Biochemical and Proteome Analysis Reveal Different Nutritional Compound Compositions and Chloroplast Development Situations between Purple-Red and White-Yellow Tea Plant Cultivars
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Determination of AA, PP, CAF and CAT
2.3. Protein Extraction, Digestion and Identification
2.4. Proteomics Database Search and Differential Protein Identification
2.5. Statistical Analysis
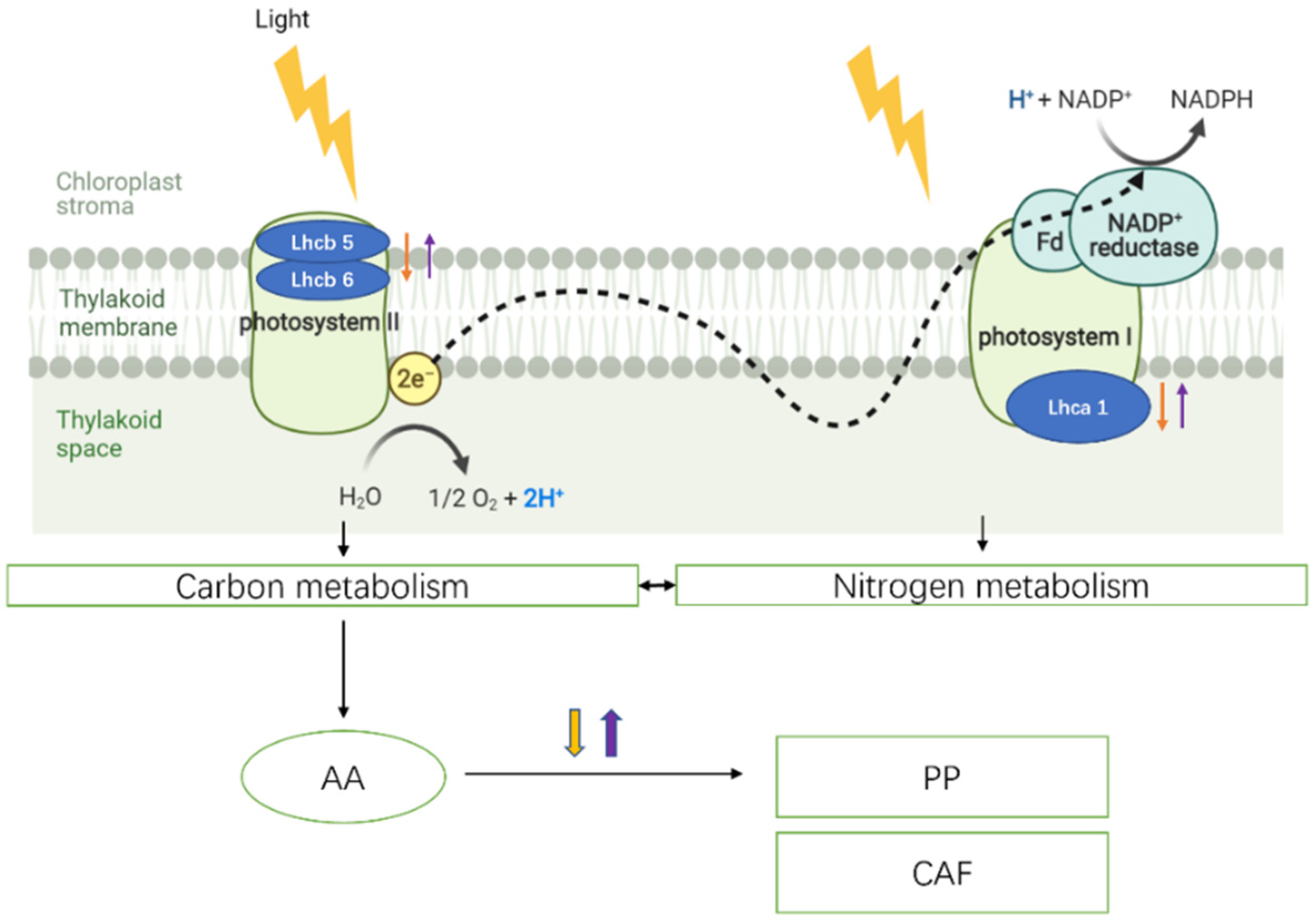
3. Results and Discussion
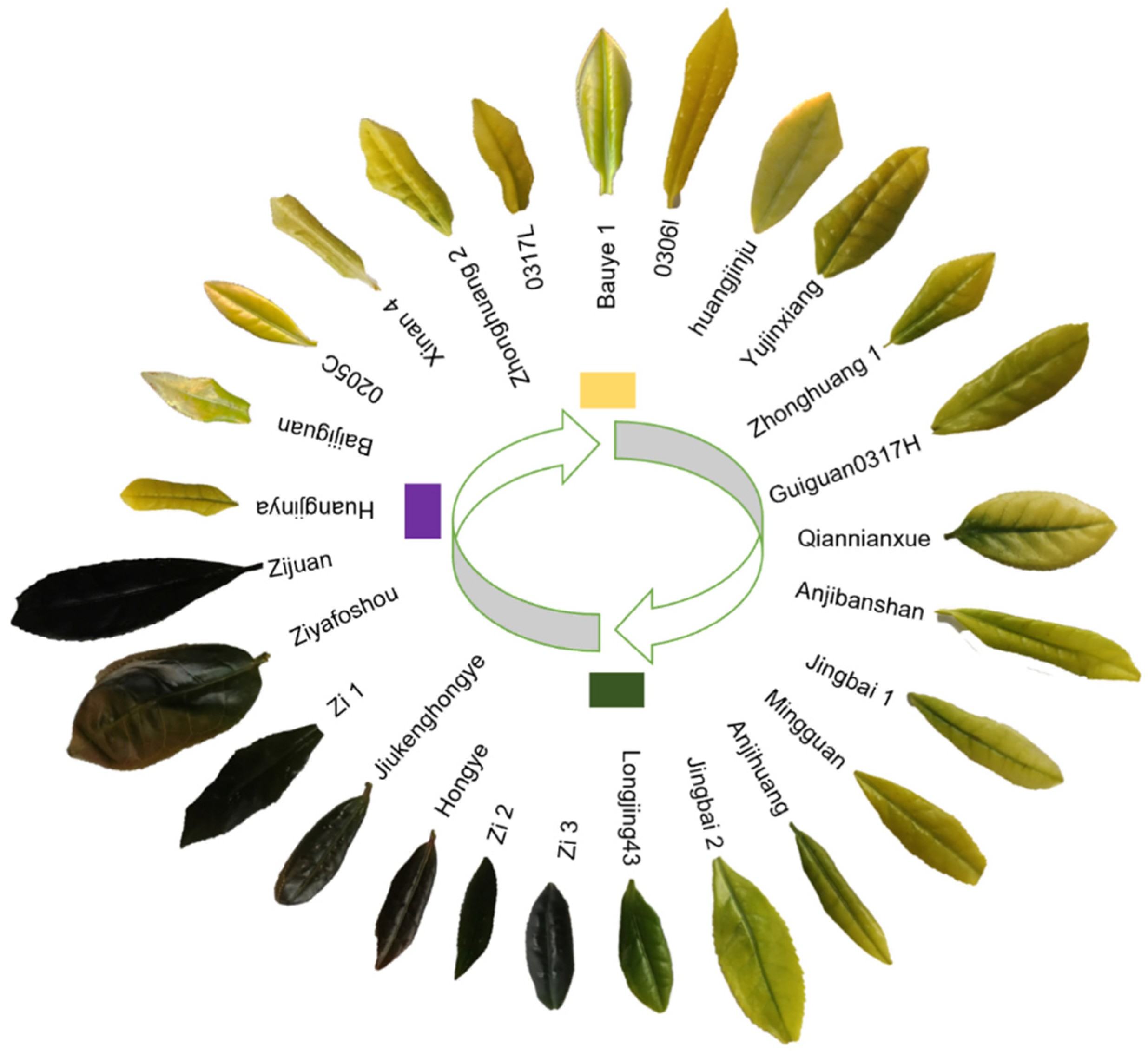
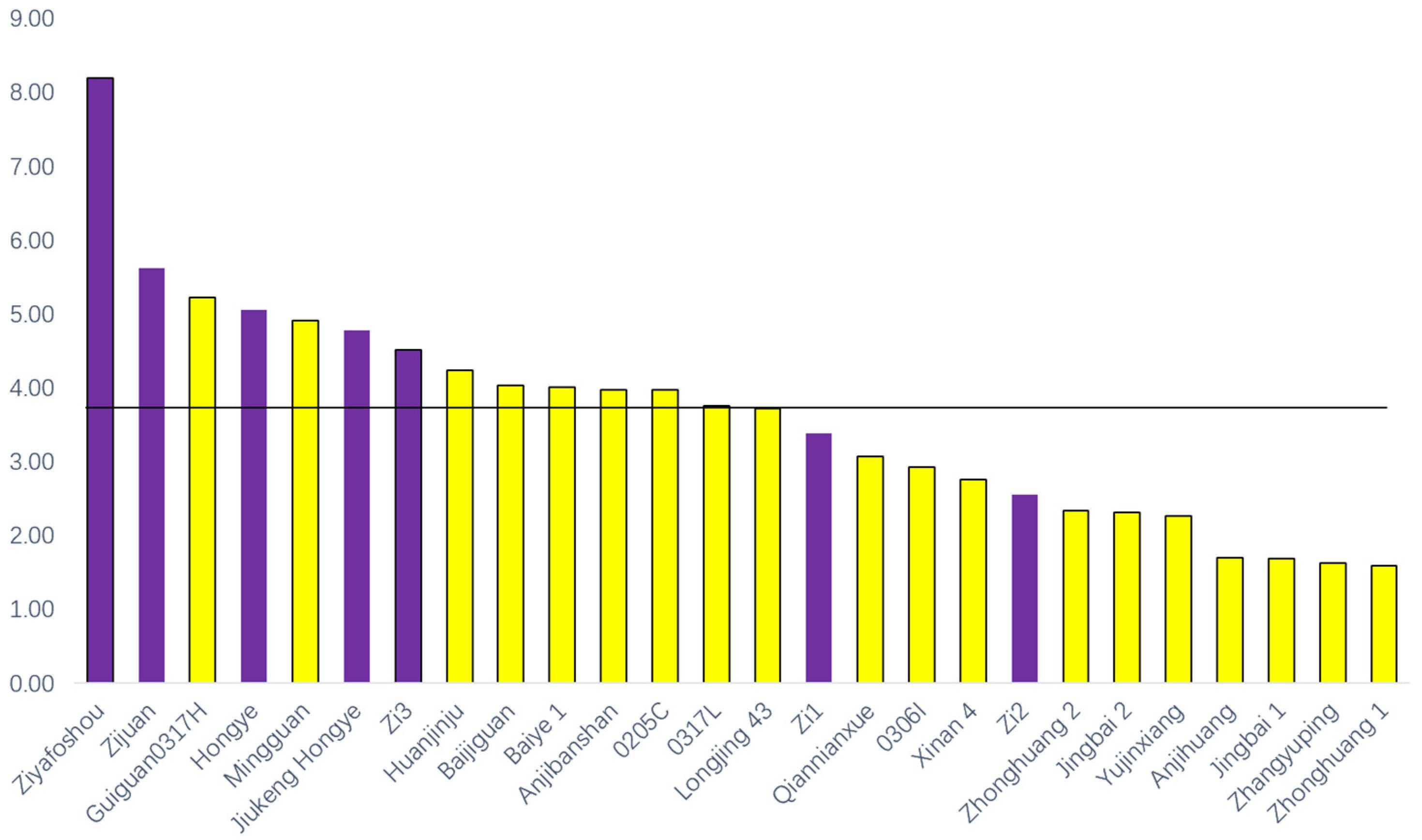
3.1. Color of Leaves in 26 Tea Cultivars
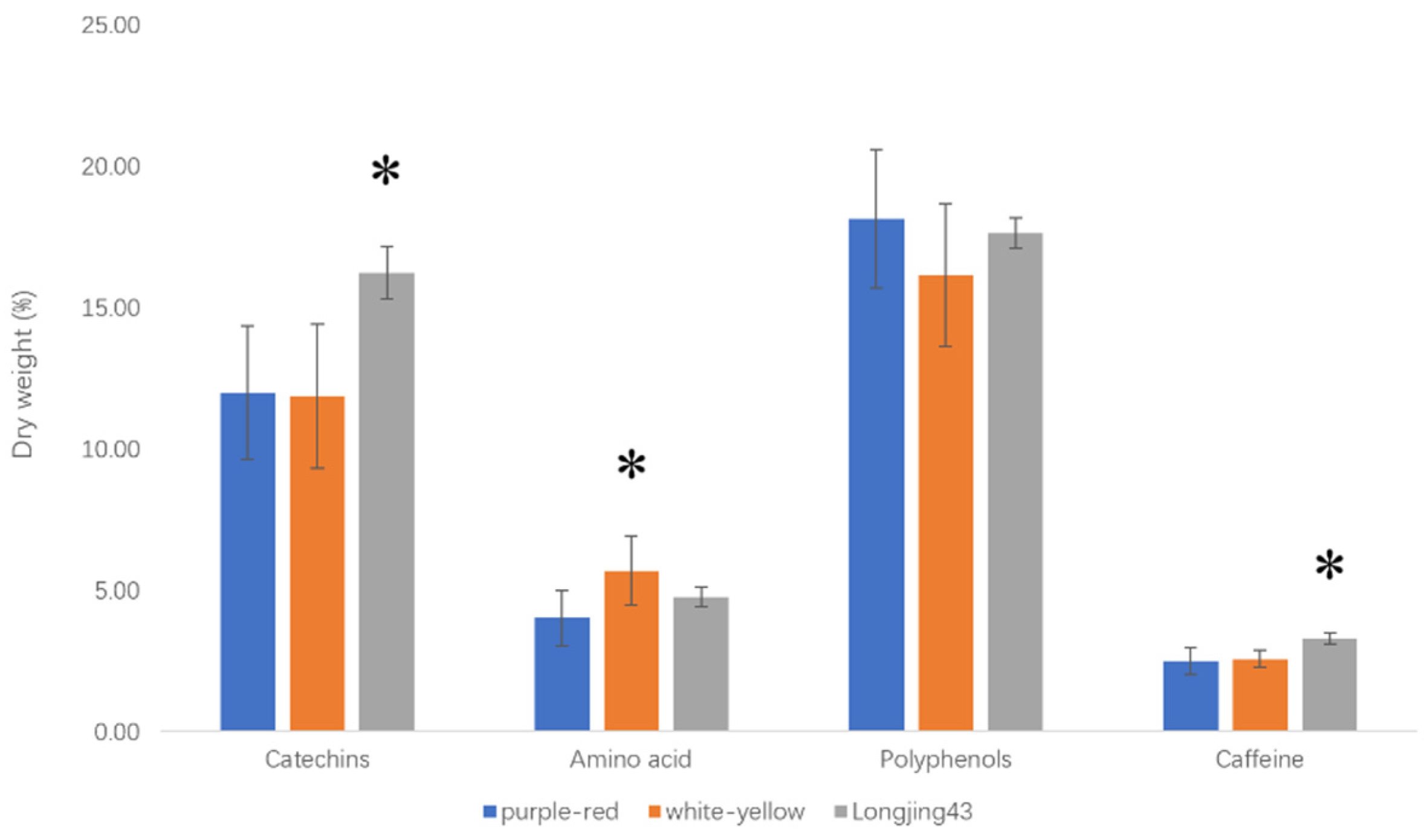
3.2. AA, PP, CAT and CAF Contents
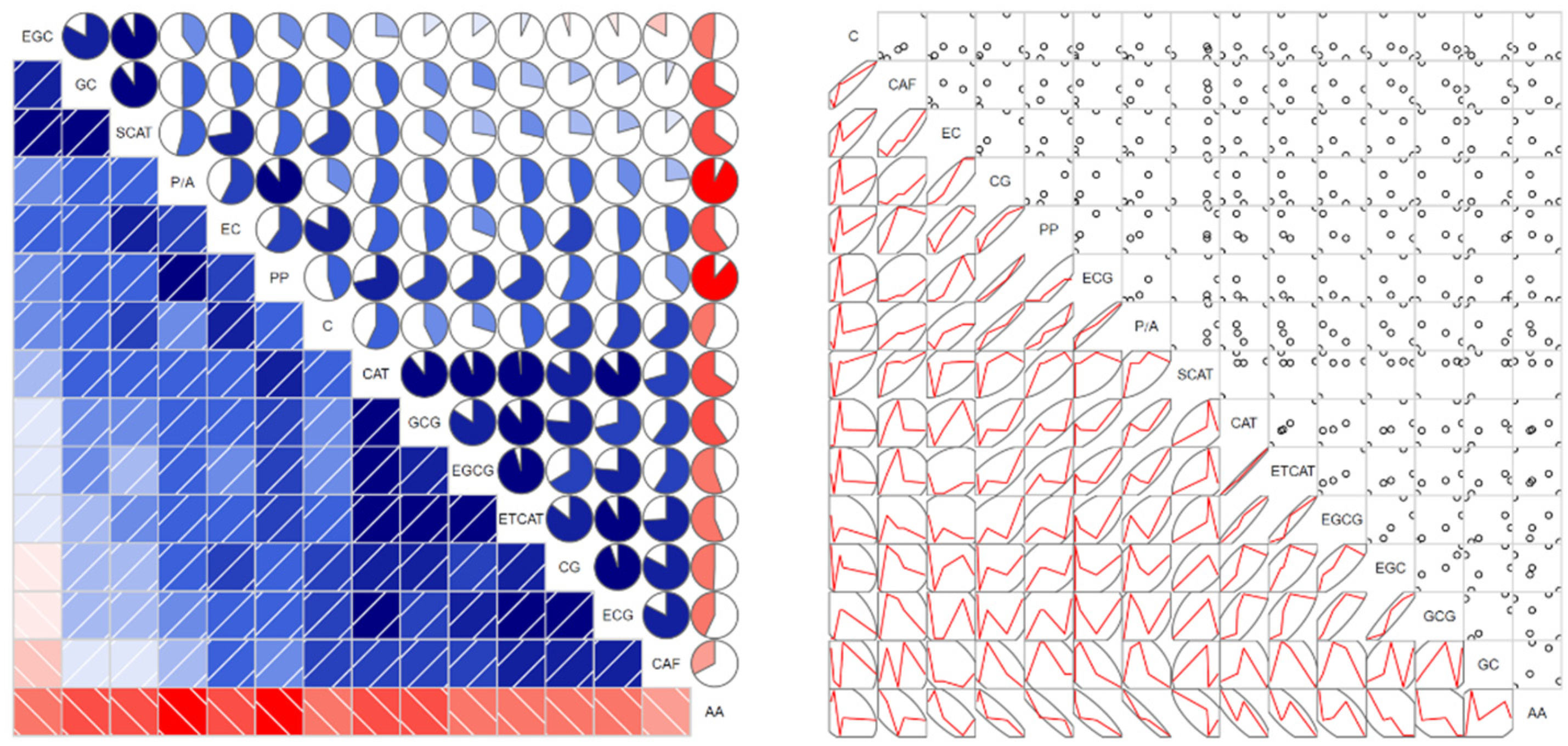
3.3. Correlation among AA, PP, CAT and CAF
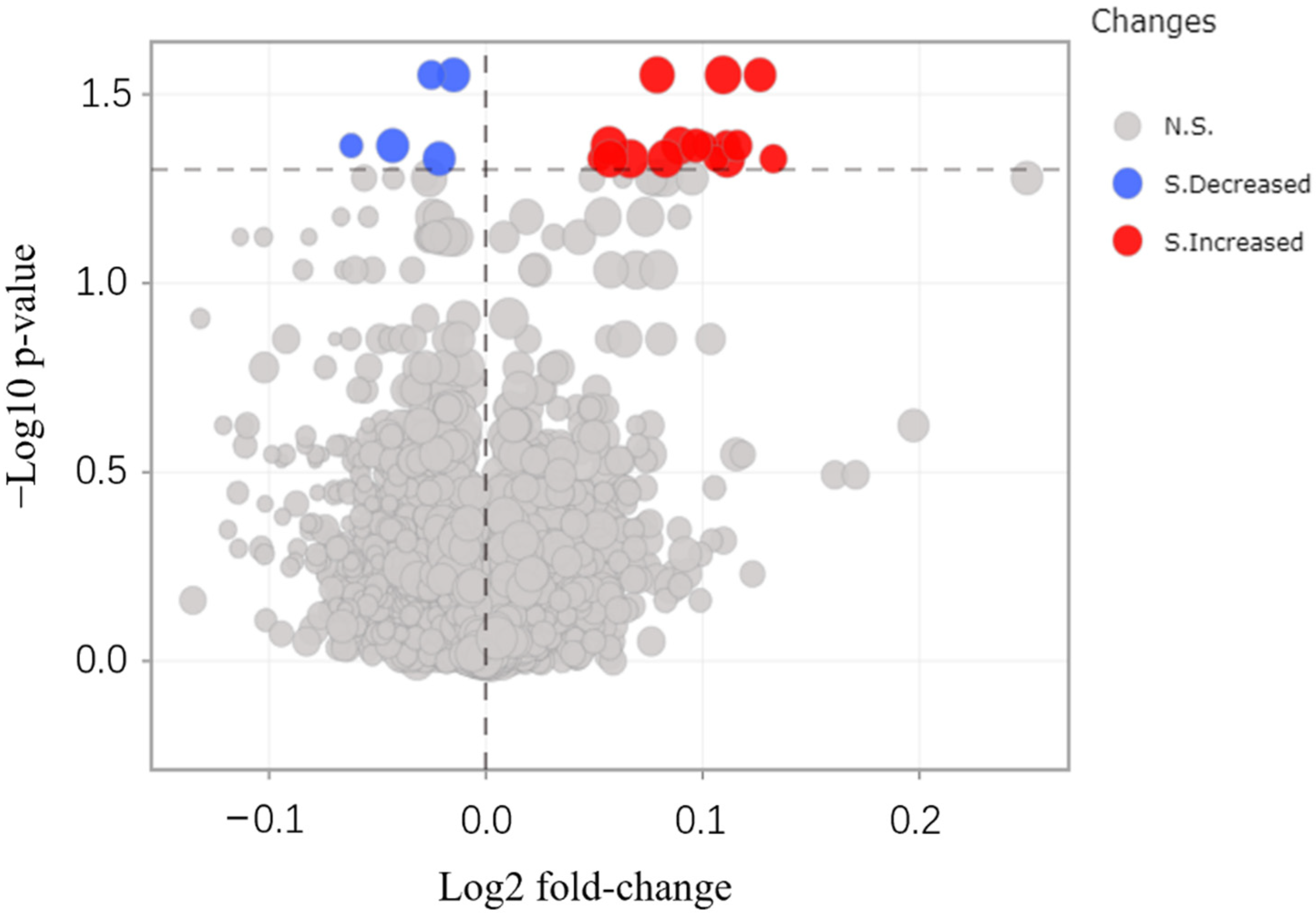
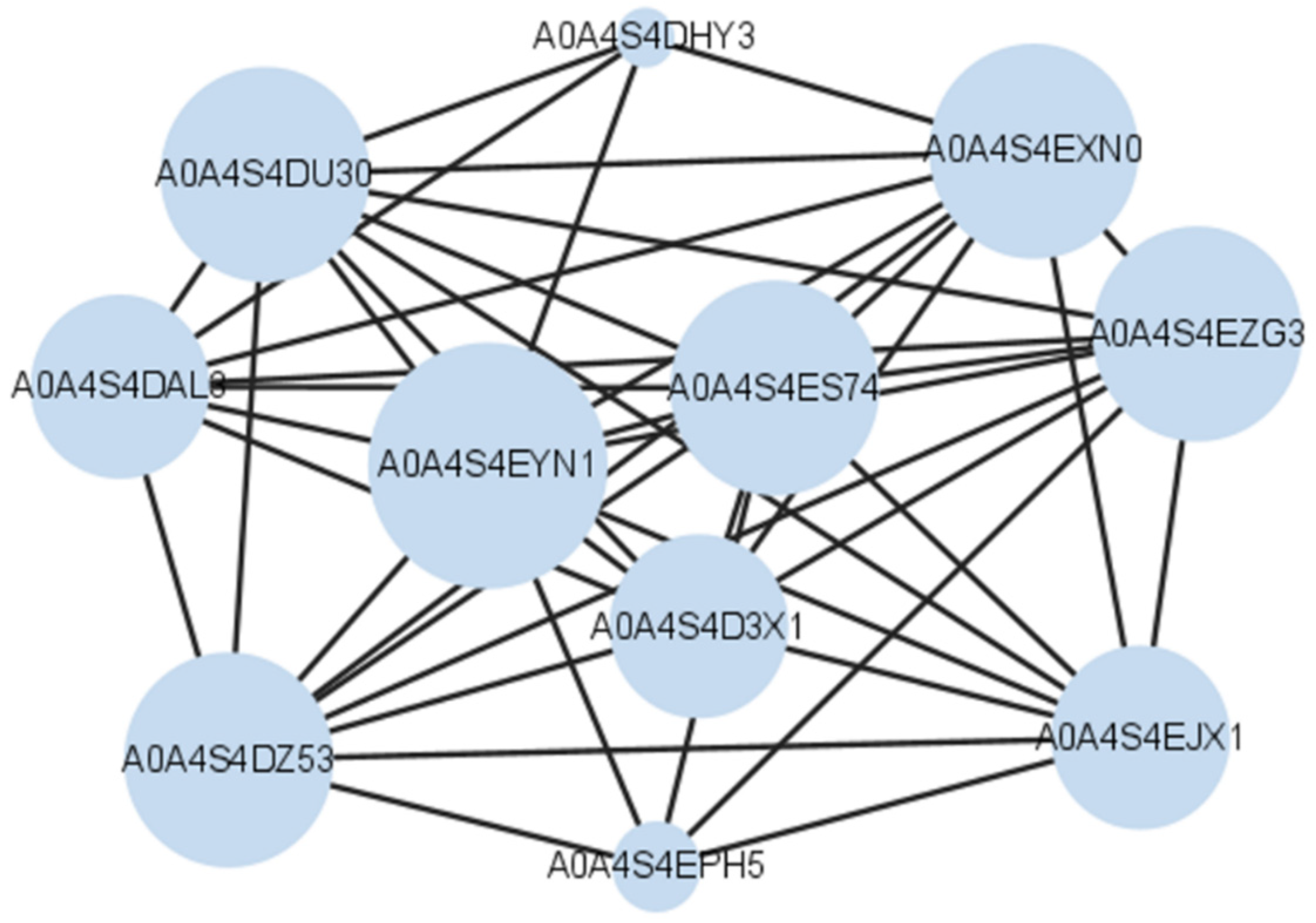
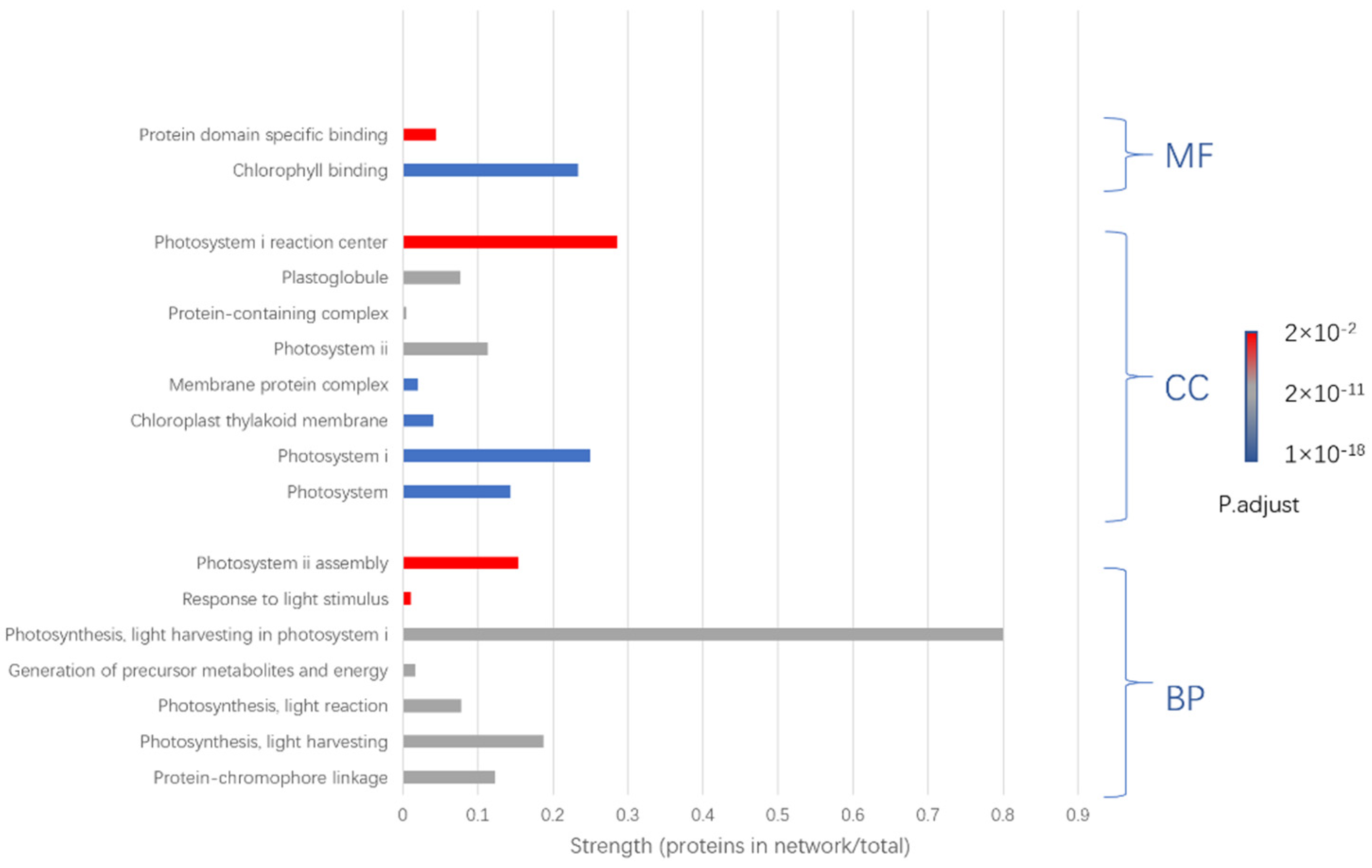
3.4. Protein Identification by MALDI-TOF/TOF-MS
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hodgson, J.M.; Croft, K. Tea flavonoids and cardiovascular health. Mol. Asp. Med. 2010, 31, 495–502. [Google Scholar] [CrossRef]
- Kao, Y.H.; Chang, H.H.; Lee, M.J.; Chen, C.L. Tea, obesity, and diabetes. Mol. Nutr. Food Res. 2006, 50, 188–210. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yue, C.; Cao, H.; Zhou, Y.; Zeng, J.; Yang, Y. Biochemical and transcriptome analyses of a novel chlorophyll-deficient chlorina tea plant cultivar. BMC Plant Biol. 2014, 14, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.F.; Xu, Y.X.; Ma, J.Q.; Jin, J.Q.; Chen, L. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘anji baicha’ (Camellia sinensis). BMC Plant Biol. 2016, 16, 195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Y.; Wang, H.; Sun, P.; Fan, Y.; Qiao, M.; Zhang, L. Response of leaf color and the expression of photoreceptor genes of camellia sinensis cv. huangjinya to different light quality conditions. Sci. Hortic. 2019, 251, 225–232. [Google Scholar] [CrossRef]
