Accurate Cultivar Authentication of Jujube Fruits Using Nano-Fluidic Genotyping of Single Nucleotide Polymorphism (SNP) Markers
Abstract
:1. Introduction
2. Materials and Methods
2.1. Jujube Fruit Samples and DNA Extraction
2.2. SNP Markers and Genotyping
2.3. Data Analysis
3. Results
3.1. DNA Extraction from the Pulp Tissue of Jujube Fruits
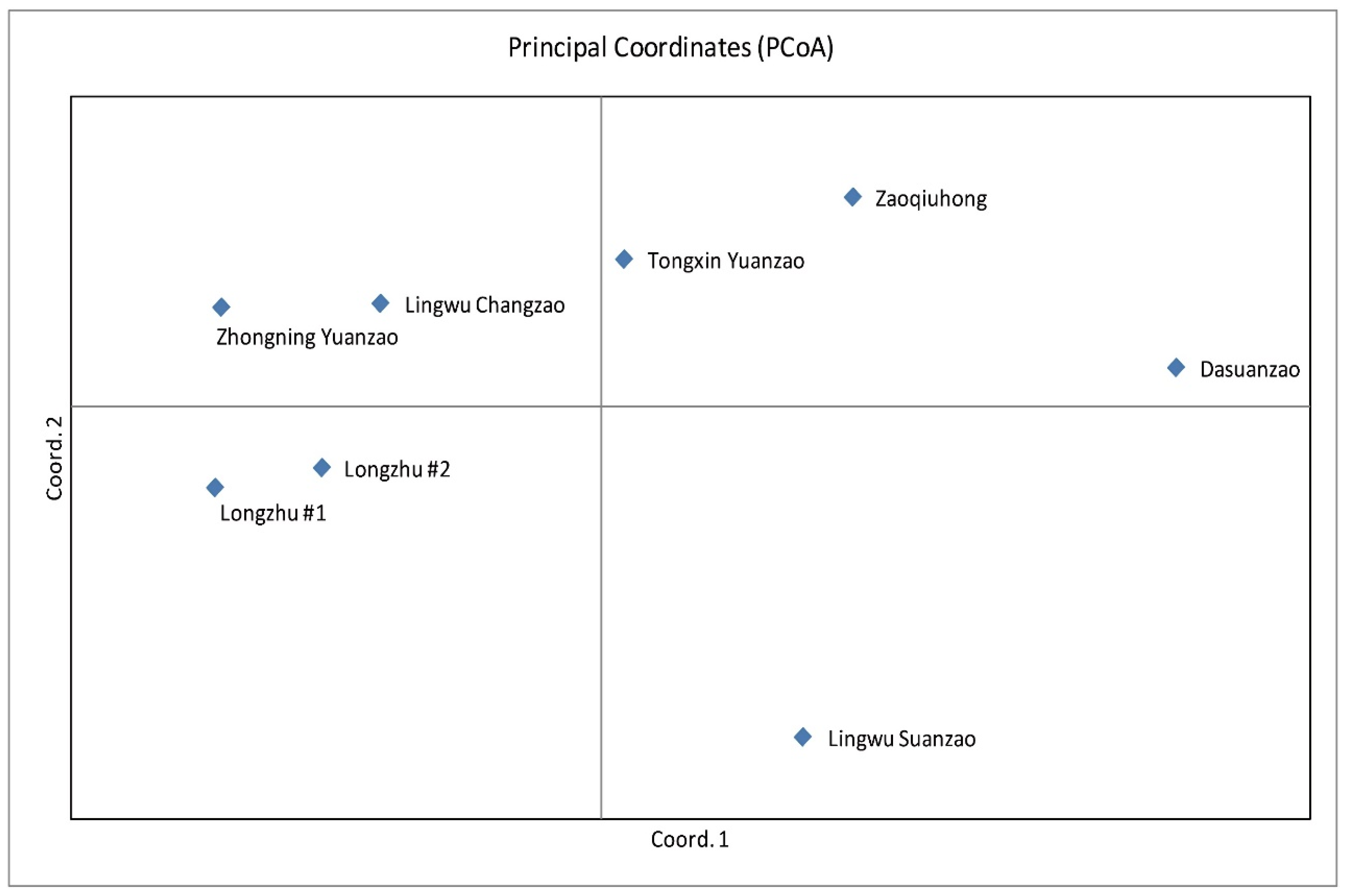
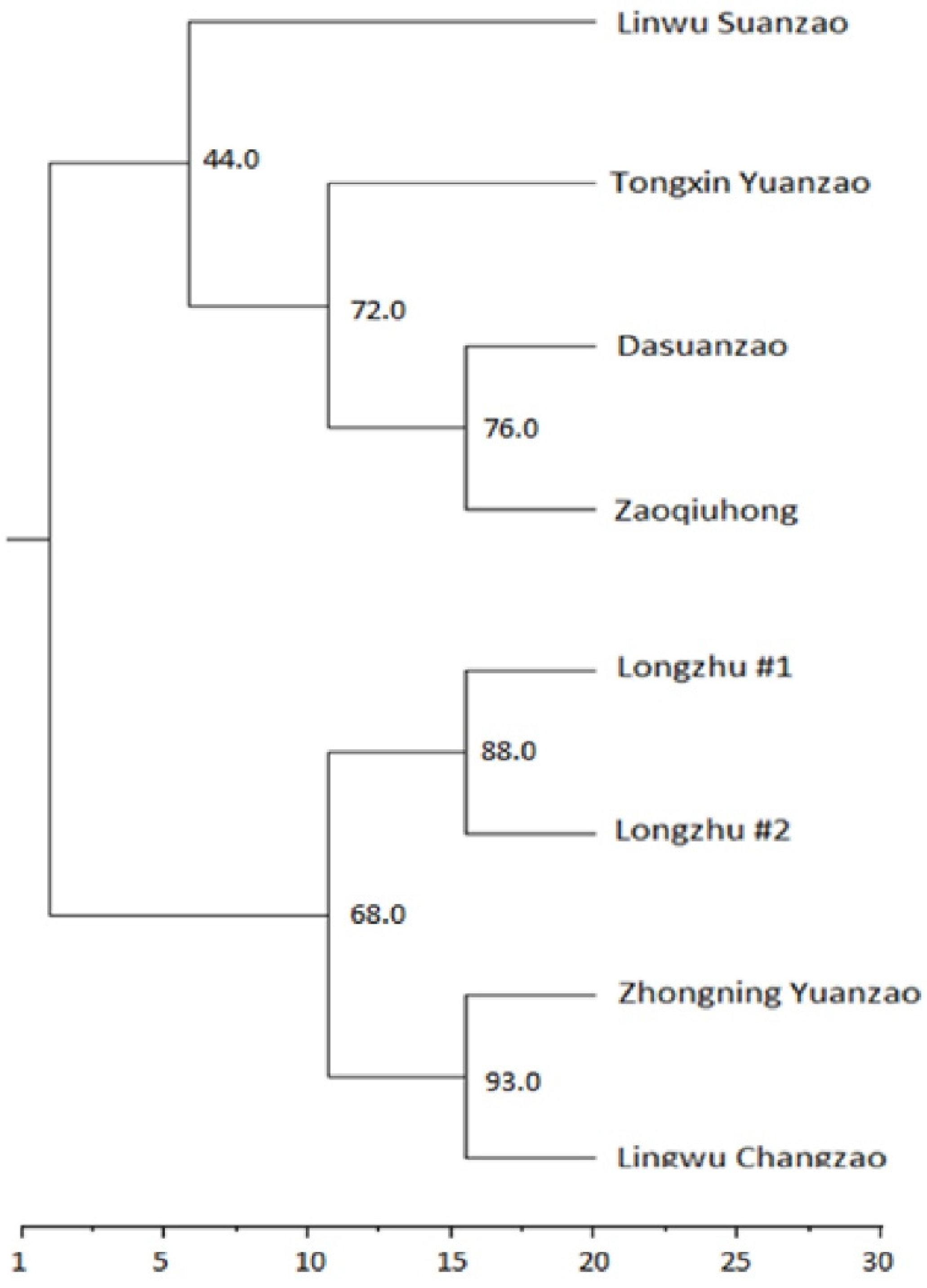
3.2. Jujube Fruit Authentication Using SNP Fingerprints
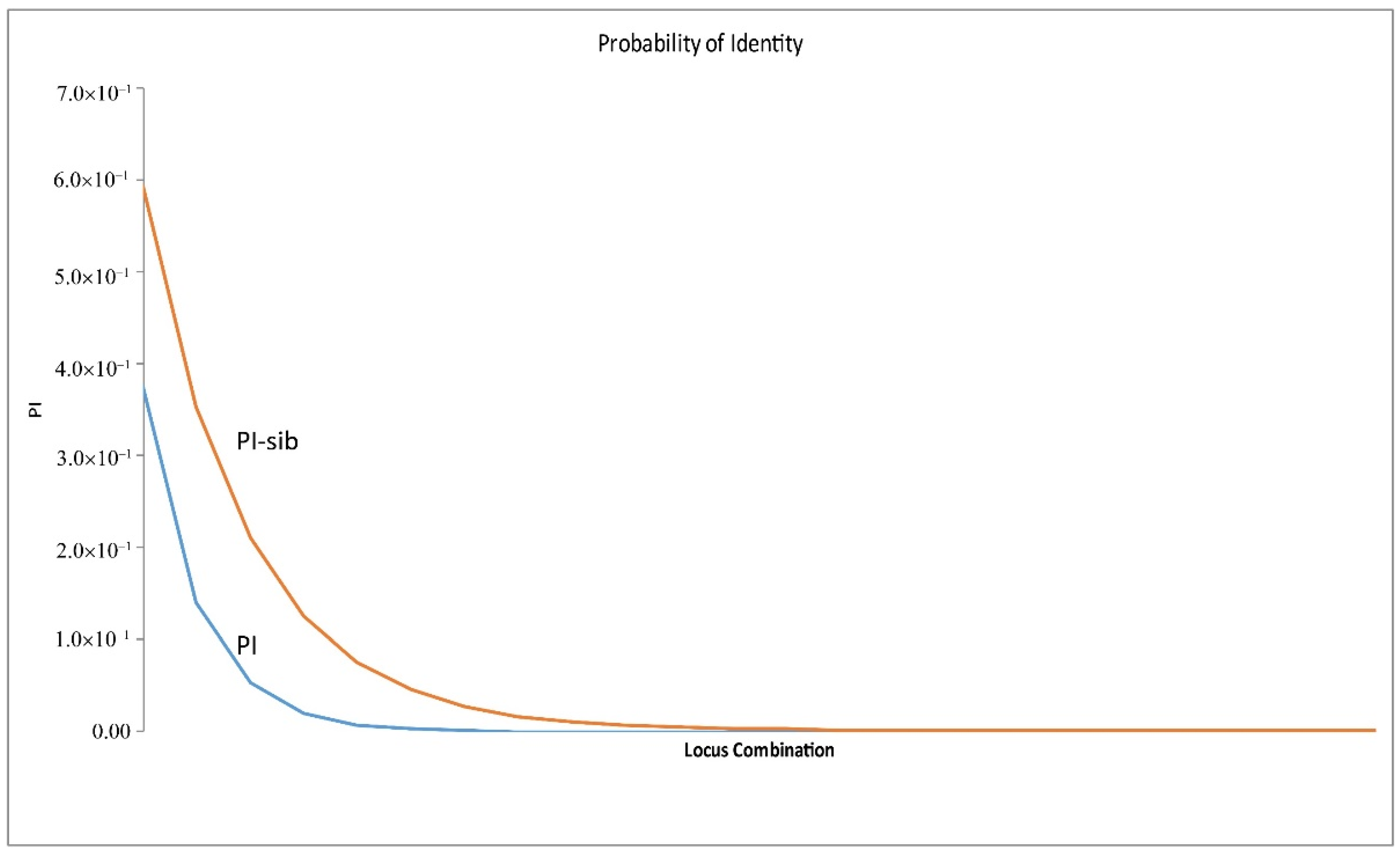
3.3. Selection of top 24 SNP Markers for Jujube Fruit Authentication
4. Discussion
4.1. DNA Quality Extracted from Jujube Fruits
4.2. The SNP Genotyping Kit for Jujube Fruit Authentication
4.3. Advantage of Using SNP Markers for Jujube Cultivar Authentication
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ma, Y.P.; Zhang, D.P.; Wang, Z.J.; Song, L.H.; Cao, B. Fruit Morphology Measurements of Jujube Cultivar ‘Lingwu Changzao’ (Ziziphus jujuba Mill. cv. Lingwuchangzao) during Fruit Development. Horticulturae 2021, 7, 26. [Google Scholar] [CrossRef]
- Liu, M.J.; Zhao, J.; Cai, Q.; Liu, G.; Wang, J.; Zhao, Z.; Liu, P.; Dai, L.; Yan, G.; Wang, W.; et al. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zhang, C.M.; Zhao, X.; Fei, Z.J.; Wan, K.K.; Zhang, Z.; Pang, X.M.; Yin, X.; Bai, Y.; Sun, X.; et al. The jujube genome provides insights into genome evolution and the domestication of sweetness/acidity taste in fruit trees. PLoS Genet. 