Metabolome and Transcriptome Analyses Unravels Molecular Mechanisms of Leaf Color Variation by Anthocyanidin Biosynthesis in Acer triflorum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Metabolite Extraction and Profiling
2.3. Qualitative and Quantitative Analysis of Metabolites
2.4. RNA Extraction, Sequencing, and Transcriptome Data Analysis
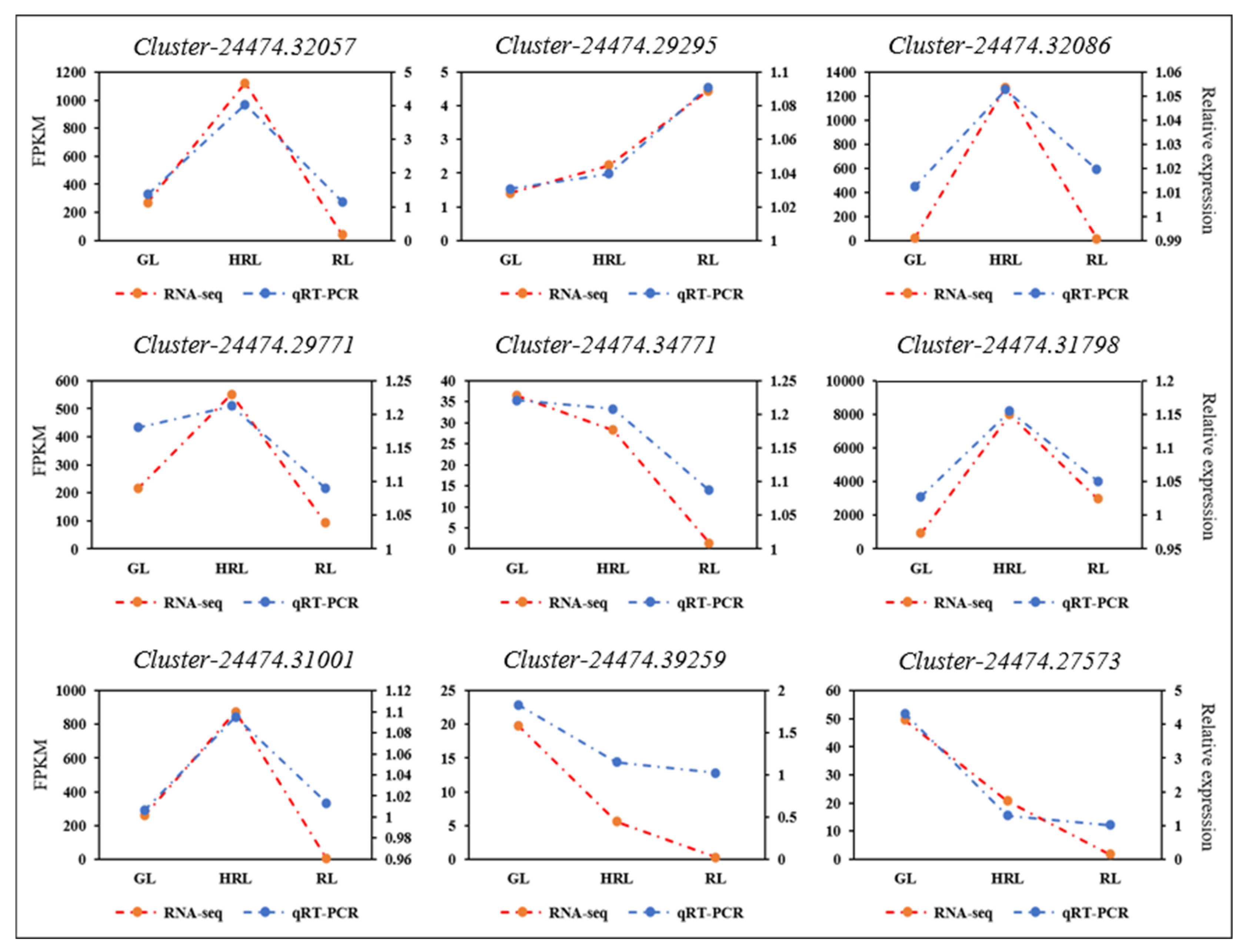
2.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Validation
2.6. Interaction Network Construction
3. Results
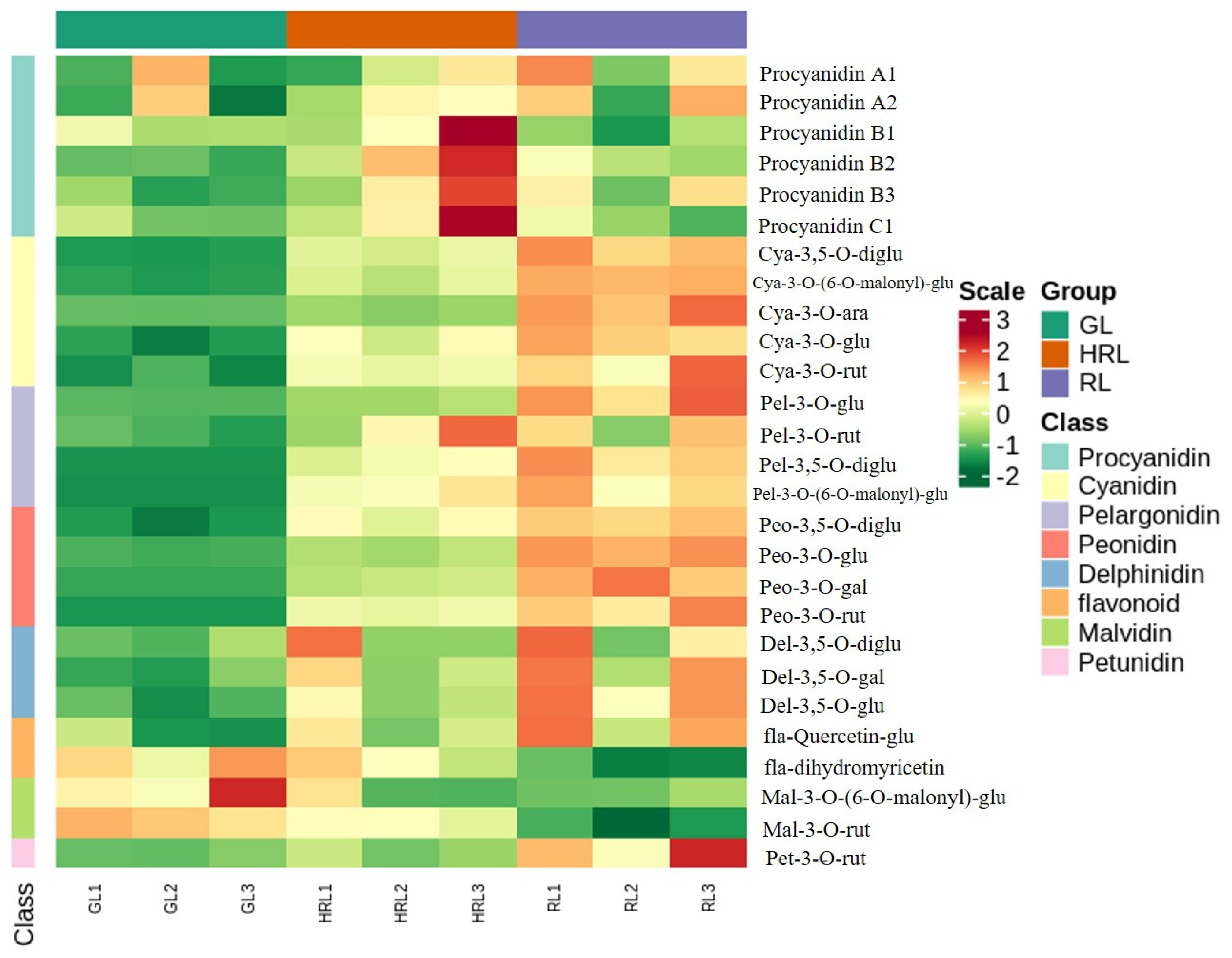
3.1. Metabolites in A. triflorum Leaves
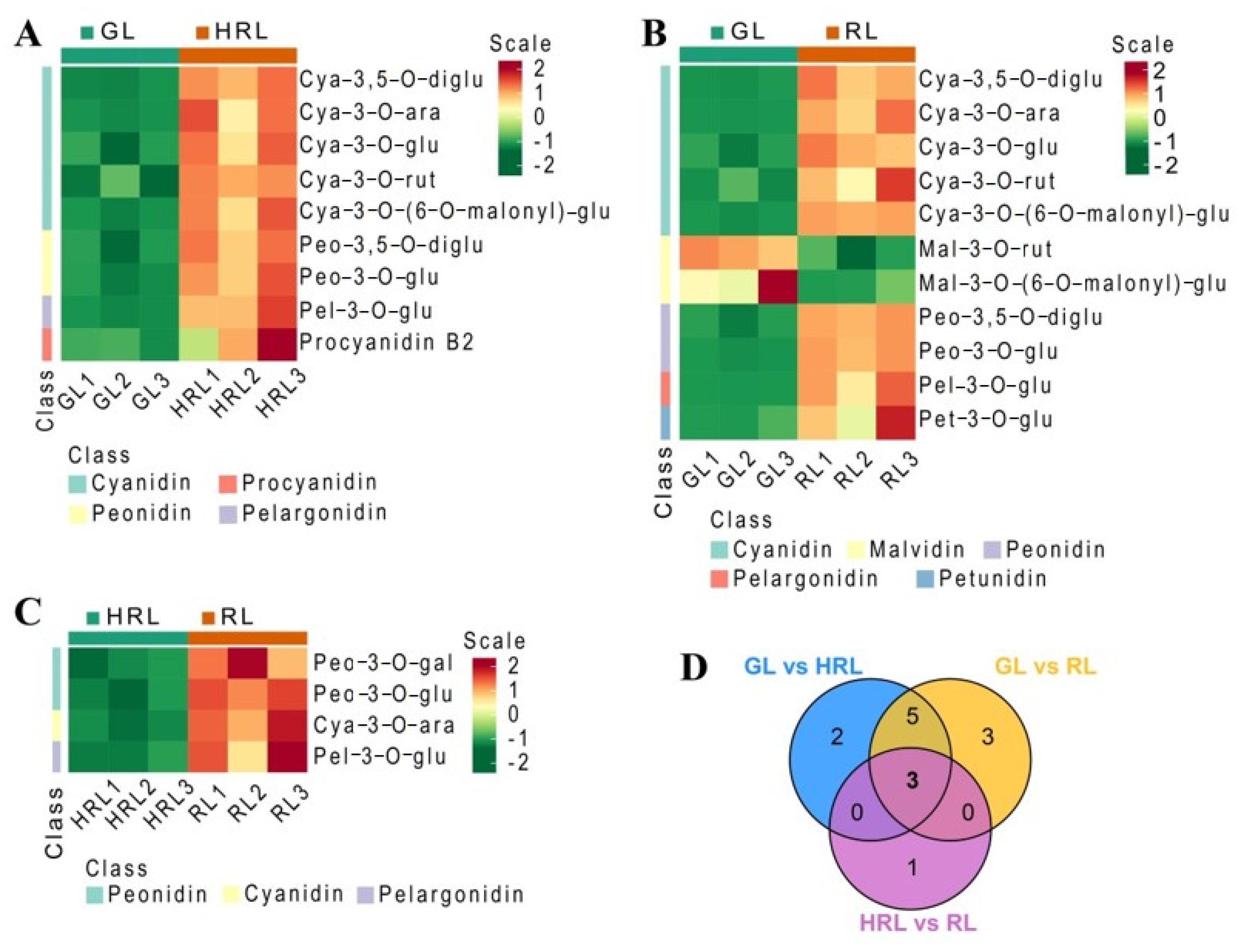
