Somatic Embryogenesis: A Tool for Fast and Reliable Virus and Viroid Elimination for Grapevine and other Plant Species
Abstract
1. Introduction
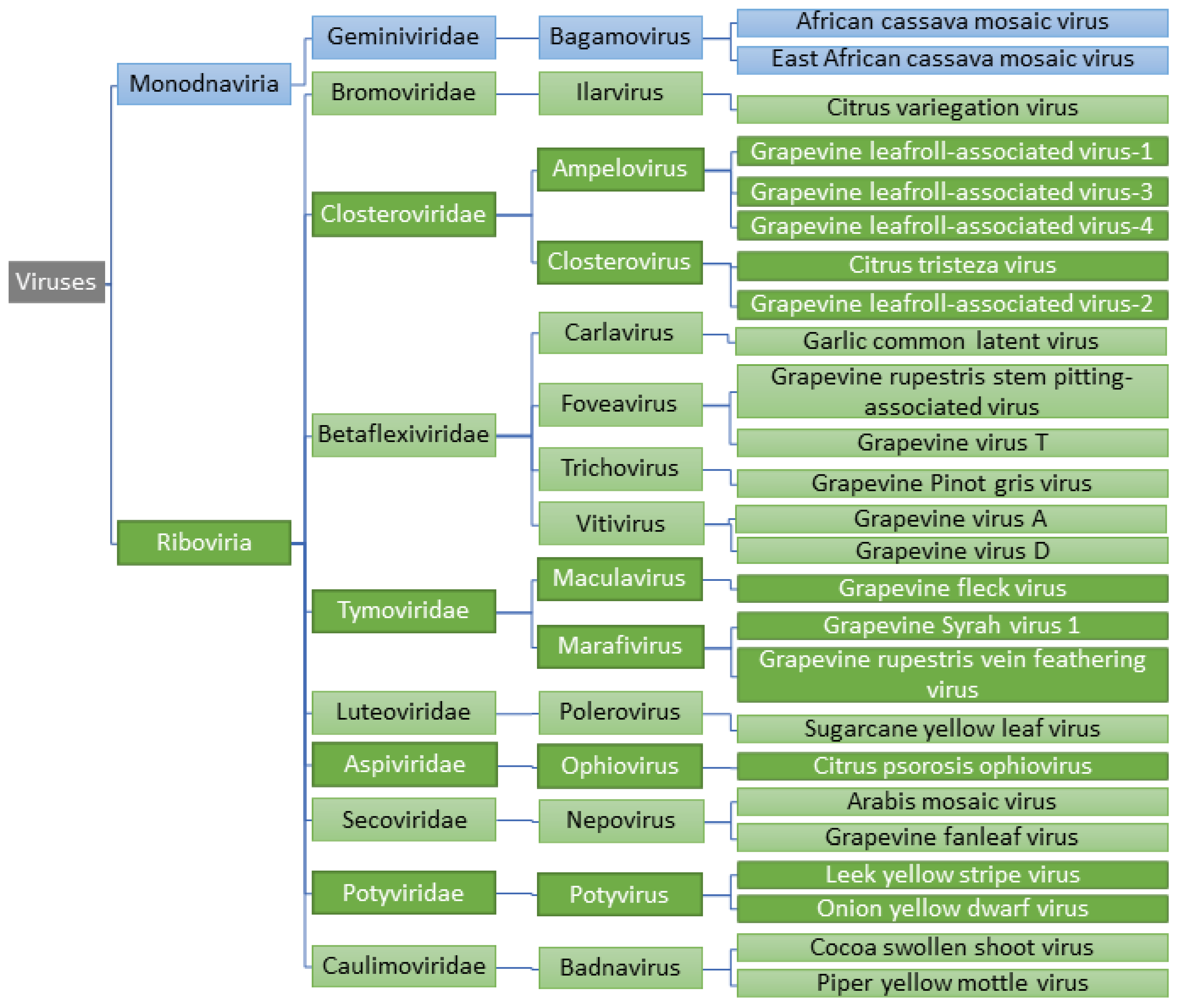
2. Detection of Viruses and Viroids in Embryogenic Callus and Somatic Embryos
2.1. DNA Viruses
2.2. Positive Sense RNA Viruses
2.2.1. Closteroviridae
