Morpho-Agronomic Characterization, Sample Size, and Plot Size for the Evaluation of Capsicum chinense Genotypes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Soil Analysis and Cultural Treatments
2.3. Experimental Design
2.4. Morpho-Agronomic Descriptors
2.5. Phenotypic and Statistical Analyzes
3. Results and Discussion
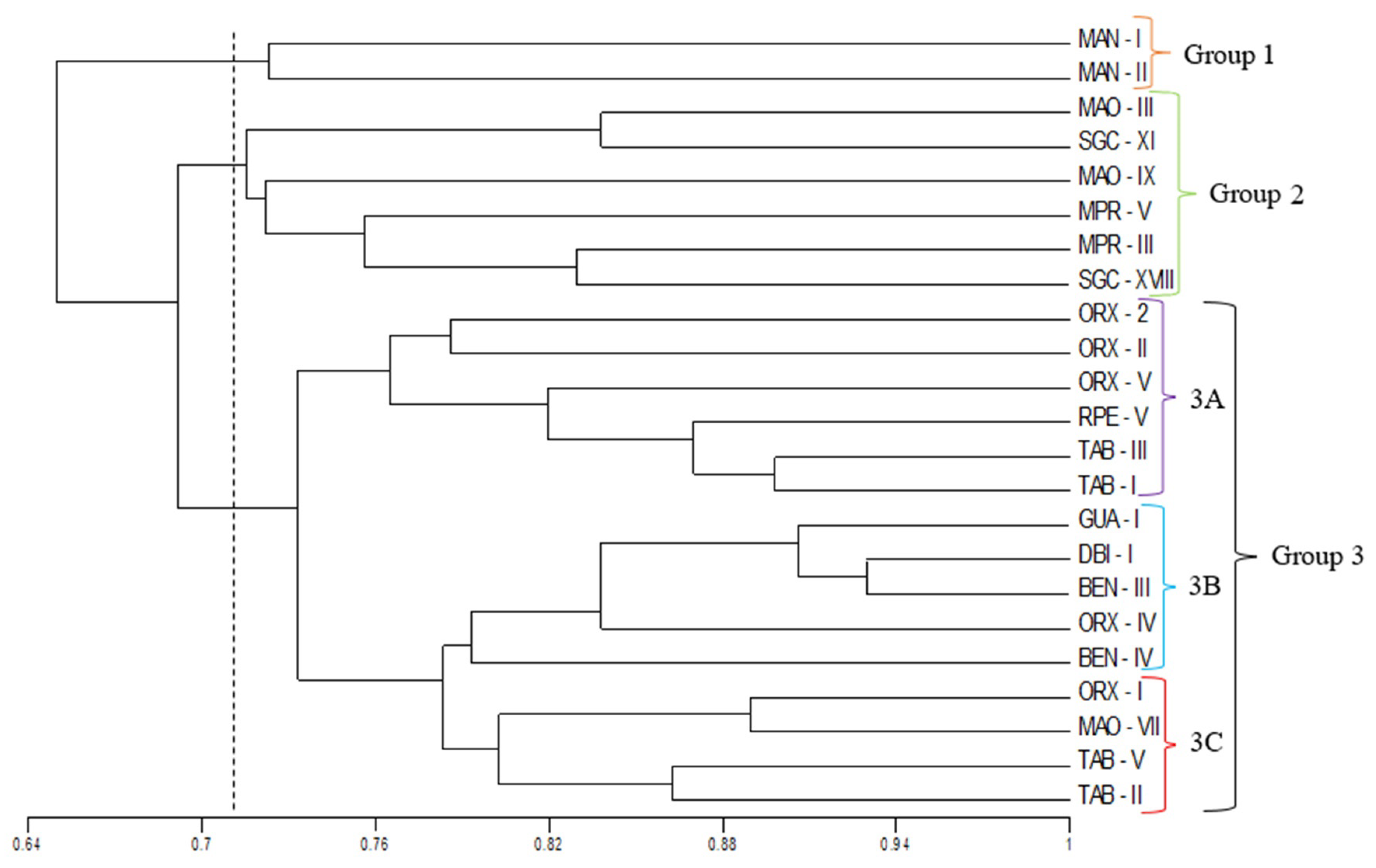
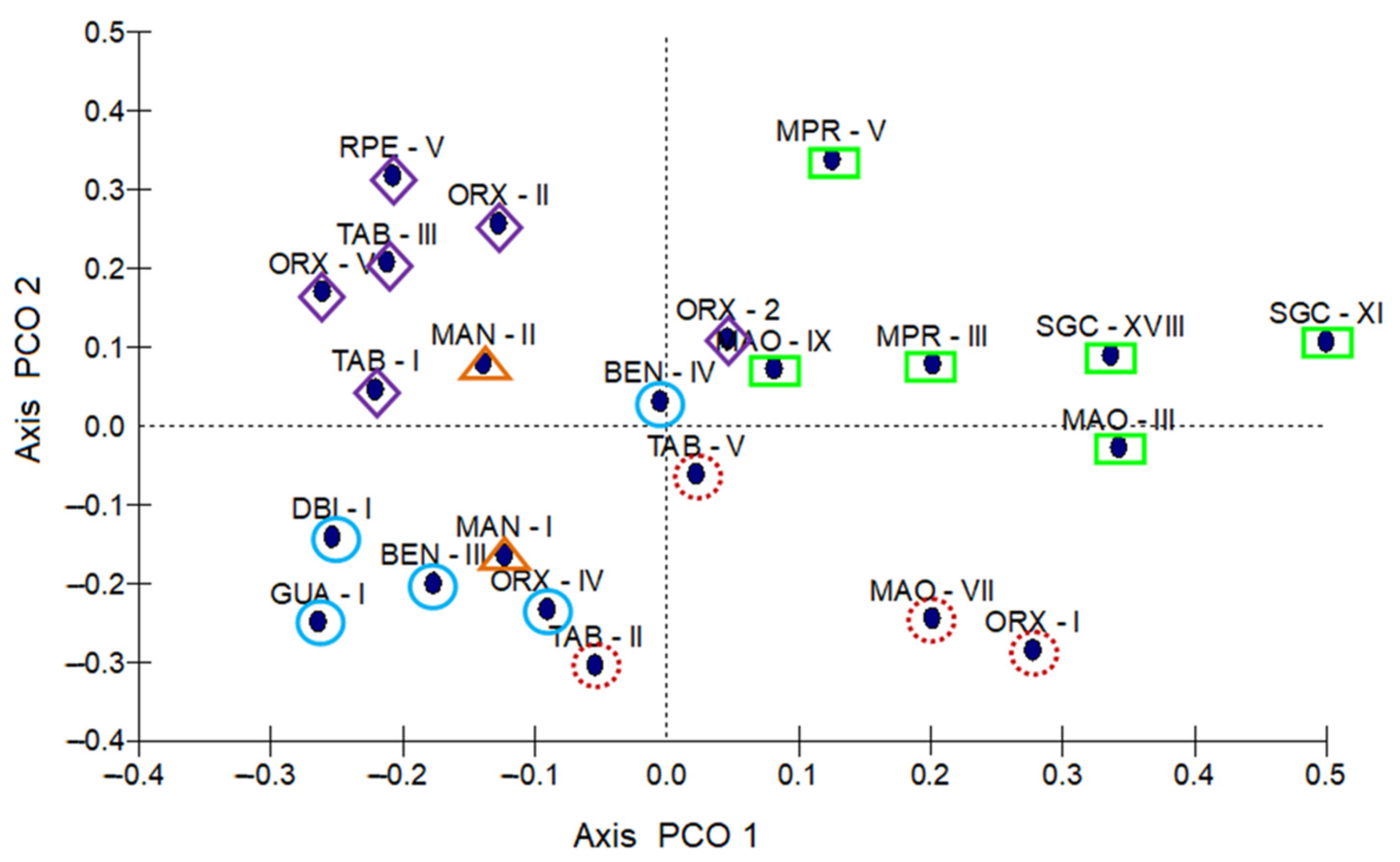
3.1. Morpho-Agronomic Characterization Data
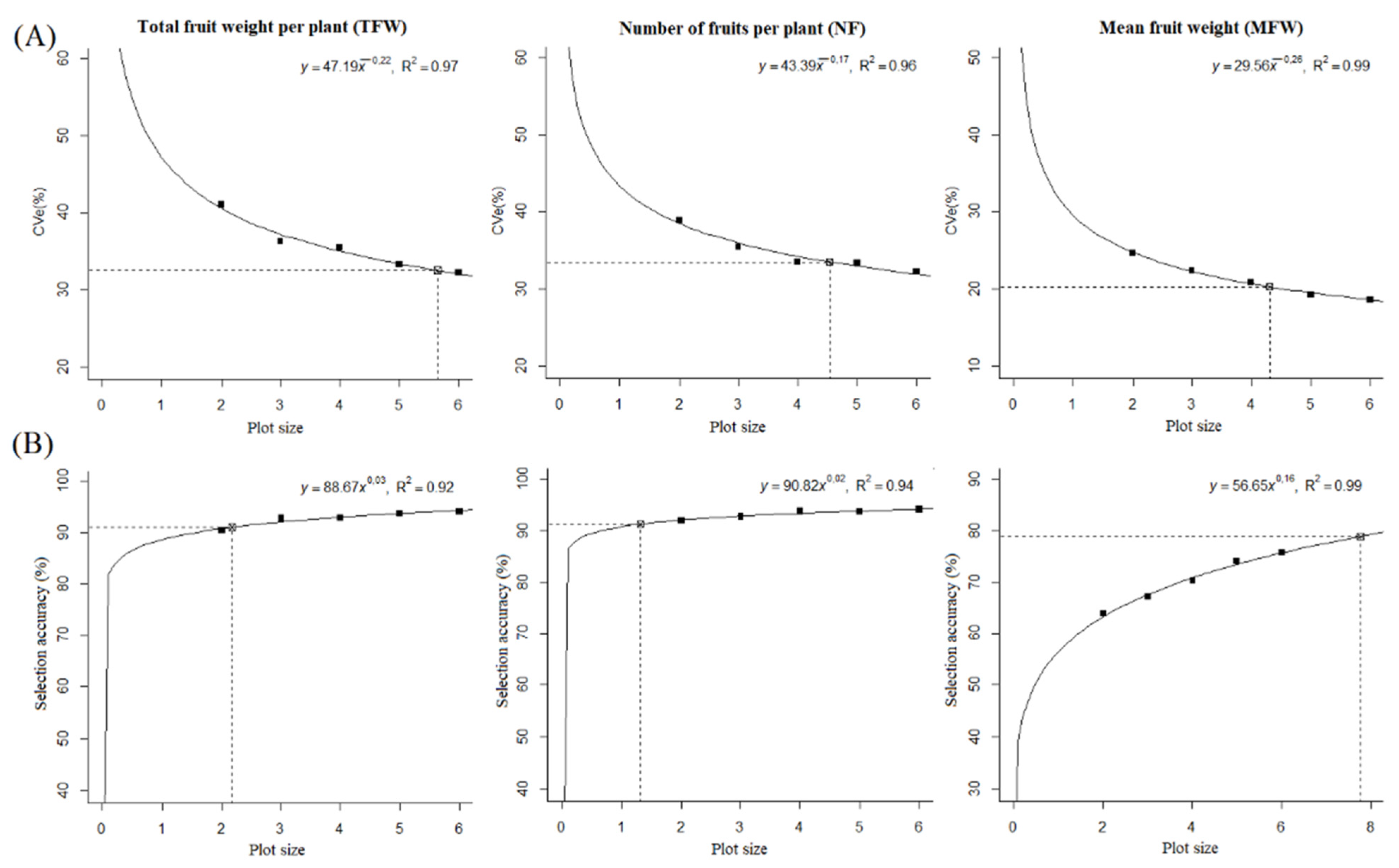
3.2. Plot Size for Evaluations off “Pimenta-de-Cheiro” C. chinense Fruits
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- García, C.C.; Sehr, E.M.; Barfuss, M.H.J.; Barboza, G.E.; Samuel, R.; Moscone, E.A.; Ehrendorfer, F. Phylogenetic relationships, diversification and expansion of chili peppers (Capsicum, Solanaceae). Ann Bot. 2016, 118, 35–51. [Google Scholar]
- Moreira, A.F.P.; Ruas, P.M.; Ruas, C.D.F.; Baba, V.Y.; Giordani, W.; Arruda, I.M.; Rodrigues, R.; Gonçalves, L.S.A. Genetic diversity, population structure and genetic parameters of fruit traits in Capsicum chinense. Sci. Hortic. 2018, 236, 1–9. [Google Scholar]
- Barboza, G.E.; García, C.C.; González, S.L.; Scaldaferro, M.; Reyes, X. Four new species of Capsicum (Solanaceae) from the tropical Andes and an update on the phylogeny of the genus. PLoS ONE 2019, 14, e0209792. [Google Scholar]
- Barboza, G.E.; de Bem Bianchetti, L.; Stehmann, J.R. Capsicum carassense (Solanaceae), a new species from the brazilian atlantic forest. PhytoKeys 2020, 140, 125–138. [Google Scholar]
- Bianchi, P.A.; da Silva, L.R.A.; da Silva Alencar, A.A.; Araújo Diniz Santos, P.H.; Pimenta, S.; Sudré, C.P.; Corte, L.E.D.; Azeredo Gonçalves, L.S.; Rodrigues, R. Biomorphological characterization of brazilian Capsicum chinense Jacq. germplasm. Agronomy 2020, 10, 447. [Google Scholar]
- de Aguiar, A.C.; Coutinho, J.P.; Barbero, G.F.; Godoy, H.T.; Martínez, J. Comparative study of capsaicinoid composition in Capsicum peppers grown in Brazil. Int. J. Food Prop. 2016, 19, 1292–1302. [Google Scholar]
- Oney-Montalvo, J.E.; Avilés-Betanzos, K.A.; Jesús Ramírez-Rivera, E.D.; Ramírez-Sucre, M.O.; Rodríguez-Buenl, I.M. Polyphenols content in Capsicum chinense fruits at different harvest times and their correlation with the antioxidant activity. Plants 2020, 9, 1394. [Google Scholar]