2016, 12, e1006433. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.P.; Han, Y.R.; Feng, X.R.; Gao, H.D.; Cao, B.; Song, L.H. Genome-wide identification of BAM (β-amylase) gene family in jujube (Ziziphus jujuba Mill.) and expression in response to abiotic stress. BMC Genom. 2022, 23, 438. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.J.; Wang, J.R.; Liu, P.; Zhao, J.; Zhao, Z.H.; Dai, L.; Li, X.S.; Liu, Z.G. Historical achievements and frontier advances in the production and research of Chinese jujube (Ziziphus jujuba) in China. Acta Hortic. Sin. 2015, 42, 1683–1689. [Google Scholar]
- Gao, Q.H.; Wu, C.S.; Wang, M. The jujube (Ziziphus jujuba Mill.) fruit: A review of current knowledge of fruit composition and health benefits. J. Agric. Food Chem. 2013, 61, 3351–3363. [Google Scholar] [CrossRef]
- Liu, M.J. The challenges and countermeasures of jujube industry during transition period. China Fruits 2018, 1, 1–4. [Google Scholar]
- Yao, S.R. Past, present, and future of jujubes—Chinese dates in the United States. HortScience 2013, 48, 672–680. [Google Scholar] [CrossRef]
- Song, L.H.; Meinhardt, L.W.; Bailey, B.; Zhang, D.P. Genetic improvement of Chinese jujube for disease resistances: Status, knowledge gaps and research needs. Crop Breed. Genet. Genom. 2019, 1, e190015. [Google Scholar]