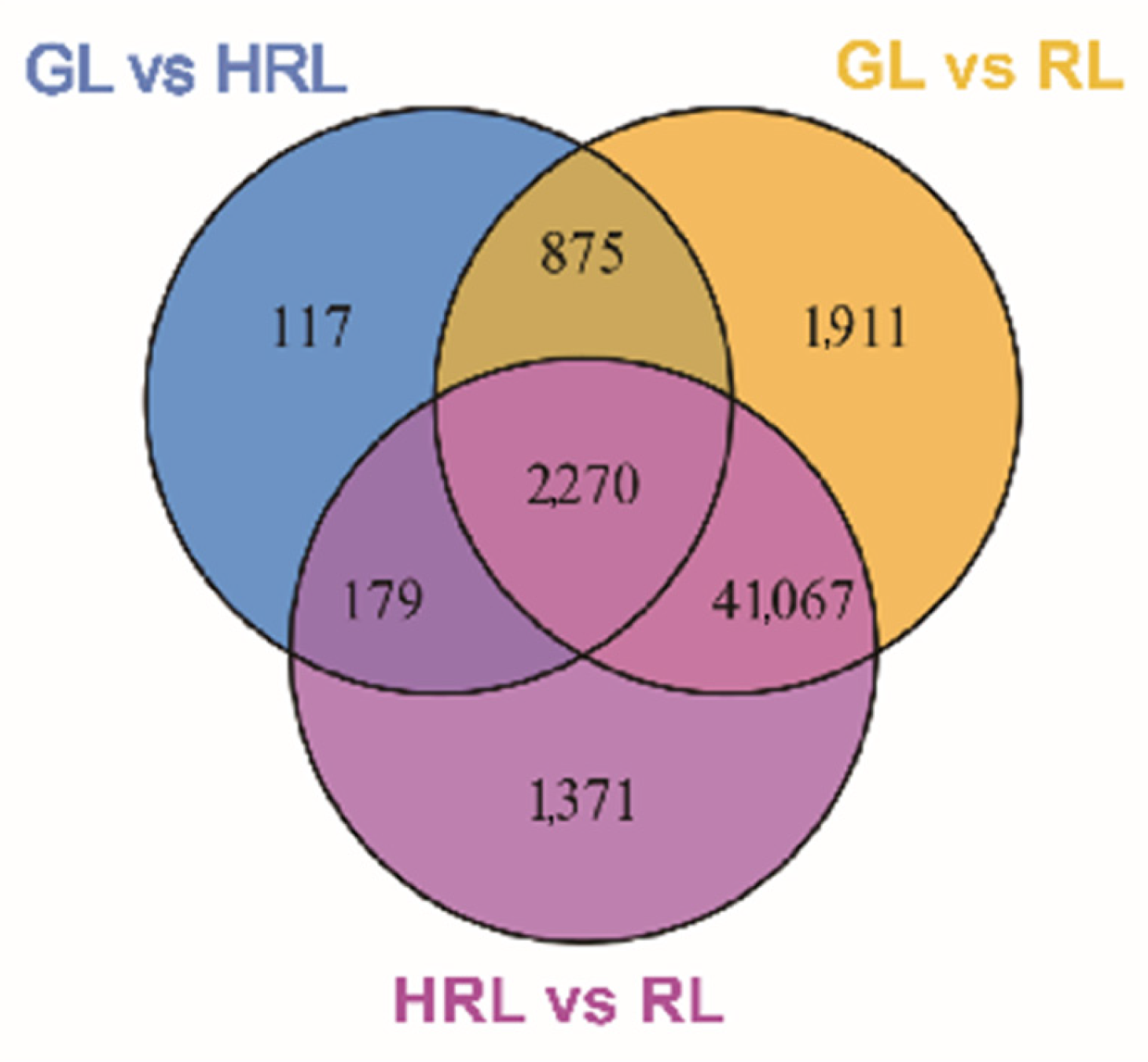
3.2. Differentially Accumulated Metabolites
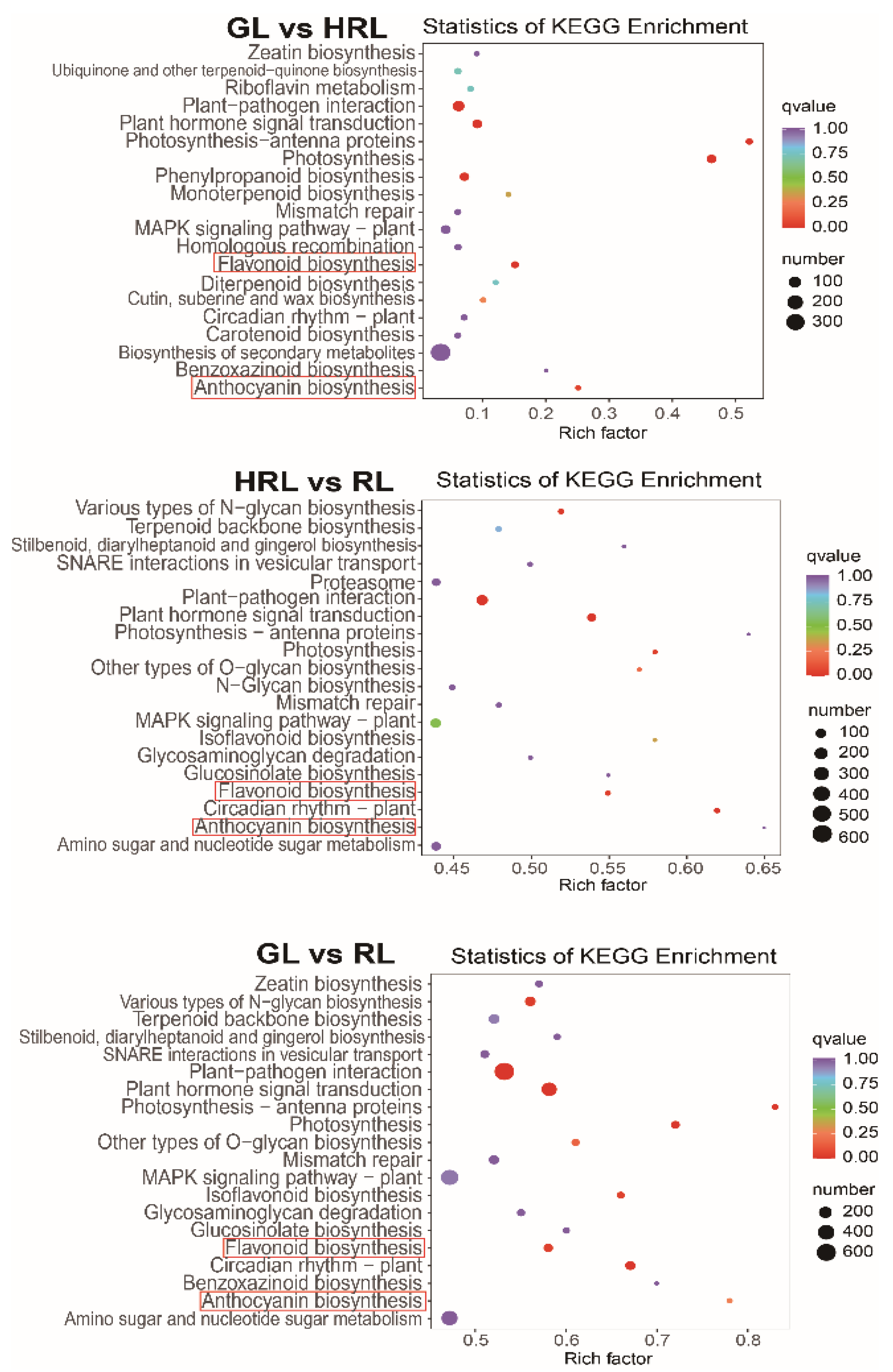
3.3. Transcriptomic Analysis of A. triflorum Leaves with Different Colors
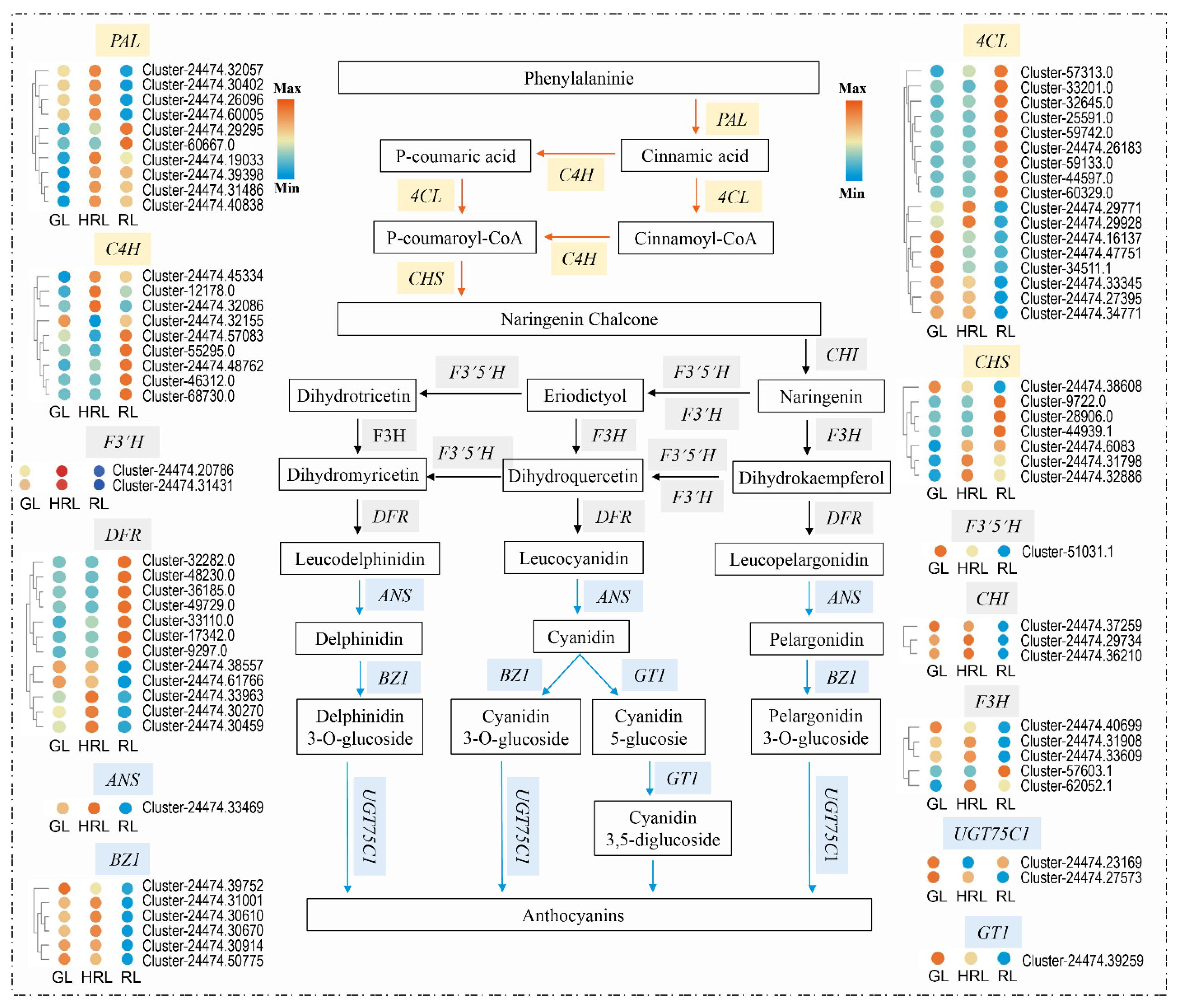
3.4. Differentially Expressed Genes in the Anthocyanin Biosynthesis Pathway