- Camposeco-Montejo, N.; Flores-Naveda, A.; Ruiz-Torres, N.; Álvarez-Vázquez, P.; Niño-Medina, G.; Ruelas-Chacón, X.; Torres-Tapia, M.A.; Rodríguez-Salinas, P.; Villanueva-Coronado, V.; García-López, J.I. Agronomic performance, capsaicinoids, polyphenols and antioxidant capacity in genotypes of habanero pepper grown in the southeast of Coahuila, Mexico. Horticulturae 2021, 7, 372. [Google Scholar]
- Martínez-Ispizua, E.; Martínez-Cuenca, M.R.; Marsal, J.I.; Díez, M.J.; Soler, S.; Valcárcel, J.V.; Calatayud, Á. Bioactive compounds and antioxidant capacity of valencian pepper landraces. Molecules 2021, 26, 1031. [Google Scholar]
- Chel-Guerrero, L.D.; Castañeda-Corral, G.; López-Castillo, M.; Scampicchio, M.; Morozova, K.; Oney-Montalvo, J.E.; Ferrentino, G.; Acevedo-Fernández, J.J.; Rodríguez-Buenfil, I.M. In vivo anti-inflammatory effect, antioxidant activity, and polyphenolic content of extracts from Capsicum chinense by-products. Molecules 2022, 27, 1323. [Google Scholar]
- Cruz, J.G.; Silveira, T.; Richter, V.; Wagner, J.G.; Neitzke, R.S.; Barbieri, R.L.; Vizzotto, M. Genetic variability of bioactive compounds in Capsicum chinense. Food Sci. Technol. 2022, 42, e123721. [Google Scholar]
- Joshi, D.D.; Somkuwar, B.G.; Kharkwal, H.; Chander, S. Aroma based varieties of Capsicum chinense Jacq., geographical distribution and scope for expansion of the species. J. Appl. Res. Med. Aromat. Plants 2022, 29, 100379. [Google Scholar]
- Van Zonneveld, M.; Ramirez, M.; Williams, D.E.; Petz, M.; Meckelmann, S.; Avila, T.; Bejarano, C.; Ríos, L.; Peña, K.; Jäger, M.; et al. Screening genetic resources of Capsicum peppers in their primary center of diversity in Bolivia and Peru. PLoS ONE 2015, 10, e0134663. [Google Scholar]
- Sudré, C.P.; Gonçalves, L.S.A.; Rodrigues, R.; Do Amaral, A.T.; Riva-Souza, E.M.; Bento, C.D.S. Genetic variability in domesticated Capsicum spp. as assessed by morphological and agronomic data in mixed statistical analysis. Genet. Mol. Res. 2010, 9, 283–294. [Google Scholar]