- Kang, L.; Zhu, J.R.; Zhao, D.Y.; Liu, H.J.; Wang, C. Strontium Isotopes to Trace the Geographical Origin of Ruoqiang Jujube. Xinjiang Agric. Sci. 2017, 54, 1066–1075. [Google Scholar]
- Wang, H.Y.; Gao, Z.F.; Fu, C.; Huang, Z.G.; Chang, Z.Q. Origin traceability of jujube in the middle of Taihang Mountain in Hebei province. J. Food Saf. Qual. 2017, 8, 2994–3000. [Google Scholar]
- Lo, Y.T.; Shaw, P.C. DNA-based techniques for authentication of processed food and food supplements. Food Chem. 2018, 240, 767–774. [Google Scholar] [PubMed]
- Corrado, G. Advances in DNA typing in the agro-food supply chain. Trends Food Sci. Technol. 2016, 52, 80–89. [Google Scholar]
- Karola, B.; Pilar, C.M.; Jorge, B.V.; Ignacio, O. Review of recent DNA-based methods for main food-authentication topics. J. Agric. Food Chem. 2019, 67, 3854–3864. [Google Scholar]
- Fang, W.; Meinhardt, L.W.; Mischke, S.; Bellato, C.M.; Motilal, L.; Zhang, D.P. Accurate determination of genetic identity for a single cacao bean, using molecular markers with a nanofluidic system, ensures cocoa authentication. J. Agric. Food Chem. 2014, 62, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Meinhardt, L.W.; Tan, H.; Zhou, L.; Mischke, S.; Wang, X.; Zhang, D.P. Identification of the varietal origin of processed loose-leaf tea based on analysis of a single leaf by SNP nanofluidic array. Crop J. 2016, 4, 304–312. [Google Scholar]
