. 2014, 56, 1–13. [Google Scholar] [CrossRef]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef]
- Xing, J.; Sun, Q.; Ni, Z. Proteomic patterns associated with heterosis. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2016, 1864, 908–915. [Google Scholar] [CrossRef]
- Mackay, I.J.; Cockram, J.; Howell, P.; Powell, W. Understanding the classics: The unifying concepts of transgressive segregation, inbreeding depression and heterosis and their central relevance for crop breeding. Plant Biotechnol. J. 2021, 19, 26–34. [Google Scholar] [CrossRef]
- Goff, S.A.; Zhang, Q. Heterosis in elite hybrid rice: Speculation on the genetic and biochemical mechanisms. Curr. Opin. Plant Biol. 2013, 16, 221–227. [Google Scholar] [CrossRef]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Cao, Y.; Ma, L. Alternative Splicing in Plant Genes: A Means of Regulating the Environmental Fitness of Plants. Int. J. Mol. Sci. 2017, 18, 432. [Google Scholar] [CrossRef] [PubMed]
- Nimeth, B.A.; Riegler, S.; Kalyna, M. Alternative Splicing and DNA Damage Response in Plants. Front. Plant Sci. 2020, 11, 91. [Google Scholar] [CrossRef]
- Swarup, R.; Crespi, M.; Bennett, M.J. One Gene, Many Proteins: Mapping Cell-Specific Alternative Splicing in Plants. Dev. Cell 2016, 39, 383–385. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Marquez, Y.; Kalyna, M.; Barta, A. Complexity of the Alternative Splicing Landscape in Plants. Plant Cell 2013, 25, 3657–3683. [Google Scholar] [CrossRef] [PubMed]
- Szakonyi, D.; Duque, P. Alternative Splicing as a Regulator of Early Plant Development. Front. Plant Sci. 2018, 9, 1174. [Google Scholar] [CrossRef]
- Chaudhary, S.; Jabre, I.; Reddy, A.S.; Staiger, D.; Syed, N.H. Perspective on Alternative Splicing and Proteome Complexity in Plants. Trends Plant Sci. 2019, 24, 496–506. [Google Scholar] [CrossRef]
- Laloum, T.; Martín, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef]
- Thatcher, S.R.; Zhou, W.; Leonard, A.; Wang, B.-B.; Beatty, M.; Zastrow-Hayes, G.; Zhao, X.; Baumgarten, A.; Li, B. Genome-Wide Analysis of Alternative Splicing in Zea mays: Landscape and Genetic Regulation. Plant Cell 2014, 26, 3472–3487. [Google Scholar] [CrossRef]
- Chamala, S.; Feng, G.; Chavarro, C.; Barbazuk, W.B. Genome-Wide Identification of Evolutionarily Conserved Alternative Splicing Events in Flowering Plants. Front. Bioeng. Biotechnol. 2015, 3, 33. [Google Scholar] [CrossRef]
- Clark, S.; Yu, F.; Gu, L.; Min, X.J. Expanding Alternative Splicing Identification by Integrating Multiple Sources of Transcription Data in Tomato. Front. Plant Sci. 2019, 10, 689. [Google Scholar] [CrossRef] [PubMed]
- Rapazote-Flores, P.; Bayer, M.; Milne, L.; Mayer, C.-D.; Fuller, J.; Guo, W.; Hedley, P.E.; Morris, J.; Halpin, C.; Kam, J.; et al. BaRTv1.0: An improved barley reference transcript dataset to determine accurate changes in the barley transcriptome using RNA-seq. BMC Genom. 2019, 20, 968. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Li, W.; Wang, S.; Zhang, X.; Coules, A.; Ding, G.; Xu, F.; Ren, J.; Lu, C.; Shi, L. Differential Alternative Splicing Genes in Response to Boron Deficiency in Brassica napus. Genes 2019, 10, 224. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Hu, T.; Li, X.; Mu, S.; Cheng, Z.; Ge, W.; Gao, J. Genome-wide analysis of shoot growth-associated alternative splicing in moso bamboo. Mol. Genet. Genom. 2016, 291, 1695–1714. [Google Scholar] [CrossRef]
- Wang, B.-B.; Brendel, V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. USA 2006, 103, 7175–7180. [Google Scholar] [CrossRef]
- Li, Y.; Mi, X.; Zhao, S.; Zhu, J.; Guo, R.; Xia, X.; Liu, L.; Liu, S.; Wei, C. Comprehensive profiling of alternative splicing landscape during cold acclimation in tea plant. BMC Genom. 2020, 21, 65. [Google Scholar] [CrossRef]
- Zorin, E.A.; Afonin, A.M.; Kulaeva, O.A.; Gribchenko, E.S.; Shtark, O.Y.; Zhukov, V.A. Transcriptome Analysis of Alternative Splicing Events Induced by Arbuscular Mycorrhizal Fungi (Rhizophagusirregularis) in Pea (Pisum sativum L.) Roots. Plants 2020, 9, 1700. [Google Scholar] [CrossRef]
- Reddy, A.S. Alternative Splicing of Pre-Messenger RNAs in Plants in the Genomic Era. Annu. Rev. Plant Biol. 2007, 58, 267–294. [Google Scholar] [CrossRef]