- Oliveira, A.C.R.; Cecon, P.R.; Nascimento, M.; Finger, F.; Pereira, G.; Puiatti, G.A. Genetic divergence between pepper accessions based on quantitative fruit traits. Científica 2019, 47, 83–90. [Google Scholar] [CrossRef]
- Uncu, A.T. Genome-wide identification of Simple Sequence Repeat (SSR) markers in Capsicum chinense Jacq. with high potential for use in pepper introgression breeding. Biologia 2019, 74, 119–126. [Google Scholar]
- Nimmakayala, P.; Lopez-Ortiz, C.; Shahi, B.; Abburi, V.L.; Natarajan, P.; Kshetry, A.O.; Shinde, S.; Davenport, B.; Stommel, J.; Reddy, U.K. exploration into natural variation for genes associated with fruit shape and size among Capsicum chinense collections. Genomics 2021, 113, 3002–3014. [Google Scholar] [PubMed]
- Koumanov, K.S. Sample size determination in horticultural research: An empirical approach. Sci. Hortic. 2017, 225, 416–421. [Google Scholar] [CrossRef]
- Krysczun, D.K.; Lúcio, A.D.; Sari, B.; Diel, M.I.; Olivoto, T.; Santana, C.S.; Ubessi, C.; Schabarum, D.E. Sample size, plot size and number of replications for trials with Solanum melongena L. Sci. Hortic. 2018, 233, 220–224. [Google Scholar]
- de Souza, R.R.; Toebe, M.; Marchioro, V.S.; Cargnelutti Filho, A.; Lúcio, A.D.C.; Benin, G.; Mello, A.C.; de Lima Tartaglia, F.; Manfio, G.L. Soybean yield variability per plant in subtropical climate: Sample size definition and prediction models for precision statistics. Eur. J. Agron. 2022, 136, 126489. [Google Scholar]
- Coelho, A.A.; Oliveira, E.M.S.; Resende, E.D.; Thiébaut, J.T.L. Dimensionamento amostral para a caracterização da qualidade pós-colheita do maracujá-amarelo. Rev. Ceres 2011, 58, 23–28. [Google Scholar] [CrossRef]
- Stockem, J.E.; Korontzis, G.; Wilson, S.E.; de Vries, M.E.; van Eeuwijk, F.A.; Struik, P.C. Optimal plot dimensions for performance testing of hybrid potato in the field. Potato Res. 2022, 65, 417–434. [Google Scholar]
- Filgueira, F.A.R. Novo Manual de Olericultura: Agrotecnologia Moderna na Produção e Comercialização de Hortaliças; Editora UFV: Viçosa. Brazil, 2000; 402p. [Google Scholar]
- IPGRI. Descriptors for Capsicum (Capsicum spp.); International Plant Genetic Resources Institute: Rome, Italy, 1995; 49p. [Google Scholar]