- Zhang, D.P.; Vega, F.E.; Infante, F.; Solano, W.; Johnson, E.S.; Meinhardt, L.W. Accurate differentiation of green beans of Arabica and Robusta coffee using nanofluidic array of Single Nucleotide Polymorphism (SNP) markers. J. AOAC Int. 2020, 103, 315–324. [Google Scholar]
- Kalogianni, D.P.; Bazakos, C.; Boutsika, L.M.; Targem, M.B.; Christopoulos, T.K.; Kalaitzis, P.; Ioannou, P.C. Olive oil DNA fingerprinting by multiplex SNP genotyping on fluorescent microspheres. J. Agric. Food Chem. 2015, 63, 3121–3128. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol. Bology Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Smouse, R.P.P.; Peakall, R. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar]
- Waits, L.P.; Luikart, G.; Taberlet, P. Estimating the probability of identity among genotypes in natural populations: Cautions and guidelines. Mol. Ecol. 2001, 10, 249–256. [Google Scholar] [CrossRef]
- Dieringer, D.; Schlotterer, C. Microsatellite analyser (MSA): A platform independent analysis tool for large microsatellite data sets. Mol. Ecol. Notes 2003, 3, 167–169. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. PHYLIP (Phylogeny Inference Package), Version 3.5 c; Joseph Felsenstein: Washington, USA, 1993. [Google Scholar]
- Rambaut, A.; FigTree Version 1.4.0. Available online: http://tree.bio.ed.ac.uk/software/figtree (accessed on 25 October 2016).