- Song, L.; Ma, Q.; Zou, Z.; Sun, K.; Yao, Y.; Tao, J.; Kaleri, N.A.; Li, X. Molecular link between leaf coloration and gene expression of flavonoid and carotenoid biosynthesis in camellia sinensis cultivar ‘huangjinya’. Front. Plant Sci. 2017, 8, 803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Liu, Y.; Cao, D.; Gong, Z.; Jin, X. Quality constituents of high amino acid content tea cultivars with various leaf colors. Turk. J. Agric. For. 2018, 42, 383–392. [Google Scholar] [CrossRef]
- Feng, L.; Gao, M.J.; Hou, R.Y.; Hu, X.Y.; Zhang, L.; Wan, X.C.; Wei, S. Determination of quality constituents in the young leaves of albino tea cultivars. Food Chem. 2014, 155, 98–104. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Yang, Y.; Ye, J.; Lu, J.; Zheng, X.; Liang, Y. Effects of sunlight on gene expression and chemical composition of light-sensitive albino tea plant. Plant Growth Regul. 2016, 78, 253–262. [Google Scholar] [CrossRef]
- Jiang, L.; Shen, X.; Shoji, T.; Kanda, T.; Zhao, L. Characterization and activity of anthocyanins in zijuan tea (Camellia sinensis var. kitamura). J. Agric. Food Chem. 2013, 61, 3306–3310. [Google Scholar] [CrossRef]
- Saito, T.; Honma, D.; Tagashira, M.; Kanda, T.; Maeda-Yamamoto, M. Anthocyanins from new red leaf tea ‘sunrouge’. J. Agric. Food Chem. 2011, 59, 4779–4782. [Google Scholar] [CrossRef]
- Lai, Y.S.; Li, S.; Tang, Q.; Li, H.X.; Chen, S.X. The Dark-Purple Tea Cultivar ‘Ziyan’ Accumulates a Large Amount of Delphinidin-Related Anthocyanins. J. Agric. Food Chem. 2016, 64, 2719–2726. [Google Scholar] [CrossRef] [PubMed]