3.5. Candidate Genes Related with Anthocyanin Accumulation
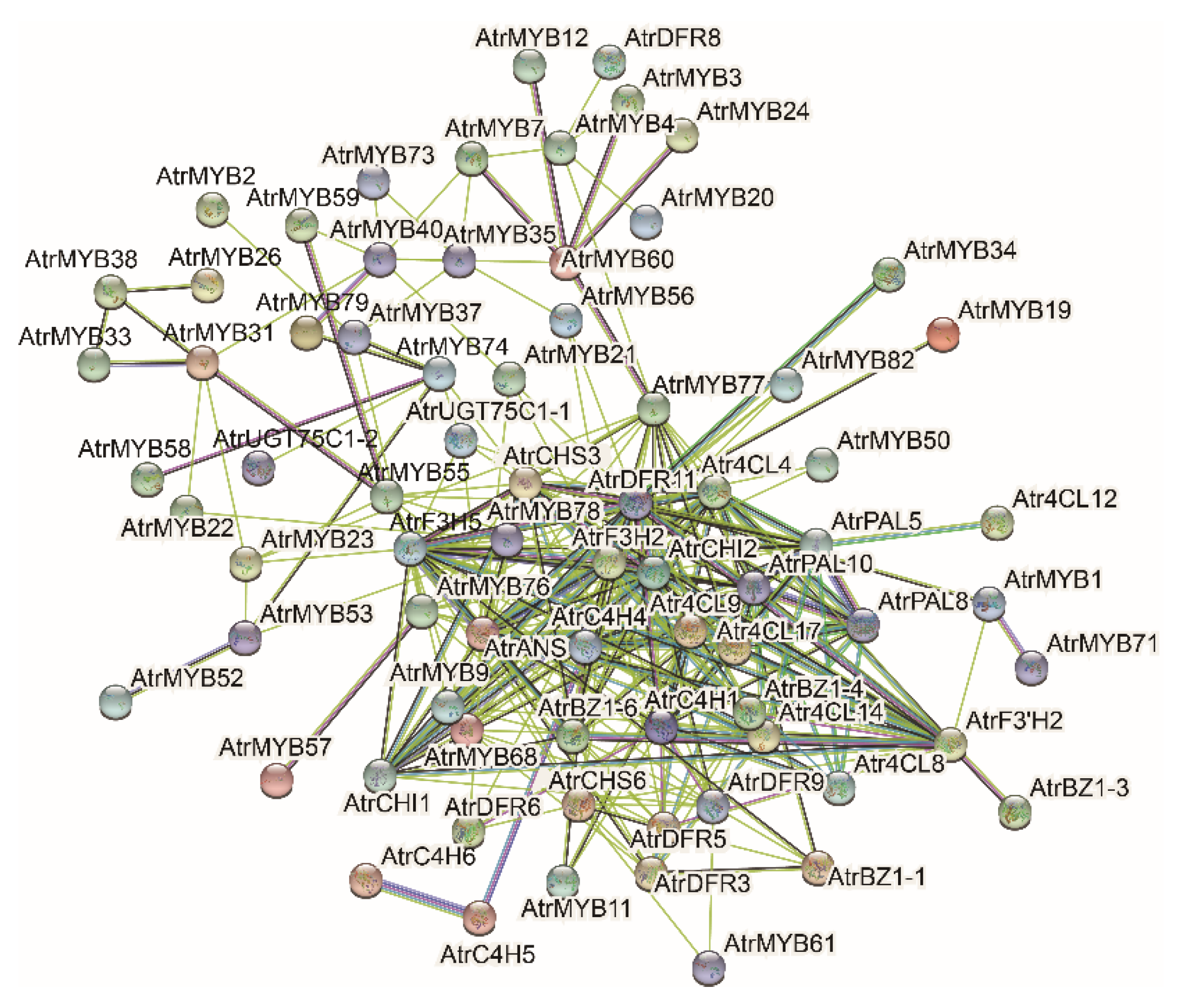
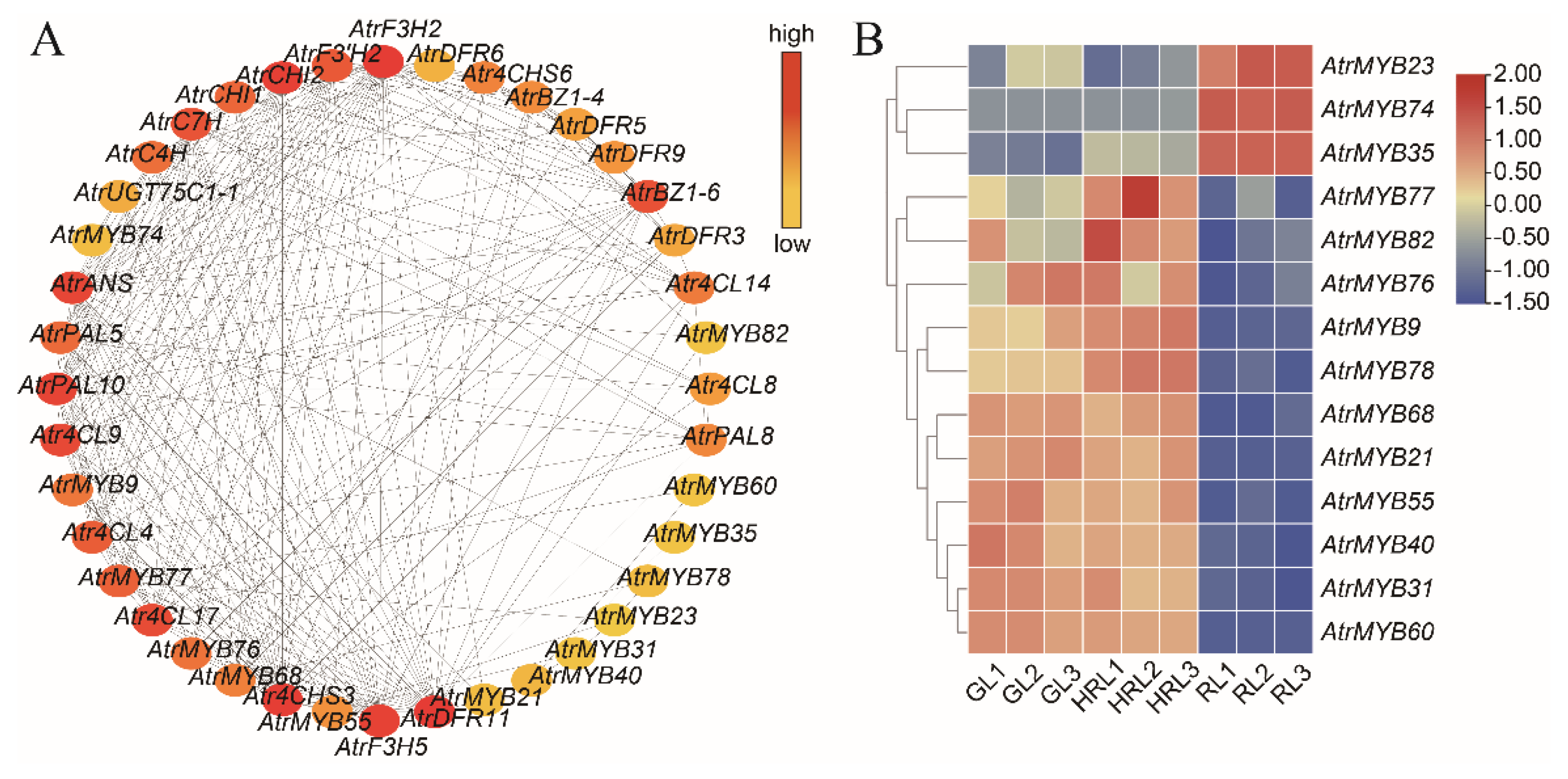
3.6. Analysis of Transcription Factors
4. Discussion
4.1. Anthocyanin Contents of Different Colored A. triflorum Leaves
4.2. Key Genes Related to Anthocyanin Synthesis in A. triflorum Leaves
4.3. Transcription Factors Associated with Anthocyanin Biosynthesis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Flora of China Editorial Committee. Flora of China; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2008; p. 94. Available online: http://www.iplant.cn/info/Acer%20triflorum?t=foc (accessed on 2 April 2022).