- Cheng, W.; He, W.Z.; Zhao, D.Y.; Liu, Z.; Fan, Y.Y.; Tian, W.N.; Wu, W.L.; Rogers, K.M. Modeling of stable isotope and multi-element compositions of jujube (Ziziphus jujuba Mill.) for origin traceability of protected geographical indication (PGI) products in Xinjiang, China. J. Food Compos. Anal. 2022, 92, 1035772. [Google Scholar]
- Kim, Y.; Shin, J.; Oh, D.R.; Kim, D.W.; Lee, H.S.; Choi, C. Complete chloroplast genome sequences of Vaccinium bracte-atum Thunb., V. vitis-idaea L. and V. uliginosum L. (Ericaceae). Mitochondrial DNA Part B 2020, 5, 1843–1844. [Google Scholar] [CrossRef]
- Lee, H.J.; Koo, H.J.; Lee, J.; Lee, S.C.; Lee, D.Y.; Giang, V.N.L.; Kim, M.; Shim, H.; Park, J.Y.; Yoo, K.O.; et al. Authentication of Zanthoxylum species based on integrated analysis of complete chloroplast genome sequences and metabolite profiles. J. Agric. Food Chem. 2017, 65, 10350–10359. [Google Scholar] [CrossRef]
- Seethapathy, G.S.; Tadesse, M.; Urumarudappa, S.K.J.; Gunaga, V.S.; Vasudeva, R.; Malterud, K.E.; Shaanker, R.U.; de Boer, H.J.; Ravikanth, G.; Wangensteen, H. Authentication of Garcinia fruits and food supplements using DNA barcoding and NMR spectroscopy. Sci. Rep. 2018, 8, 10561. [Google Scholar] [CrossRef]
- Li, J.W.; Fan, L.P.; Ding, S.D.; Ding, X.L. Nutritional composition of five cultivars of Chinese jujube. Food Chem. 2007, 103, 454–460. [Google Scholar] [CrossRef]
- Liu, C.T.; Lu, K.H.; Tobin, M.; Sheen, L.Y. Chinese Dates (Jujubes): A Traditional Functional Food. In Chinese Dates; CRC Press: Boca Raton, FL, USA, 2016; pp. 257–270. [Google Scholar]
- Peng, J.H.; Bai, Y.; Haley, S.D.; Lapitan, N.L.V. Microsatellite-based molecular diversity of bread wheat germplasm and association mapping of wheat resistance to the Russian wheat aphid. Genetica 2009, 135, 95–122. [Google Scholar] [CrossRef]
- Mammadov, J.; Aggarwal, R.; Buyyarapu, R.; Kumpatla, S. SNP markers and their impact on plant breeding. Int. J. Plant Genom. 2012, 2012, 728398. [Google Scholar]

| # | Cultivars | Type | Origin | No. of Samples Analyzed Fruits |
|---|---|---|---|---|
| 1 | Lingwu Changzao | Traditional cultivar | Ningxia, China | 3 |
| 2 | Zhongning Yuanzao | Traditional cultivar | Ningxia, China | 3 |
| 3 | Lingwu Suanzao | Traditional cultivar | Ningxia, China | 3 |
| 4 | Longzhu #1 | Bred cultivar | Ningxia, China | 3 |
| 5 | Longzhu #2 | Bred cultivar | Ningxia, China | 3 |
| 6 | Suanzao | Traditional cultivar | Ningxia, China | 3 |
| 7 | Zaoqiuhong | Bred cultivar | Ningxia, China | 3 |
| 8 | Tongxin Yuanzao | Traditional cultivar | Ningxia, China | 3 |
| Total | 24 |
| # | SNP Code | Chr. | SNPs and Flanking Sequences |