- Gower, J.C. A general coefficient of similarity and some of its properties. Biometrics 1971, 27, 857. [Google Scholar]
- Kovach, W.L. A Multivariate Statistical Package for Windows, ver 3.1; Kovach Computing Services: Pentraeth, UK, 1999. [Google Scholar]
- Cruz, C.D. GENES—Software para análise de dados em estatística experimental e em genética quantitativa. Acta Sci.-Agron. 2013, 35, 271–276. [Google Scholar]
- Meier, V.D.; Lessman, K.J. Estimation of optimum field plot shape and size for testing yield in Crambe abyssinica Hochst. Crop Sci. 1971, 11, 648–650. [Google Scholar] [CrossRef]
- Moses, M.; Umaharan, P.; Dayanandan, S. Microsatellite Based Analysis of the Genetic structure and diversity of Capsicum chinense in the neotropics. Genet. Resour. Crop Evol. 2014, 61, 741–755. [Google Scholar]
- Brilhante, B.D.G.; Santos, T.d.O.; Santos, P.H.A.D.; Kamphorst, S.H.; Neto, J.D.S.; Rangel, L.H.; Valadares, F.V.; de Almeida, R.N.; Rodrigues, R.; Júnior, A.C.S.; et al. Phenotypic and molecular characterization of brazilian Capsicum germplasm. Agronomy 2021, 11, 854. [Google Scholar]
- Jarret, R.L.; Berke, T. Variation for fruit morphological characteristics in a Capsicum chinense Jacq. Germplasm Collection. HortScience 2008, 43, 1694–1697. [Google Scholar]
- Luitel, B.P.; Ro, N.Y.; Ko, H.C.; Sung, J.S.; Rhee, J.H.; Hur, O.S. Phenotypic variation in a germplasm collection of pepper (Capsicum chinense Jacq.) from Korea. J. Crop Sci. Biotechnol. 2018, 21, 499–506. [Google Scholar]
- Silva, R.; Silva, C.; Francisco, C.; Isabel, S.; Carvalho, C.; De Rodrigues, I.; Da Silva Filho, J.G.; Trevisan Braz, L.; Beckercd, F.J.; Reifschneider, B. New Brazilian lines of Habanero pepper Capsicum chinense: Morpho-Agronomic and biochemical characterization in different environments. Sci. Hortic. 2019, 261, 108941. [Google Scholar]
- Toebe, M.; Machado, L.N.; Tartaglia, F.L.; de Carvalho, J.O.; Bandeira, C.T.; Filho, C.A. Sample size for estimating mean and coefficient of variation in species of Crotalarias. An. Acad. Bras. 2018, 90, 1705–1715. [Google Scholar] [CrossRef] [PubMed]