- Kilel, E.C.; Faraj, A.K.; Wanyoko, J.K.; Wachira, F.N.; Mwingirwa, V. Green tea from purple leaf coloured tea clones in kenya—Their quality characteristics. Food Chem. 2013, 141, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Kumari, M.; Thakur, S.; Kumar, A.; Joshi, R.; Kumar, R. Regulation of color transition in purple tea (Camellia sinensis). Planta 2019, 251, 35. [Google Scholar] [CrossRef]
- Shen, J.; Zou, Z.; Zhang, X.; Zhou, L.; Wang, Y.; Fang, W.; Zhu, X. Metabolic analyses reveal different mechanisms of leaf color change in two purple-leaf tea plant (Camellia sinensis L.) cultivars. Hortic. Res. 2018, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Kerio, L.C.; Wachira, F.N.; Wanyoko, J.K.; Rotich, M.K. Total polyphenols, catechin profiles and antioxidant activity of tea products from purple leaf coloured tea cultivars. Food Chem. 2013, 136, 1405–1413. [Google Scholar] [CrossRef]
- Hsu, C.P.; Shih, Y.T.; Lin, B.R.; Chiu, C.F.; Lin, C.C. Inhibitory effect and mechanisms of an anthocyanins- and anthocyanidins-rich extract from purple-shoot tea on colorectal carcinoma cell proliferation. J. Agric. Food Chem. 2012, 60, 3686–3692. [Google Scholar] [CrossRef]
- Li, Y.; Yuan, P.; Wang, C.; Wu, Z. The research progress of main chemical constituents and functional activity in purple tea. Int. J. Nutr. Food Sci. 2021, 10, 24. [Google Scholar] [CrossRef]
- Joshi, R.; Rana, A.; Gulati, A.A. Studies on quality of orthodox teas made from anthocyanin-rich tea clones growing in Kangra Valley, India. Food Chem. 2015, 176, 357–366. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Shen, Y.; Ling, T.; Ho, C.T.; Xie, Z. Analysis of differentiated chemical components between zijuan purple tea and yunkang green tea by uhplc–orbitrap–ms/ms combined with chemometrics. Foods 2021, 10, 1070. [Google Scholar] [CrossRef]
- Peng, G.; Xie, X.L.; Jiang, Q.; Song, S.; Xu, C.J. Chlorophyll a/b binding protein plays a key role in natural and ethylene-induced degreening of ponkan (Citrus reticulata blanco). Sci. Hortic. 2013, 160, 37–43. [Google Scholar] [CrossRef]