|---|---|---|---|
| 1 | Zj002 | 1 | GCCTTGTTCCTGACGAGTTAGAGATTCCCTGGTAAATTAGGGTAGTGGAATGAGACCTAGGCAAACCAATATGCATCGGC[T/C]AGCAGGGCCAGAGAGCGGGTTCCCCACCTCGAAAAACACACACACACACATACATATATATATATATATATATATGTAAT |
| 2 | Zj018 | 1 | GTCGAGCTTTAGTTCCAAAAGTTATTCGGATTCAGATGATTGGGCATCCTATAATATTACCAATTGTAGATATGCCTTTG[T/C]GATGGAAGAAAGCAAATTCAAGTTCTCTACAAGTTTCAAAGAAAAGGTTCCAATGGTTATAAATTGGGCAATTGGGAAAG |
| 3 | Zj028 | 2 | TCTTTTTCTTTTTAAACTAAGTATAACCTTAAGAAACCTACCTTCTTATTATAAAAACAAATAAATAAAAATATATATAA[C/T]TTTTTTAAAAACACACACACACACACACACACACACACACACACACACATATATTGGTTTATAATTTAAAATATCCAAAT |
| 4 | Zj041 | 2 | AAAATAAAAAAAACCAAAATTTTCTTTTCTTTTTCAAAAACAATAAAAAAGTTTTCCAAATGCTTCAAAAAAAAATAAAA[A/C]AAATCAAGCACCACCATGATCAAAACAAAATGGGTTATCTTTCAATGTTCCAAAAAAAACTTCACCAAGAGAAAAAATGT |
| 5 | Zj056 | 3 | AATTTTTGAAGTAATCGAAAGGCTAATAAGGGAGGTTTTGAACTAAGAATTTATAAATTAGTTTGGAAGAATCATGCTGA[T/C]ATAACAAATGCTTTTGGTGATATCTCATGAAATATTTAAAAACTAAAAAATTAGTGGGACAAGTAACAACCATTAGCACT |
| 6 | Zj062 | 3 | CTGAAATTGTAAGATACTTCAACTTTTGTTTCAGATTCTCTGCATTCCATGGTTTAAATCTTTTGTTGAGATCATTACGT[T/C]GTGTTGTTGTTGTTATATTTATTTATTATTTTTTTAAATGTATTCTGGTTGTAGGGTTGTTCATTGGTTGTACTTGATCA |
| 7 | Zj089 | 4 | TTCTCCCTAACAATCTAAAACCTAAACAAATAAAAAGGGGGCATCCAAGTTCAATTCTTGTTAATACGTGTACTTCATAC[C/T]TATGCTGCTTTTTAATTATATTTTCAATTAAGTAAAATAAATTTACATGCTATTGCAAGGCGTTATCGATGATTTAAATT |
| 8 | Zj090 | 4 | TAATATATATAAATCTTCATGGTGTATCTAATTAATTACCATCTAATCATGATCATGATCAACTATAATGAGCGACATCT[C/A]CCACCATATTGTGCCAAACAGAGCTAGATTCTTCGTCCCTTCTTGTGTGTCCCCAAGTCTATCTCATCAACCAATTAATG |
| 9 | Zj106 | 5 | TTCTTTATATATTTATTGGAAGAGTTTTTTCATTTTTTGTCTGAATATGGTTGTTATGGTTTTGTGCTCTTATTTGTTTT[T/C]CTTAACCCTGCACGTTTCTTTTTTCTTTAAAAAGAGACCCTGCATGTTTAGTTGCATATGAATTCCCTAGCTTTAGTGAT |
| 10 | Zj124 | 5 | CTGCTTCGTTTGATGCTGCAGAAATTTTAAAAGTAGAGACATGTTGAGGAGGTCTGGTACTTGGTGTGAATATATCTATC[T/G]CTTGTGGAGCTGTAGAGACTGAAAGCCATATTGTTCATCATCATTGTCAAAGACAACAATGTTCAAGGCAAATACAATAA |
| 11 | Zj151 | 6 | AATTTGAGAAAATTGAGTGCTTGAATTTTGCTATCTCATATCTTATTAATTAAAAGTCCCTAGAAAGATAGCTATAGTAG[T/C]ATTGTAAATAAACATTTTTTTTCGATTTTAATATAGTTCTTCACTGGTAAAGCGAGGAATTGGAAACAACATTTATTGAA |
| 12 | Zj157 | 6 | CACCACATACCAATAATATGCAAAGCACAGAATACACACATTTCTAAAGAAAAATAAAGCAATCCAAGGCGATTAGGGGG[C/T]TATCGATATCCTCCATAGGAATAGAGTCGATGTCATGGTCATGAAGTGGAGCCAGCGCTTCGCCAACCGCAATCTCGTCG |
| 13 | Zj171 | 7 | GACACATTCGGTCATATGCTTACAATCTTTAAAATTTTCTGTTCAAAGTTTCAAGAACTAATATTCTGATGCAGAATCAA[C/T]AACATAACAATTTTATGTTGGTCTTAAACAACAAAGATTCAGCAGACCTTGCATTTTAAATGCTTATATAAAATTATGAA |
| 14 | Zj179 | 7 | TGGATCAAAATATGCAATGATCCAATATTTTTTTGACTGGTGTTGCTTAAAAACACATTTTTGCAAGTGAACTGAAATTT[A/G]CCCAAGGTGGTCACTTGATAGTTTACTGCTCTTTCTTCTTGCTTCAACTGAACAAGTTTACCGGGGAATATAATCAAGCA |