- Schmildt, E.R.; Schmildt, O.; Cruz, C.D.; Cattaneo, L.F.; Ferreguetti, G.A. Optimum plot size and number of replications in papaya field experiment. Rev. Bras. Frutic. 2016, 38. [Google Scholar] [CrossRef][Green Version]
- Storck, L.; Lúcio, A.D.; Krause, W.; De Araújo, D.V.; Silva, C.A. Scaling the number of plants per plot and number of plots per genotype of yellow passion fruit plants. Acta Sci. Agron. 2014, 36, 73. [Google Scholar] [CrossRef][Green Version]
- Toebe, M.; Cargnelutti Filho, A.; Lopes, S.J.; Burin, C.; Silveira TR, D.; Casarotto, G. Sample size in the estimation of correlation coefficients for corn hybrids in crops and accuracy levels. Bragantia 2015, 74, 16–24. [Google Scholar] [CrossRef][Green Version]
- Toebe, M.; Cargnelutti Filho, A.; Storck, L.; Lúcio, A.D. Sample size for estimation of direct effects in path analysis of corn. Genet. Mol. Res. 2017, 16, 11018–11756. [Google Scholar] [CrossRef] [PubMed]
- da Silva, A.R.; Hilário, R.F.; do Rêgo, E.R.; do Nascimento, N.F.F.; dos Santos Dias, C.T.; de Lima, R.P. A Multivariate Approach to determine sample size for morphological characterization of pepper fruits. Aust. J. Crop Sci. 2015, 9, 1064–1068. [Google Scholar]
- Lorentz, L.H.; Lúcio, A.D. Plot size and shape for chili pepper in plastic greenhouse. Cienc. Rural 2009, 39, 2380–2387. [Google Scholar] [CrossRef]
- Lohmor, N.; Khan, M.; Kapoor, K.; Bishnoi, S. Estimation of optimum plot size and shape from a uniformity trial for field experiment with sunflower (Helianthus Annuus) crop in soil of hisar. Int. J. Plant Soil Sci. 2017, 15, 1–5. [Google Scholar] [CrossRef]

| Genotypes | Trait 1 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CM | CAC | IFC | MFC | FSh | FL | FD | FW | PL | FWTh | OFIP | NFB | FTSh | AFT | CCS | NL | FS | PUG | ARM | SS | NS | |
| BEN-III | 3 | 1 | 8 | 5 | 4 | 4 | 2 | 4 | 2 | 2 | 3 | 0 | 1 | 0 | 5 | 3 | 3 | 1 | 3 | 5 | 2 |
| BEN-IV | 2 | 1 | 7 | 4 | 4 | 4 | 2 | 4 | 2 | 2 | 4 | 0 | 2 | 0 | 7 | 3 | 3 | 2 | 2 | 5 | 2 |
| DBI-I | 3 | 1 | 8 | 8 | 4 | 4 | 2 | 4 | 2 | 2 | 4 | 0 | 1 | 0 | 7 | 3 | 3 | 1 | 2 | 5 | 2 |
| GUA-I | 3 | 1 | 9 | 5 | 1 | 4 | 2 | 4 | 2 | 2 | 4 | 0 | 1 | 0 | 5 | 3 | 4 | 1 | 1 | 5 | 2 |
| MAN-I | 3 | 0 | 4 | 7 | 5 | 3 | 3 | 4 | 2 | 2 | 3 | 0 | 1 | 0 | 5 | 3 | 2 | 1 | 2 | 5 | 2 |