- Wang, H.Y.; Wei, B.Y.; Xiao, H.X. Development of 15 microsatellite markers in Acer triflorum (Aceraceae) and cross-amplification in congeneric species. Appl. Plant Sci. 2018, 6, e01166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, X.H.; Yu, X.Y.; Fu, Q.D.; Zheng, Y.Q.; Zhang, C.H. Complete chloroplast genome sequence of the three-flowered maple, Acer triflorum (Sapindaceae). Mitochondrial DNA Part B 2020, 5, 1859–1860. [Google Scholar] [CrossRef] [Green Version]
- Qiao, Q.; Hu, K.R.; Wu, C.; Wang, J.W.; Liu, Q.Z.; Tao, J.H.; Liu, Y.; Lu, Y.Z.; Feng, Z. Acer triflorum as a new nervonic acid-containing oilseed resource for the future: Fruit-and-seed weight, oil content and fatty acid composition. Ind. Crops Prod. 2022, 179, 114684. [Google Scholar] [CrossRef]
- Su, Y.J.; Xue, W.X.; Jia, B.Y.; Chu, J.T.; Wang, L.B.; Feng, Z.; Yu, X.Y. Acer triflorum: A maple species that serves as a resource for nervonic acid and that has an ultralow erucic acid content. Food Sci. Technol. 2021, 41, 633–636. [Google Scholar] [CrossRef]
- Shi, Y.; Liu, W.H. Cultivation and application of seedlings of color-leaf tree species Acer triflorum. Spec. Econ. Anim. Plants 2017, 20, 31–32. [Google Scholar]
- Lin, S.J.; Xu, D.; Cui, Z.; Yang, H.X.; Xie, P.; Zhang, Z.H.; Gou, T.B.; Yang, C.Y.; Zhao, S.S.; Bao, G.D. Determination of biological activity of methanol extract from Acer triflorum seeds. Chin. Agric. Sci. Bull. 2014, 30, 19–22. [Google Scholar]
- Williams, C.A.; Grayer, R.J. Anthocyanins and other flavonoids. Nat. Prod. Rep. 2004, 21, 539–573. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhai, J.; Shao, L.; Lin, W.; Peng, C. Accumulation of Anthocyanins: An Adaptation Strategy of Mikania micrantha to Low Temperature in Winter. Front. Plant Sci. 2019, 10, 1049. [Google Scholar] [CrossRef] [Green Version]
- Song, X.H.; Duan, X.J.; Chang, X.C.; Xian, L.H.; Yang, Q.S.; Liu, Y. Molecular and metabolic insights into anthocyanin biosynthesis during leaf coloration in autumn. Environ. Exp. Bot. 2021, 190, 101584. [Google Scholar] [CrossRef]
- Zhang, S.K.; Zhan, W.; Sun, A.R.; Xie, Y.; Han, Z.M.; Qu, X.B.; Wang, J.Y.; Zhang, L.F.; Tian, M.S.; Pang, X.H.; et al. Combined transcriptome and metabolome integrated analysis of Acer mandshuricum to reveal candidate genes involved in anthocyanin accumulation. Sci. Rep. 2021, 11, 23148. [Google Scholar] [CrossRef]
- Delgado-Vargas, F.; Jim’enez, A.R.; Paredes-L’opez, O. Natural pigments: Carotenoids, anthocyanins, and betalains-characteristics, biosynthesis, processing, and stability. Crit. Rev. Food Sci. Nutr. 2000, 40, 173–289. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Ohmiya, A. Seeing is believing: Engineering anthocyanin and carotenoid biosynthetic pathways. Curr. Opin. Biotechnol. 2008, 19, 190–197. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A Transcriptional Network Promotes Anthocyanin Biosynthesis in Tomato Flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, S.; Liang, D.Y.; Qu, G.Z.; Chen, S.; Zhao, X.Y. Transcriptomic analyses of Pinus koraiensis under different cold stresses. BMC Genom. 2020, 21, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiao, F.C.; Zhao, L.; Wu, X.F.; Song, Z.B.; Li, Y.P. Metabolome and transcriptome analyses of the molecular mechanisms of flower color mutation in tobacco. BMC Genom. 2020, 21, 611. [Google Scholar] [CrossRef]
- Ge, W.; Wang, X.X.; Li, J.Y.; Zhu, W.P.; Cui, J.T.; Zhang, K.Z. Regulatory mechanisms of leaf color change in Acer pictum subsp. mono. Genome 2019, 62, 793–805. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lu, X.; Xuan, Y.; Tang, F.; Wang, J.; Shi, D.; Fu, S.; Ren, J. Transcriptome analysis based on a combination of sequencing platforms provides insights into leaf pigmentation in Acer rubrum. BMC Plant Biol. 2019, 19, 240. [Google Scholar] [CrossRef] [Green Version]
- Quattrocchio, F.; Wing, J.F.; Woude, K.; Mol, J.N.; Koes, R. Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 1998, 13, 475–488. [Google Scholar] [CrossRef]
- Su, Y.K.; Zhang, J.; Xu, Z.C.; Li, J.Y.; Wang, P.F.; Song, Z.Q.; Tian, G.Q.; Li, L.; Song, J.Y.; Wang, J.H. Integrative analysis of metabolome and transcriptome reveals the mechanism of color formation in white root (Salvia miltiorrhiza). Ind. Crops Prod. 2021, 170, 113784. [Google Scholar] [CrossRef]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410. [Google Scholar] [PubMed] [Green Version]
- Varet, H.; Brillet-Guéguen, L.; Coppée, J.Y.; Dillies, M.A. SARTools: A DESeq2- and EdgeR-Based R pipeline for comprehensive differential analysis of RNA-Seq data. PLoS ONE 2016, 11, e0157022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, 14. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Mao, X.; Huang, J.; Yang, D.; Wu, J.; Dong, S.; Lei, K.; Ge, G.; Li, C.Y.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, 316–322. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522. [Google Scholar] [CrossRef]
- Liu, C.Q.; Yao, X.Q.; Li, G.Q.; Huang, L.; Xie, Z.J. Transcriptomic profiling of purple broccoli reveals light-induced anthocyanin biosynthetic signaling and structural genes. PeerJ 2020, 8, e8870. [Google Scholar] [CrossRef]