| 15 | Zj190 | 8 | ACTTATAGTATATGCTACAAACCAGAAAAGTTCTAAGTAGTTTAATGAAAATTTAGGACCTTACATAACTTTTTAGATTA[T/C]ATTTAAATCATTTTGAGAAGCAATGCCAATATCAGCATATTTCTTACTATTTCCTAAAAATTGATTTTTATTTTTATTTA |
| 16 | Zj204 | 8 | AAAATAGTAACTTTCTTAAACTTATTTGTCCAGCAAACAAATAAATAAAAACCAAAAGTAAAGGTAATTTCCTAAAACAA[A/G]AAGTAGAAATCAAATTAGAAAACTAAGATACTATCTATTTTACTAATTTGACTTTGACCAATATTTGATCATGAAGTCCC |
| 17 | Zj216 | 9 | ATTGATTGTAGTTTATTGACTAGTAGTCCACCAAAATCGGAGCTGCAATTAAAAAGTTATGATCAAAACAATTTGTGATC[T/C]AGTCTAATTGAGCAAGCTCGAATTTGATTTTTTATTTGCATGAATTTTGGTTTTGAATTGTATACCATTATATAGTACTC |
| 18 | Zj220 | 9 | TTTAGTGATTGTTAGATTTGTTTATTTAAGTTGCAAGATTTTTAATACCCCAAATCAGAATTTTTTCGATTTTTAGATCC[A/G]AGTCTTGTCTTAAAGGGAGAGATTAAGATATATGAGCCATTTACATGTCTGAATCTTGACACTCTGAGAAGTTGTAGAGC |
| 19 | Zj225 | 10 | GAAAGACATGCAAATTTTGTAACGAATAAATGTGTACTTGAAAAACAGAAAAATAAACGGCAATCTTCAATATTTTGCTT[C/T]GCATTTTTAAGAAATATTTTCAGTTTTTATTTGTAGATGAAAAGTCAAAAATTAATCAAAGAAAAAAAAATGTGCGTTTT |
| 20 | Zj242 | 10 | AGTTTAGAAACAATCTTGGTGAAGCCTATGAAGAAGTGATCATAAAGAGTCATCTTGTTGTAGCTCTCCATCCAAGACCT[G/A]GTAACTAGCTTGACACTGTCACTGAATGCCTTTTCCAATGTATTGAAGAGATCATTTCATAGCATTGATGGTTATGTTGC |
| 21 | Zj271 | 11 | TCATCATTGGATGTCACACCGACACGACACTCGTAACCATTCATTTCTTTGCAACTTTCTTCTTCATTTTCTACTATGGC[T/C]GCGTGCTTTAATGATGTTGACATGGCCTGAGACAATACCACTGGATTCTTTTCCACACATCTTACACCATCTTCTGTAGG |
| 22 | Zj275 | 11 | AGTATGAGAATCCACAGAATTGCAAGCAGCTCTGTAGCAGACATGTAAACGGCAGAAAGCATTGCAGTGTGTATGTTCTG[C/T]AATAACCAGCCAGACCAGGATTATTTTTTCATCTCTTGTAATACTTATATGCAGTTTCCCATCCAAAAAAAGATTTAATT |
| 23 | Zj290 | 12 | GTTCATGTGTCACTGTTCACACGTACTGTACACTATTCATTATCACTGTTCATTGCCGGGTGTCGCACTAAAGACCGACA[C/T]GTGGCACGTGCCACACTCTCGAACGGCCACGTGTCATCTGCTACAGACGACATGTGGATCATCCTTAGCACATTTGAACG |
| 24 | Zj304 | 12 | TTGGTCAAAGAATGGTCAAAGTCAACTTAGTCAAAAAGCTAGTATTTTAGTTTCCTAATTTGATTTTACTTTTTGTTTTA[A/G]GAAATTACCATTACTTTTTGGATTCGATTTACTTATTTTATGGACAAATAAGTTGAGGAAAATTATTATTTTATTATTTC |
| Name of Cultivar | Concentration (ng µL−1) | A260/280 | ||
|---|---|---|---|---|
| Range | Mean | Range | Mean | |
| Lingwu Changzao | 29.3–57.6 | 44.4 | 1.75–1.96 | 1.86 |
| Zhongning Yuanzao | 32.5–65.8 | 48.8 | 1.74–1.89 | 1.81 |
| Lingwu Suanzao | 42.7–57.5 | 49.4 | 1.82–2.01 | 1.95 |
| Longzhu #1 | 40.4–58.6 | 52.6 | 1.77–1.89 | 1.81 |
| Longzhu #2 | 38.8–67.5 | 46.5 | 1.79–1.99 | 1.85 |
| Suanzao | 32.6–55.4 | 38.8 | 1.83–2.00 | 1.93 |
| Zhaoqiuhong | 44.2–62.3 | 54.5 | 1.75–1.95 | 1.85 |
| Tongxin Yuanzao | 42.1–47.9 | 44.1 | 1.78–1.88 | 1.80 |
| Mean | 47.4 | 1.86 | ||
| # | Cultivar | Zj002 | zj018 | zj028 | zj041 | zj056 | zj062 | zj089 | zj090 | zj106 | zj124 | zj151 | zj157 | zj171 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Lingwu Changzao | TT | CT | CT | AC | CT | CT | CT | CC | CT | GT | CC | CT | CT |