2.2.2. Betaflexiviridae
2.2.3. Tymoviridae
2.2.4. Secoviridae
2.3. Negative Sense RNA Viruses
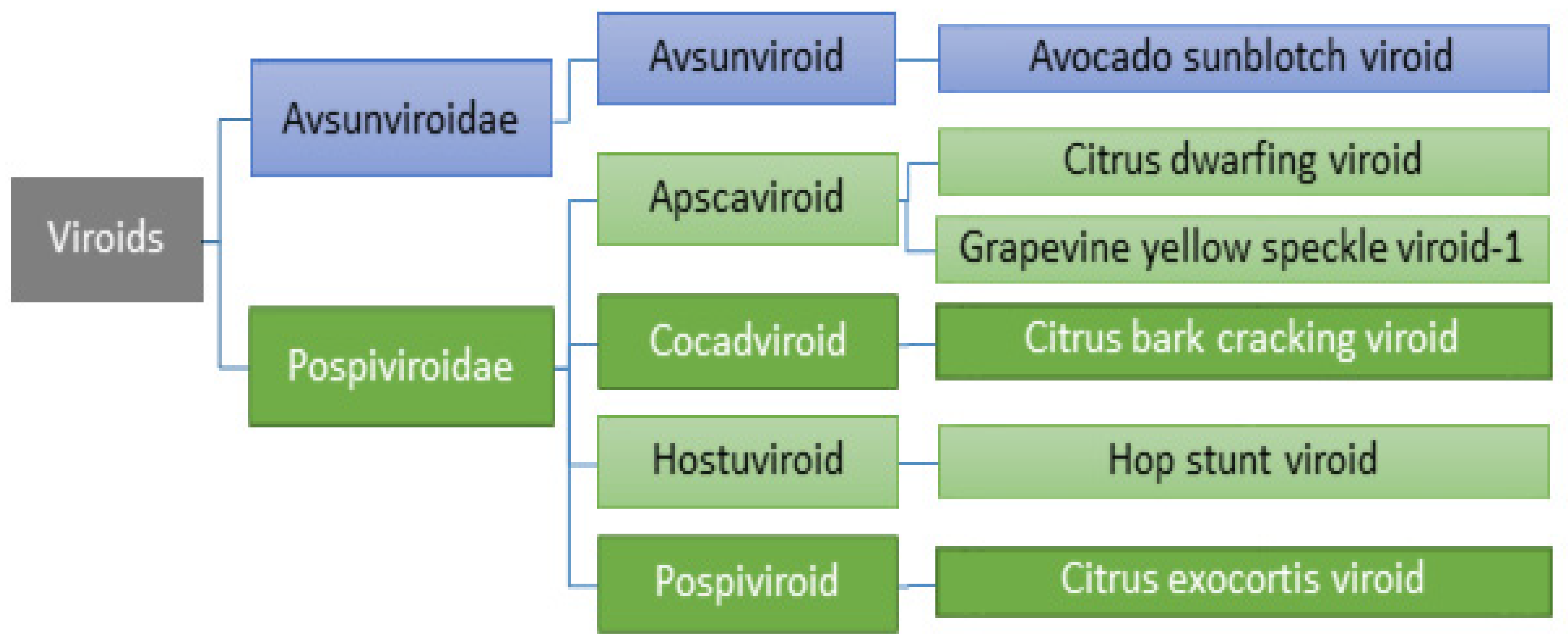
2.4. Viroids