| MAN-II | 3 | 0 | 8 | 5 | 6 | 4 | 3 | 4 | 2 | 2 | 3 | 0 | 2 | 1 | 5 | 4 | 4 | 1 | 2 | 5 | 3 |
| MAO-III | 2 | 1 | 8 | 5 | 4 | 4 | 2 | 5 | 2 | 3 | 3 | 0 | 3 | 0 | 5 | 3 | 2 | 1 | 3 | 5 | 3 |
| MAO-VII | 2 | 1 | 8 | 8 | 4 | 4 | 2 | 4 | 2 | 2 | 3 | 0 | 1 | 0 | 5 | 3 | 2 | 1 | 3 | 5 | 3 |
| MAO-IX | 3 | 1 | 2 | 13 | 5 | 4 | 2 | 4 | 2 | 3 | 3 | 0 | 2 | 0 | 3 | 3 | 2 | 2 | 2 | 5 | 2 |
| MPR-III | 3 | 1 | 4 | 9 | 3 | 4 | 2 | 4 | 2 | 2 | 3 | 0 | 2 | 0 | 7 | 3 | 2 | 2 | 3 | 5 | 3 |
| MPR-V | 2 | 0 | 4 | 8 | 4 | 4 | 2 | 4 | 2 | 2 | 3 | 0 | 3 | 1 | 7 | 3 | 3 | 2 | 3 | 5 | 2 |
| ORX-2 | 3 | 1 | 4 | 6 | 1 | 4 | 2 | 4 | 2 | 2 | 2 | 0 | 3 | 1 | 5 | 3 | 2 | 1 | 3 | 5 | 3 |
| ORX-I | 2 | 1 | 4 | 8 | 1 | 4 | 2 | 4 | 2 | 2 | 1 | 0 | 1 | 0 | 5 | 3 | 3 | 1 | 3 | 5 | 3 |
| ORX-II | 3 | 1 | 4 | 8 | 1 | 4 | 2 | 5 | 2 | 2 | 3 | 1 | 3 | 1 | 7 | 3 | 3 | 1 | 2 | 5 | 2 |
| ORX-IV | 3 | 1 | 4 | 8 | 1 | 4 | 2 | 4 | 2 | 2 | 2 | 0 | 1 | 0 | 5 | 2 | 3 | 1 | 2 | 5 | 2 |
| ORX-V | 3 | 1 | 7 | 5 | 4 | 4 | 2 | 3 | 1 | 2 | 3 | 0 | 3 | 1 | 3 | 3 | 1 | 1 | 2 | 5 | 2 |
| RPE-V | 3 | 1 | 9 | 3 | 4 | 4 | 2 | 4 | 2 | 2 | 3 | 0 | 3 | 1 | 7 | 3 | 3 | 2 | 2 | 5 | 2 |
| SGC-XI | 2 | 1 | 4 | 8 | 4 | 4 | 3 | 5 | 1 | 3 | 3 | 0 | 3 | 0 | 5 | 3 | 3 | 2 | 3 | 5 | 3 |
| SGC-XVIII | 2 | 1 | 4 | 8 | 4 | 4 | 2 | 4 | 2 | 2 | 1 | 0 | 3 | 0 | 7 | 3 | 2 | 2 | 1 | 5 | 3 |
| TAB-I | 3 | 1 | 9 | 5 | 4 | 4 | 2 | 4 | 3 | 2 | 3 | 0 | 3 | 0 | 7 | 3 | 2 | 1 | 2 | 5 | 2 |
| TAB-II | 2 | 1 | 9 | 3 | 3 | 4 | 2 | 4 | 1 | 2 | 1 | 0 | 1 | 0 | 5 | 3 | 2 | 1 | 2 | 5 | 2 |
| TAB-III | 3 | 1 | 8 | 5 | 4 | 4 | 2 | 4 | 2 | 2 | 3 | 0 | 3 | 1 | 7 | 3 | 2 | 1 | 3 | 5 | 2 |
| TAB-V | 2 | 1 | 8 | 5 | 4 | 4 | 2 | 4 | 1 | 2 | 3 | 0 | 3 | 0 | 5 | 3 | 2 | 1 | 3 | 5 | 2 |
| Genotypes | TFW (g) | NF (n) | FW (g) | FL (mm) | FD (mm) | DLR | FWTh (mm) | SS (mm) |
|---|---|---|---|---|---|---|---|---|
| BEN-III | 1504.7 c | 208.5 d | 7.2 b | 49.5 c | 20.1 c | 2.5 b | 1.3 c | 3.4 a |
| BEN-IV | 1850.7 c | 275.7 d | 6.7 b | 49.4 c | 20.7 c | 2.4 b | 1.5 c | 3.4 a |
| DBI-I | 1760.1 c | 242.0 d | 7.4 b | 58.9 b | 22.3 c | 2.6 b | 1.5 c | 3.5 a |
| GUA-I | 6077.4 a | 1123.0 a | 5.4 b | 57.6 b | 19.6 c | 2.9 a | 1.8 b | 3.5 a |
| MAN-I | 3893.0 b | 547.2 c | 7.2 b | 32.1 e | 32.4 a | 1.0 e | 1.8 b | 3.5 a |