- Mastrangelo, A.M.; Marone, D.; Laidò, G.; De Leonardis, A.M.; De Vita, P. Alternative splicing: Enhancing ability to cope with stress via transcriptome plasticity. Plant Sci. 2012, 185–186, 40–49. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative Splicing at the Intersection of Biological Timing, Development, and Stress Responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef]
- Tu, Z.; Shen, Y.; Wen, S.; Zong, Y.; Li, H. Alternative Splicing Enhances the Transcriptome Complexity of Liriodendron chinense. Front. Plant Sci. 2020, 11, 578100. [Google Scholar] [CrossRef] [PubMed]
- Gorlach, J.; Raesecke, H.-R.; Abel, G.; Wehrli, R.; Amrhein, N.; Schmid, J. Organ-specific differences in the ratio of alternatively spliced chorismate synthase (LeCS2) transcripts in tomato. Plant J. 1995, 8, 451–456. [Google Scholar] [CrossRef] [PubMed]
- Kopriva, S.; Cossu, R.; Bauwe, H. Alternative splicing results in two different transcripts for H-protein of the glycine cleavage system in the C4 species Flaveriatrinervia. Plant J. 1995, 8, 435–441. [Google Scholar] [CrossRef]
- Slotte, T.; Huang, H.-R.; Holm, K.; Ceplitis, A.; Onge, K.S.; Chen, J.; Lagercrantz, U.; Lascoux, M. Splicing Variation at a FLOWERING LOCUS C Homeolog Is Associated With Flowering Time Variation in the Tetraploid Capsella bursa-pastoris. Genetics 2009, 183, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Nanaei, H.A.; Wei, B.; Wang, Y.; Wang, X.; Li, Z.; Dai, X.; Wang, Z.; Jiang, Y.; Shao, J. Comparative Transcriptome Profiling Analysis Uncovers Novel Heterosis-Related Candidate Genes Associated with Muscular Endurance in Mules. Animals 2020, 10, 980. [Google Scholar] [CrossRef] [PubMed]
- Mei, C.; Li, S.; Abbas, S.H.; Tian, W.; Wang, H.; Li, Y.; Gui, L.; Zhang, Y.; Wu, X.; Zan, L. Performance Measurement and Comparative Transcriptome Analysis Revealed the Efforts on Hybrid Improvement of Qinchuan Cattle. Anim. Biotechnol. 2018, 30, 13–20. [Google Scholar] [CrossRef]
- Lu, H.; Deng, Q.; Wu, M.; Wang, Z.; Wei, D.; Wang, H.; Xiang, H.; Zhang, H.; Tang, Q. Mechanisms of alternative splicing in regulating plant flowering: A review. Sheng Wu Gong Cheng Xue Bao 2021, 37, 2991–3004. [Google Scholar] [CrossRef]
- Punzo, P.; Grillo, S.; Batelli, G. Alternative splicing in plant abiotic stress responses. Biochem. Soc. Trans. 2020, 48, 2117–2126. [Google Scholar] [CrossRef]
- Lin, J.; Zhu, Z. Plant responses to high temperature: A view from pre-mRNA alternative splicing. Plant Mol. Biol. 2021, 105, 575–583. [Google Scholar] [CrossRef]
- Ling, Y.; Mahfouz, M.M.; Zhou, S. Pre-mRNA alternative splicing as a modulator for heat stress response in plants. Trends Plant Sci. 2021, 26, 1153–1170. [Google Scholar] [CrossRef]
- Yu, K.; Feng, M.; Yang, G.; Sun, L.; Qin, Z.; Cao, J.; Wen, J.; Li, H.; Zhou, Y.; Chen, X.; et al. Changes in Alternative Splicing in Response to Domestication and Polyploidization in Wheat. Plant Physiol. 2020, 184, 1955–1968. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Hu, F.; Zhang, X.; Wei, Q.; Dong, J.; Bo, C.; Cheng, B.; Ma, Q. Comparative transcriptome analysis reveals important roles of nonadditive genes in maize hybrid An’nong 591 under heat stress. BMC Plant Biol. 2019, 19, 273. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yamada, M.; Han, X.; Ohler, U.; Benfey, P.N. High-Resolution Expression Map of the Arabidopsis Root Reveals Alternative Splicing and lincRNA Regulation. Dev. Cell 2016, 39, 508–522. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Qin, J.; Tian, X.; Xu, S.; Wang, Y.; Li, H.; Wang, X.; Peng, H.; Yao, Y.; Hu, Z.; et al. Global profiling of alternative splicing landscape responsive to drought, heat and their combination in wheat (Triticum aestivum L.). Plant Biotechnol. J. 2017, 16, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Zhang, Y.; Yan, T.; Li, Y.; Jiang, N.; Zhou, Y.; Zhou, Q.; Qin, P.; Fu, C.; Lin, H.; et al. Transcriptome profiling of two super hybrid rice provides insights into the genetic basis of heterosis. BMC Plant Biol. 2022, 22, 314. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, A.; Song, Q.; Chen, H.Y.; Harmon, F.G.; Chen, Z.J. Temporal Regulation of the Metabolome and Proteome in Photosynthetic and Photorespiratory Pathways Contributes to Maize Heterosis. Plant Cell 2020, 32, 3706–3722. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).