- Green, B.R.; Pichersky, E.; Kloppstech, K. Chlorophyll a/b-binding proteins: An extended family. Trends Biochem. Sci. 1991, 16, 181. [Google Scholar] [CrossRef] [Green Version]
- Wei, X.; Su, X.; Cao, P.; Liu, X.; Chang, W.; Li, M. Structure of spinach photosystem II–LHCII supercomplex at 3.2 resolution. Nature 2016, 534, 69. [Google Scholar] [CrossRef] [PubMed]
- Li, X.W.; Zhu, Y.L.; Chen, C.Y.; Geng, Z.J.; Du, F. Cloning and characterization of two chlorophyll a/b binding protein genes and analysis of their gene family in Camellia sinensis. Sci. Rep. 2020, 10, 4602. [Google Scholar] [CrossRef] [Green Version]
- Amarie, S.; Wilk, L.; Barros, T.; Kühlbrandt, W.; Dreuw, A.; Wachtveitl, J. Properties of zeaxanthin and its radical cation bound to the minor light-harvesting complexes cp24, cp26 and cp29. Biochim. Biophys. Acta—Bioenerg. 2009, 1787, 747–752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, N.; Zhang, C.; Zhou, P.; Gao, X.; Wang, M.; Tian, S.; Lu, C.; Wang, K.; Shen, C. Transcriptomic analyses reveal variegation-induced metabolic changes leading to high L-theanine levels in albino sectors of variegated tea (Camellia sinensis). Plant Physiol. Biochem. 2021, 169, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Chen, Z.; Lee, J.; Li, X.; Sun, W. Proteomic analysis of tea plants (Camellia sinensis) with purple young shoots during leaf development. PLoS ONE 2017, 12, e0177816. [Google Scholar]
- Ye, Y.; Yan, J.; Cui, J.; Mao, S.; Li, M.; Liao, X.; Tong, H. Dynamic changes in amino acids, catechins, caffeine and gallic acid in green tea during withering. J. Food Compos. Anal. 2018, 66, 98–108. [Google Scholar] [CrossRef]