| 2 | Zhongning Yuanzao | CC | TT | CC | CC | CT | CT | TT | CC | CT | GT | CC | CT | TT |
| 3 | Lingwu Suanzao | CC | TT | CT | AC | CC | TT | TT | TT | AC | CT | GT | CT | CT |
| 4 | Longzhu#1 | TT | CC | CC | CC | CT | TT | TT | AC | CT | GT | CC | CT | CT |
| 5 | Longzhu#2 | CC | TT | CC | CC | CT | CT | CT | AC | CT | GG | CC | CT | CT |
| 6 | Dasuanzao | CC | TT | CC | CC | CT | TT | TT | AC | TT | GG | TT | CT | CC |
| 7 | Zaoqoiuhong | TT | CT | CT | CT | CT | TT | TT | CC | TT | GT | TT | CT | CT |
| 8 | Tongxin Yuanzao | TT | CT | CT | CT | CT | TT | TT | CC | CT | GT | CC | CT | CT |
| Locus | zjoo2 | zj018 | zjo38 | zj041 | zj056 | zj062 | zj089 | zj090 | zj106 | zj124 | zj151 | zj157 | zj171 | zj179 | zj190 | zj204 | zj216 | zj220 | zj225 | zj242 | zj271 | zj275 | zj290 | zj304 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI | 9.0000 | 7.0760 | 2.9547 | 1.1198 | 0.4199 | 0.1592 | 0.0665 | 0.0249 | 0.0098 | 0.0038 | 0.0015 | 0.0008 | 0.0003 | 0.0001 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 | 0.0000 |
| PI-Sib | 14.2500 | 12.6535 | 8.0925 | 4.8446 | 2.8765 | 1.7220 | 1.1013 | 0.6539 | 0.4013 | 0.2462 | 0.1511 | 0.1103 | 0.0677 | 0.0405 | 0.0243 | 0.0149 | 0.0095 | 0.0058 | 0.0035 | 0.0021 | 0.0012 | 0.0007 | 0.0004 | 0.0003 |
| PIC | 0.37 | 0.29 | 0.26 | 0.37 | 0.38 | 0.23 | 0.38 | 0.31 | 0.37 | 0.37 | 0.37 | 0.36 | 0.37 | 0.37 | 0.29 | 0.37 | 0.30 | 0.38 | 0.37 | 0.38 | 0.22 | 0.37 | 0.37 | 0.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Ma, Y.; Meinhardt, L.W.; Zhang, D.; Cao, B.; Song, L. Accurate Cultivar Authentication of Jujube Fruits Using Nano-Fluidic Genotyping of Single Nucleotide Polymorphism (SNP) Markers. Horticulturae 2022, 8, 792. https://doi.org/10.3390/horticulturae8090792
Zhang Y, Ma Y, Meinhardt LW, Zhang D, Cao B, Song L. Accurate Cultivar Authentication of Jujube Fruits Using Nano-Fluidic Genotyping of Single Nucleotide Polymorphism (SNP) Markers. Horticulturae. 2022; 8(9):792. https://doi.org/10.3390/horticulturae8090792
Chicago/Turabian StyleZhang, Yue, Yaping Ma, Lyndel W. Meinhardt, Dapeng Zhang, Bing Cao, and Lihua Song. 2022. "Accurate Cultivar Authentication of Jujube Fruits Using Nano-Fluidic Genotyping of Single Nucleotide Polymorphism (SNP) Markers" Horticulturae 8, no. 9: 792. https://doi.org/10.3390/horticulturae8090792
APA StyleZhang, Y., Ma, Y., Meinhardt, L. W., Zhang, D., Cao, B., & Song, L. (2022). Accurate Cultivar Authentication of Jujube Fruits Using Nano-Fluidic Genotyping of Single Nucleotide Polymorphism (SNP) Markers. Horticulturae, 8(9), 792. https://doi.org/10.3390/horticulturae8090792