- Song, B.; Xu, H.; Chen, L.Z.; Fan, X.X.; Jing, Z.G.; Chen, S.; Xu, Z.G. Study of the relationship between leaf color formation and anthocyanin metabolism among different purple pakchoi lines. Molecules 2020, 25, 4809. [Google Scholar] [CrossRef]
- Liu, C.X.; Yu, Q.X.; Li, Z.Q.; Jin, X.L.; Xing, W. Metabolic and transcriptomic analysis related to flavonoid biosynthesis during the color formation of Michelia crassipes tepal. Plant Physiol. Biochem. 2020, 155, 938–951. [Google Scholar] [CrossRef]
- Chen, Z.; Lu, X.Y.; Li, Q.Z.; Li, T.-C.; Zhu, L.; Ma, Q.Y.; Wang, J.J.; Lan, W.; Ren, J. Systematic analysis of MYB gene family in Acer rubrum and functional characterization of ArMYB89 in regulating anthocyanin biosynthesis. J. Exp. Bot. 2021, 72, 6319–6335. [Google Scholar] [CrossRef] [PubMed]
- Li, S.S.; Li, Q.Z.; Rong, L.P.; Tang, L.; Wang, J.J.; Zhang, B. Analysis of the transcriptome of green and mutant golden-yellow leaves of Acer palmatum Thunb. using high-throughput RNA sequencing. J. Hortic. Sci. Biotechnol. 2015, 90, 388–394. [Google Scholar] [CrossRef]
- Dong, H.Y.; Xu, B.; Ji, K.S. Comparative transcriptome analysis of genes involved in response to thermal stress and leaf colour change of Acer palmatum. Sci. Hortic. 2019, 255, 77–85. [Google Scholar] [CrossRef]
- Gao, Y.F.; Zhao, D.H.; Zhang, J.Q.; Chen, J.S.; Li, J.L.; Weng, Z.; Rong, L.P. De novo transcriptome sequencing and anthocyanin metabolite analysis reveals leaf color of Acer pseudosieboldianum in autumn. BMC Genom. 2021, 22, 383. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.J.; Zhang, Q.; Lang, L.X.; Jiang, C.; Wang, X.F.; Sun, H.M. Integrated metabolome and transcriptome analysis of the anthocyanin biosynthetic pathway in relation to color mutation in miniature roses. BMC Plant Biol. 2021, 21, 257. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.Y.; Yang, X.; Zheng, J.R.; Wang, L.; Yang, X.Y.; Tu, Y.; Ye, J.B.; Zhang, W.W.; Liao, Y.L.; Cheng, S.Y.; et al. Unraveling the regulatory mechanism of color diversity in Camellia japonica petals by integrative transcriptome and metabolome analysis. Front. Plant Sci. 2021, 12, 685136. [Google Scholar] [CrossRef]
- Kumar, D.; Sarmah, B.K.; Das, P.K. Ethylene mediates repression of anthocyanin accumulation in black rice pericarps in the absence of light. J. Plant Physiol. 2019, 236, 34–38. [Google Scholar] [CrossRef]
- Guo, Y.Q.; Chang, X.J.; Zhu, C.; Zhang, S.T.; Li, X.Z.; Fu, H.F.; Chen, C.S.; Lin, Y.L.; Lai, Z.X. De novo transcriptome combined with spectrophotometry and gas chromatography-mass spectrometer (GC-MS) reveals differentially expressed genes during accumulation of secondary metabolites in purple-leaf tea (Camellia sinensis cv Hongyafoshou). J. Pomol. Hortic. Sci. 2018, 94, 349–367. [Google Scholar] [CrossRef]
- Kiselev, K.V.; Suprun, A.R.; Aleynova, O.A.; Ogneva, Z.V.; Kalachev, A.V.; Dubrovina, A.S. External dsRNA downregulates anthocyanin biosynthesis-related genes and affects anthocyanin accumulation in Arabidopsis thaliana. Int. J. Mol. Sci. 2021, 23, 6749. [Google Scholar] [CrossRef]
- Han, Y.P.; Vimolmangkang, S.; Soria-Guerra, R.E.; Rosales-Mendoza, S.; Zheng, D.; Lygin, A.V.; Korban, S.S. Ectopic expression of apple F3‘H genes contributes to anthocyanin accumulation in the Arabidopsis tt7 mutant grown under nitrogen stress. Plant Physiol. 2010, 153, 806–820. [Google Scholar] [CrossRef] [Green Version]
- Sundaramoorthy, J.; Park, G.T.; Chang, J.H.; Lee, J.D.; Kim, J.H.; Seo, H.S.; Chung, G.; Song, J.T. Identification and molecular analysis of four new alleles at the W1 locus associated with flower color in soybean. PLoS ONE 2016, 11, e0159865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.Y.; Shi, Y.F.; Li, K.Y.; Yang, D.; Liu, N.N.; Zhang, L.J.; Zhao, L.; Zhang, X.F.; Liu, Y.J.; Gao, L.P.; et al. Roles of the 2-Oxoglutarate-Dependent Dioxygenase Superfamily in the Flavonoid Pathway: A Review of the Functional Diversity of F3H, FNS I., FLS, and LDOX/ANS. Molecules 2021, 26, 6745. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Lee, J.R.; Hong, S.T.; Yoo, Y.K.; An, G.; Kim, S.R. Molecular cloning and analysis of anthocyanin biosynthesis genes preferentially expressed in apple skin. Plant Sci. 2003, 165, 403–413. [Google Scholar] [CrossRef]
- Wu, M.; Liu, J.L.; Song, L.Y.; Li, X.Y.; Cong, L.; Yue, R.R.; Yang, C.Q.; Liu, Z.; Xu, L.F.; Wang, Z.G. Differences among the Anthocyanin Accumulation Patterns and Related Gene Expression Levels in Red Pears. Plants 2019, 8, 100. [Google Scholar] [CrossRef] [Green Version]
- Han, Y.P.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.G.; Li, S.J.; Kang, Y.X.; Liu, J.J. Molecular characterization and expression analyses of an anthocyanin synthase gene from Magnolia sprengeri pamp. Appl. Biochem. Biotechnol. 2015, 175, 477–488. [Google Scholar] [CrossRef]
- Qin, S.J.; Liu, Y.T.; Cui, B.Q.; Cheng, J.L.; Liu, S.Y.; Liu, H.Z. Isolation and functional diversification of dihydroflavonol 4-Reductase gene HvDFR from Hosta ventricosa indicate its role in driving anthocyanin accumulation. Plant Signal. Behav. 2021, 17, 2010389. [Google Scholar] [CrossRef]