- Yu, C.; Hao, D.; Chu, Q.; Wang, T.; Liu, S.; Lan, T.; Pan, C. A one adsorbent QuEChERS method coupled with LC-MS/MS for simultaneous determination of 10 organophosphorus pesticide residues in tea. Food Chem. 2020, 321, 126657. [Google Scholar] [CrossRef]
- Zuo, Y.; Chen, H.; Deng, Y. Simultaneous determination of catechins, caffeine and gallic acids in green, Oolong, black and pu-erh teas using HPLC with a photodiode array detector. Talanta 2002, 57, 307–316. [Google Scholar] [CrossRef]
- Simpson, D.M.; Beynon, R.J. Acetone precipitation of proteins and the modification of peptides. J. Proteome Res. 2010, 9, 444–450. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, D.; Ma, L.; Jin, X.; Yang, P.; Ye, F.; Wei, C. TMT-based quantitative proteomics analysis reveals the response of tea plant (Camellia sinensis) to fluoride. J. Proteom. 2018, 176, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chaturvedi, P.; Fu, J.; Cai, Q.; Weckwerth, W.; Yang, P. Induction and quantitative proteomic analysis of cell dedifferentiation during callus formation of lotus (Nelumbo nucifera Gaertn. spp. baijianlian). J. Proteom. 2016, 131, 61–70. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Deker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, M.; Jianyun, R. Integrated Transcriptome and metabolic analyses reveals novel insights into free amino acid metabolism in Huangjinya tea cultivar. Front. Plant Sci. 2017, 8, 891. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Tang, D.; Liu, M.; Ruan, J. Integrated analyses of the transcriptome and metabolome of the leaves of albino tea cultivars reveal coordinated regulation of the carbon and nitrogen metabolism. Sci. Hortic. 2017, 231, 272–281. [Google Scholar] [CrossRef]
- Wei, K.; Wang, L.Y.; Zhou, J.; He, W.; Zeng, J.M.; Jiang, Y.W.; Cheng, H. Comparison of catechins and purine alkaloids in albino and normal green tea cultivars (Camellia sinensis L.) by HPLC. Food Chem. 2012, 130, 720–724. [Google Scholar] [CrossRef]