| MAN-II | 3790.4 b | 530.3 c | 7.3 b | 42.3 d | 26.5 b | 1.6 d | 1.8 b | 3.5 a |
| MAO-III | 5244.0 a | 469.2 c | 11.2 a | 58.5 b | 24.1 c | 2.4 b | 2.1 a | 3.7 a |
| MAO-VII | 4351.8 b | 450.7 c | 9.6 a | 52.8 c | 23.8 c | 2.2 c | 1.8 b | 3.7 a |
| MAO-IX | 4795.3 b | 467.7 c | 10.4 a | 51.3 c | 23.0 c | 2.2 c | 2.1 a | 3.7 a |
| MPR-III | 6404.9 a | 679.6 b | 9.4 a | 56.0 b | 24.9 b | 2.2 c | 1.7 b | 3.8 a |
| MPR-V | 6717.4 a | 779.0 b | 8.6 a | 58.2 b | 22.8 c | 2.6 b | 1.8 b | 3.5 a |
| ORX-2 | 3995.9 b | 488.8 c | 8.2 a | 68.7 a | 22.5 c | 3.0 a | 1.6 c | 3.6 a |
| ORX-I | 4338.8 b | 508.0 c | 8.6 a | 68.5 a | 21.7 c | 3.2 a | 1.4 c | 3.4 a |
| ORX-II | 7029.9 a | 750.7 b | 9.3 a | 75.1 a | 23.1 c | 3.2 a | 1.7 b | 3.8 a |
| ORX-IV | 2306.9 c | 284.1 d | 8.0 b | 62.2 b | 20.6 c | 3.1 a | 1.6 c | 3.8 a |
| ORX-V | 1284.1 c | 190.0 d | 7.4 b | 49.2 c | 19.9 c | 2.5 b | 1.3 c | 3.7 a |
| RPE-V | 1933.0 c | 342.1 d | 6.3 b | 46.0 c | 21.3 c | 2.2 c | 1.4 c | 3.1 a |
| SGC-XI | 5530.6 a | 664.0 b | 8.6 a | 49.1 c | 25.5 b | 1.9 c | 2.1 a | 3.6 a |
| SGC-XVIII | 5342.3 a | 617.9 b | 8.8 a | 47.3 c | 24.8 b | 1.9 c | 1.7 b | 3.4 a |
| TAB-I | 2062.7 c | 267.8 d | 8.6 a | 62.9 b | 22.8 c | 2.8 b | 1.6 c | 3.4 a |
| TAB-II | 1734.6 c | 196.5 d | 8.3 a | 42.2 d | 21.8 c | 1.9 c | 1.4 c | 3.5 a |
| TAB-III | 1243.5 c | 182.4 d | 7.1 b | 41.8 d | 22.2 c | 1.9 c | 1.5 c | 3.6 a |
| TAB-V | 2793.6 c | 340.0 d | 7.8 b | 53.0 c | 21.9 c | 2.4 b | 1.3 c | 3.5 a |
| Mean | 3738.5 | 461.1 | 8.2 | 53.6 | 23.0 | 2.4 | 2.4 | 3.5 |
| Statistic | MPR-V Genotype | MAN-I Genotype | TAB-V Genotype | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FW | FL | FD | DLR | FW | FL | FD | DLR | FW | FL | FD | DLR | |
| Minimum | 5.81 | 48.67 | 17.27 | 1.69 | 4.23 | 21.64 | 19.73 | 0.79 | 4.63 | 36.22 | 18.62 | 1.70 |
| Mean | 8.04 | 60.92 | 23.74 | 2.61 | 6.43 | 27.72 | 31.39 | 0.88 | 6.52 | 56.15 | 21.78 | 2.59 |
| Maximum | 9.88 | 74.57 | 29.34 | 4.07 | 8.60 | 35.40 | 36.80 | 1.00 | 8.79 | 67.61 | 25.44 | 3.36 |
| SD | 0.98 | 6.44 | 2.96 | 0.44 | 1.34 | 4.29 | 3.43 | 0.07 | 0.90 | 7.30 | 1.36 | 0.39 |
| CV (%) | 12.21 | 10.57 | 12.45 | 17.05 | 20.83 | 15.46 | 10.93 | 8.30 | 13.81 | 12.99 | 6.25 | 15.13 |