- Yoshihara, N.; Imayama, T.; Fukuchi-Mizutani, M.; Okuhara, H.; Tanaka, Y.; Ino, I.; Yabuya, T. cDNA cloning and characterization of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase in Iris hollandica. Plant Sci. 2005, 169, 496–501. [Google Scholar] [CrossRef]
- Hiromoto, T.; Honjo, E.; Noda, N.; Tamada, T.; Kazuma, K.; Suzuki, M.; Blaber, M.; Kuroki, R. Structural basis for acceptor-substrate recognition of UDP-glucose: Anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea. Protein Sci. 2015, 24, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, Y.; Sasaki, N.; Ohmiya, A. Biosynthesis of plant pigments: Anthocyanins, betalains and carotenoids. Plant J. 2008, 54, 733–749. [Google Scholar] [CrossRef]
- Sun, T.H.; Yuan, H.; Cao, H.B.; Yazdani, M.; Tadmor, Y.; Li, L. Carotenoid Metabolism in Plants: The Role of Plastids. Mol. Plant 2018, 11, 58–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, L.; Yuan, Y.W. Transcriptional Regulation of Carotenoid Biosynthesis in Plants: So Many Regulators, So Little Consensus. Front. Plant Sci. 2019, 10, 1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.R.; Shi, Q.Q.; Niu, L.X.; Zhang, Y.L. Transcriptomic analysis of leaf in tree peony reveals differentially expressed pigments genes. Molecules 2017, 22, 324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shan, X.T.; Li, Y.Q.; Yang, S.; Gao, R.F.; Zhou, L.D.; Bao, T.T.; Han, T.T.; Wang, S.C.; Gao, X.; Wang, L. A functional homologue of Arabidopsis TTG1 from Freesia interacts with bHLH proteins to regulate anthocyanin and proanthocyanidin biosynthesis in both Freesia hybrida and Arabidopsis thaliana. Plant Physiol. Biochem. 2019, 141, 60–72. [Google Scholar] [CrossRef]
- Yan, H.L.; Pei, X.N.; Zhang, H.; Li, X.; Zhang, X.X.; Zhao, M.H.; Chiang, V.L.; Sederoff, R.R.; Zhao, X.Y. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef]
- Gil-Muoz, F.; Sánchez-Navarro, J.A.; Besada, C.; Salvador, A.; Badenes, M.L.; Naval, M.D.M.; Ríos, G. MBW complexes impinge on anthocyanidin reductase gene regulation for proanthocyanidin biosynthesis in persimmon fruit. Sci. Rep. 2020, 10, 3543. [Google Scholar] [CrossRef]
- Zhang, Y.Z.; Xu, S.Z.; Ma, H.P.; Duan, X.J.; Gao, S.X.; Zhou, X.J.; Cheng, Y.W. The R2R3-MYB gene PsMYB58 positively regulates anthocyanin biosynthesis in tree peony flowers. Plant Physiol. Biochem. 2021, 164, 279–288. [Google Scholar] [CrossRef]
- Deng, G.M.; Zhang, S.; Yang, Q.S.; Gao, H.J.; Sheng, O.; Bi, F.C.; Li, C.Y.; Dong, T.; Yi, G.J.; He, W.D.; et al. MaMYB4, an R2R3-MYB Repressor Transcription Factor, Negatively Regulates the Biosynthesis of Anthocyanin in Banana. Front. Plant Sci. 2021, 11, 600704. [Google Scholar] [CrossRef]
- Ning, Z.Y.; Hu, K.D.; Zhou, Z.L.; Zhao, D.L.; Tang, J.; Wang, H.; Li, L.X.; Ding, C.; Chen, X.Y.; Yao, G.F.; et al. IbERF71, with IbMYB340 and IbbHLH2, coregulates anthocyanin accumulation by binding to the IbANS1 promoter in purple-fleshed sweet potato (Ipomoea batatas L.). Plant Cell Rep. 2021, 40, 157–169. [Google Scholar] [CrossRef]
- Cui, D.L.; Zhao, S.X.; Xu, H.N.; Allan, A.C.; Zhang, X.D.; Fan, L.; Chen, L.M.; Su, J.; Shu, Q.; Li, K.Z. The interaction of MYB, bHLH and WD40 transcription factors in red pear (Pyrus pyrifolia) peel. Plant Mol. Biol. 2021, 106, 407–417. [Google Scholar] [CrossRef]
- Chen, X.; Li, M.H.; Ni, J.; Hou, J.Y.; Shu, X.; Zhao, W.W.; Su, P.F.; Wang, D.C.; Shah, F.A.; Huang, S.W.; et al. The R2R3-MYB transcription factor SsMYB1 positively regulates anthocyanin biosynthesis and determines leaf color in Chinese tallow (Sapium sebiferum Roxb.). Ind. Crops Prod. 2021, 164, 113335. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, A.; Pei, X.; Zhang, S.; Han, Z.; Xie, Y.; Qu, G.; Hu, X.; Tigabu, M.; Zhao, X. Metabolome and Transcriptome Analyses Unravels Molecular Mechanisms of Leaf Color Variation by Anthocyanidin Biosynthesis in Acer triflorum. Horticulturae 2022, 8, 635. https://doi.org/10.3390/horticulturae8070635
Sun A, Pei X, Zhang S, Han Z, Xie Y, Qu G, Hu X, Tigabu M, Zhao X. Metabolome and Transcriptome Analyses Unravels Molecular Mechanisms of Leaf Color Variation by Anthocyanidin Biosynthesis in Acer triflorum. Horticulturae. 2022; 8(7):635. https://doi.org/10.3390/horticulturae8070635
Chicago/Turabian StyleSun, Anran, Xiaona Pei, Shikai Zhang, Zhiming Han, Ying Xie, Guanzheng Qu, Xiaoqing Hu, Mulualem Tigabu, and Xiyang Zhao. 2022. "Metabolome and Transcriptome Analyses Unravels Molecular Mechanisms of Leaf Color Variation by Anthocyanidin Biosynthesis in Acer triflorum" Horticulturae 8, no. 7: 635. https://doi.org/10.3390/horticulturae8070635
APA StyleSun, A., Pei, X., Zhang, S., Han, Z., Xie, Y., Qu, G., Hu, X., Tigabu, M., & Zhao, X. (2022). Metabolome and Transcriptome Analyses Unravels Molecular Mechanisms of Leaf Color Variation by Anthocyanidin Biosynthesis in Acer triflorum. Horticulturae, 8(7), 635. https://doi.org/10.3390/horticulturae8070635