- Du, Y.Y.; Liang, Y.R.; Wang, H.; Wang, K.R.; Lu, J.L.; Zhang, G.H.; Fang, Q.Y. A study on the chemical composition of albino tea cultivars. J. Hortic. Sci. Biotechnol. 2006, 81, 809–812. [Google Scholar] [CrossRef]
- Qin, X.; Suga, M.; Kuang, T.; Shen, J.R. Structural basis for energy transfer pathways in the plant PSI-LHCI supercomplex. Science 2015, 348, 989–995. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Toporik, H.; Borovikova, A.; Nelson, N. Structure determination and improved model of plant photosystem I. J. Biol. Chem. 2010, 285, 3478–3486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazor, Y.; Borovikova, A.; Caspy, I.; Nelson, N. Structure of the plant photosystem I supercomplex at 2.6 Å resolution. Nat. Plants 2017, 3, 17014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, X.; Ma, J.; Wei, X.; Cao, P.; Zhu, D.; Chang, W.; Li, M. Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 2017, 357, 815–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, X.; Ma, J.; Su, X.; Cao, P.; Chang, W.; Liu, Z.; Li, M. Structure of the maize photosystem I supercomplex with light-harvesting complexes I and II. Science 2018, 360, 1109–1113. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Li, J.; Yu, Y.; Kou, X.; Periakaruppan, R.; Chen, X.; Li, X. STAY-GREEN and light-harvesting complex II chlorophyll a/b binding protein are involved in albinism of a novel albino tea germplasm ‘Huabai 1’. Sci. Hortic. 2022, 293, 110653. [Google Scholar] [CrossRef]
- Zhang, Q.; Ma, C.; Wang, X.; Ma, Q.; Fan, S.; Zhang, C. Genome-wide identification of the light-harvesting chlorophyll a/b binding (Lhc) family in Gossypium hirsutum reveals the influence of GhLhcb 2.3 on chlorophyll a synthesis. Plant Biol. 2021, 23, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Cao, H.; Chen, C.; Yue, C.; Hao, X.; Yang, Y.; Wang, X. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. J. Proteom. 2016, 130, 160–169. [Google Scholar] [CrossRef] [PubMed]