| AS (1) | −0.19 ns | −0.03 ns | −0.27 ns | 0.73 ** | −0.36 ns | 0.16 ns | −1.06 ** | 0.39 ns | 0.03 ns | −0.57 ns | 0.28 ns | −0.13 ns |
| CT (2) | 2.73 ns | 2.18 ns | 2.50 ns | 4.85 ** | 2.41 ns | 2.12 ns | 5.15 ** | 1.71 ns | 2.38 ns | 2.90 ns | 3.57 ns | 3.00 ns |
| Lilliefors (3) | 0.04 ns | 0.06 ns | 0.04 ns | 0.03 ns | 0.07 ns | 0.09 ns | 0.06 ns | 0.23 ** | 0.09 ns | 0.06 ns | 0.03 ns | 0.05 ns |
| n1 | 19 | 23 | 22 | 21 | 24 | 22 | 16 | 18 | 20 | 25 | 23 | 20 |
| n2 | 24 | 18 | 25 | 48 | 71 | 39 | 20 | 11 | 31 | 28 | 6 | 37 |
| Error (%) | 3.90 | 3.38 | 3.98 | 5.45 | 6.66 | 4.95 | 3.49 | 2.66 | 4.42 | 4.16 | 2.00 | 4.84 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, S.R.M.; Lopes, R.; Meneses, C.; Valente, M.S.F.; Martins, C.C.; Ramos, S.F.; Oliveira, I.; de Jesus Pinto Fraxe, T.; Costa, L.; Lopes, M.T.G. Morpho-Agronomic Characterization, Sample Size, and Plot Size for the Evaluation of Capsicum chinense Genotypes. Horticulturae 2022, 8, 785. https://doi.org/10.3390/horticulturae8090785
Alves SRM, Lopes R, Meneses C, Valente MSF, Martins CC, Ramos SF, Oliveira I, de Jesus Pinto Fraxe T, Costa L, Lopes MTG. Morpho-Agronomic Characterization, Sample Size, and Plot Size for the Evaluation of Capsicum chinense Genotypes. Horticulturae. 2022; 8(9):785. https://doi.org/10.3390/horticulturae8090785
Chicago/Turabian StyleAlves, Silfran Rogério Marialva, Ricardo Lopes, Carlos Meneses, Magno Sávio Ferreira Valente, Cibele Chalita Martins, Santiago Ferreyra Ramos, Izamara Oliveira, Therezinha de Jesus Pinto Fraxe, Lucifrancy Costa, and Maria Teresa Gomes Lopes. 2022. "Morpho-Agronomic Characterization, Sample Size, and Plot Size for the Evaluation of Capsicum chinense Genotypes" Horticulturae 8, no. 9: 785. https://doi.org/10.3390/horticulturae8090785
APA StyleAlves, S. R. M., Lopes, R., Meneses, C., Valente, M. S. F., Martins, C. C., Ramos, S. F., Oliveira, I., de Jesus Pinto Fraxe, T., Costa, L., & Lopes, M. T. G. (2022). Morpho-Agronomic Characterization, Sample Size, and Plot Size for the Evaluation of Capsicum chinense Genotypes. Horticulturae, 8(9), 785. https://doi.org/10.3390/horticulturae8090785