| Purple-Red | White-Yellow | Green (Longjing43) | |
|---|---|---|---|
| GC | 0.90 ± 0.15 Aa | 0.70 ± 0.25 Ab | 0.80 ± 0.10 Aab |
| EGC | 1.03 ± 0.21 Aa | 0.99 ± 0.43 Aa | 0.89 ± 0.19 Aa |
| C | 0.25 ± 0.11 ABb | 0.25 ± 0.78 Bb | 0.32 ± 0.05 Aa |
| EGCG | 4.92 ± 1.47 Bb | 5.36 ± 1.35 Bb | 6.89 ± 0.34 Aa |
| EC | 0.61 ± 0.24 AaB | 0.55 ± 0.14 aB | 0.65 ± 0.11 Aa |
| GCG | 2.45 ± 0.43 Bb | 2.41 ± 0.56 Bb | 3.01 ± 0.23 Aa |
| ECG | 1.45 ± 0.76 Bb | 1.36 ± 0.41 Bb | 3.05 ± 0.17 Aa |
| CG | 0.38 ± 0.19 Bb | 0.32 ± 0.11 Bb | 0.63 ± 0.06 Aa |
| Simple catechins | 2.79 ± 0.44 Aa | 2.50 ± 0.83 Aa | 2.66 ± 0.44 Aa |
| Ester type catechins | 9.20 ± 2.24 Bb | 9.46 ± 2.30 Bb | 13.58 ± 0.67 Aa |
| No. | Change | Protein Name | Gene Name |
|---|---|---|---|
| 1 | Increased | Photosystem I iron-sulfur center (PSI-C) | psaC |
| 2 | Increased | Germin-like protein (A0A4S4EA81) | TEA_002437 |
| 3 | Increased | Chlorophyll a-b binding protein (A0A4S4EJX1) | TEA_020597 |
| 4 | Increased | Chlorophyll a-b binding protein (A0A4S4D3X1) | TEA_001699 |
| 5 | Increased | Chlorophyll a-b binding protein (A0A4S4DAL3) | TEA_001579 |
| 6 | Increased | Rhodanese domain-containing protein (A0A4S4EPH5) | TEA_012572 |
| 7 | Increased | Photosystem I P700 chlorophyll a apoprotein A1 (A0A4S4DHY3) | TEA_001227 |
| 8 | Increased | A0A4S4DU30 | TEA_014123 |
| 9 | Increased | Chlorophyll a-b binding protein (A0A4S4DZ53) | TEA_027100 |
| 10 | Increased | A0A4S4E5H4 | TEA_005661 |
| 11 | Increased | A0A4S4EM82 | TEA_030166 |
| 12 | Increased | A0A4S4ES74 | TEA_019755 |
| 13 | Increased | Chlorophyll a-b binding protein (A0A4S4EXN0) | TEA_015110 |
| 14 | Increased | Photosystem I subunit V (A0A4S4EYN1) | TEA_018047 |
| 15 | Increased | Chlorophyll a-b binding protein (A0A4S4EZG3) | TEA_025056 |
| 16 | Increased | Oxygen-evolving enhancer protein | OEP1 |
| 17 | Decreased | Nop domain-containing protein (A0A4S4D679) | TEA_025749 |
| 18 | Decreased | Proteasome subunit beta (A0A4S4DXI4) | TEA_014823 |
| 19 | Decreased | Derlin (A0A4S4E641) | TEA_007225 |
| 20 | Decreased | Peptidylprolyl isomerase (A0A4S4F203) | TEA_026953 |
| 21 | Decreased | Ribosomal_L14e domain-containing protein (A0A4S4F275) | TEA_019930 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Wang, X.; Cui, H.; Zhao, Y. Biochemical and Proteome Analysis Reveal Different Nutritional Compound Compositions and Chloroplast Development Situations between Purple-Red and White-Yellow Tea Plant Cultivars. Horticulturae 2022, 8, 685. https://doi.org/10.3390/horticulturae8080685
Ding Y, Wang X, Cui H, Zhao Y. Biochemical and Proteome Analysis Reveal Different Nutritional Compound Compositions and Chloroplast Development Situations between Purple-Red and White-Yellow Tea Plant Cultivars. Horticulturae. 2022; 8(8):685. https://doi.org/10.3390/horticulturae8080685
Chicago/Turabian StyleDing, Yi, Xinchao Wang, Hongchun Cui, and Yun Zhao. 2022. "Biochemical and Proteome Analysis Reveal Different Nutritional Compound Compositions and Chloroplast Development Situations between Purple-Red and White-Yellow Tea Plant Cultivars" Horticulturae 8, no. 8: 685. https://doi.org/10.3390/horticulturae8080685
APA StyleDing, Y., Wang, X., Cui, H., & Zhao, Y. (2022). Biochemical and Proteome Analysis Reveal Different Nutritional Compound Compositions and Chloroplast Development Situations between Purple-Red and White-Yellow Tea Plant Cultivars. Horticulturae, 8(8), 685. https://doi.org/10.3390/horticulturae8080685