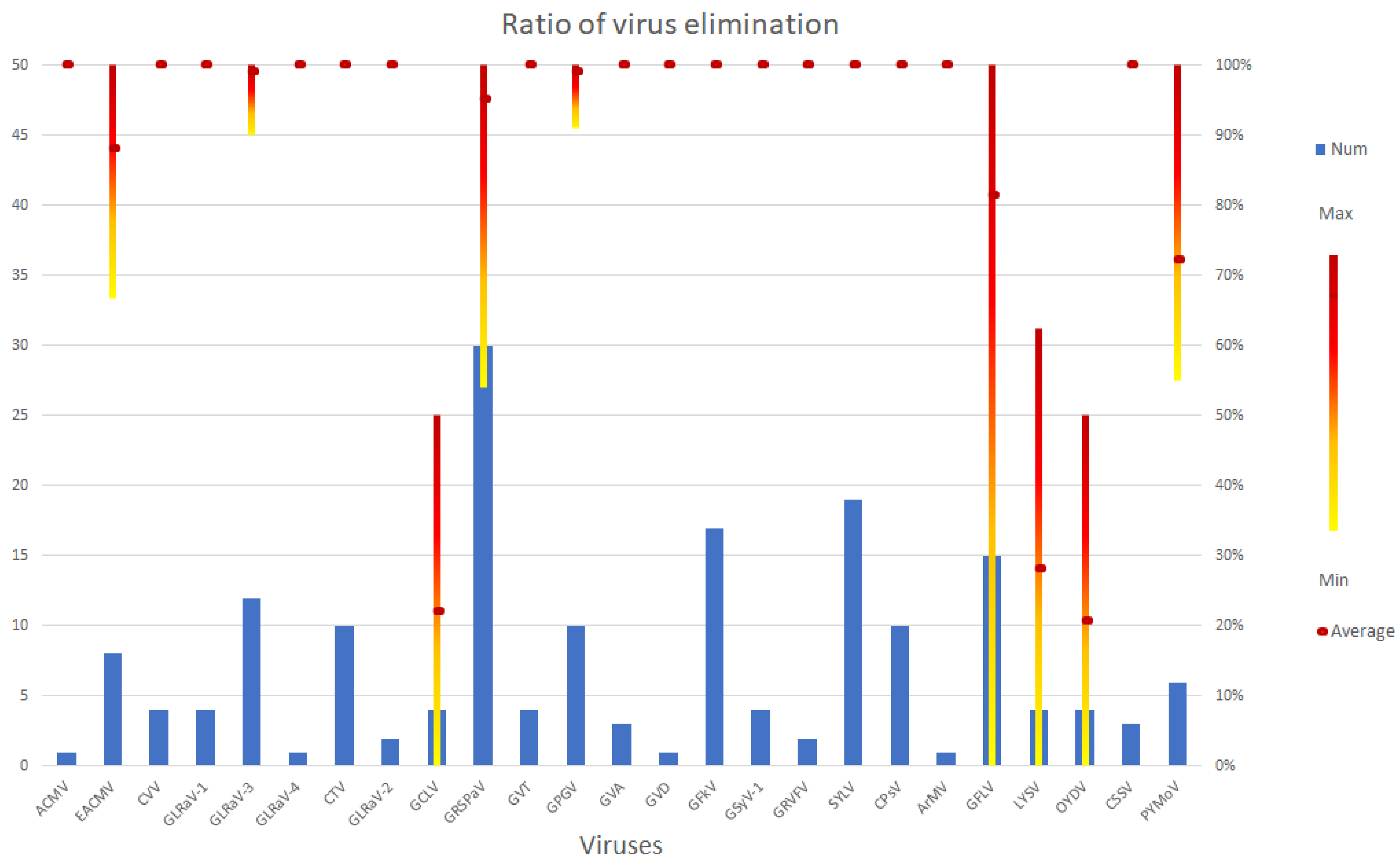
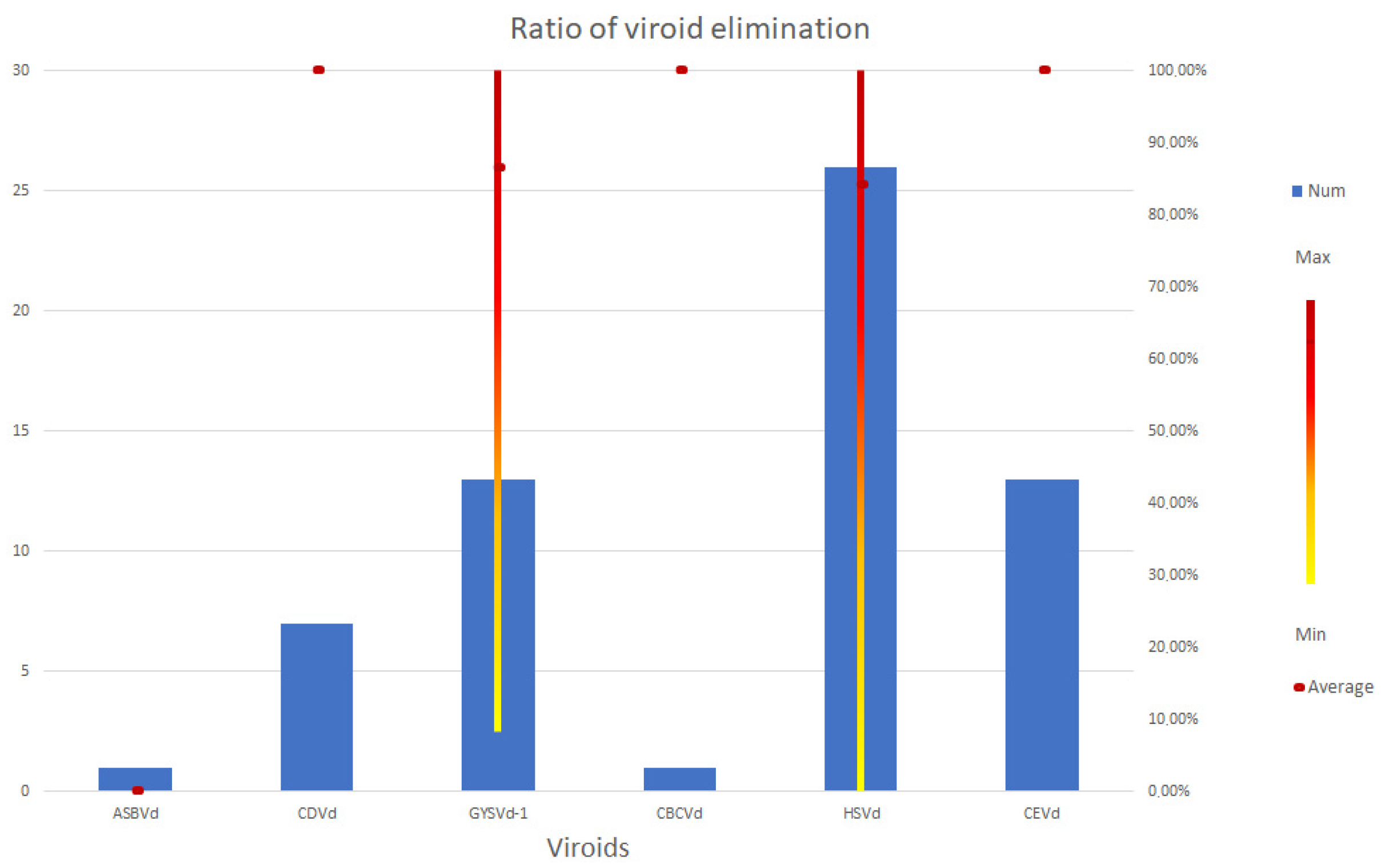
3. Healthy Plant Production: Viruses and Viroids in SE Based Elimination Experiments
3.1. DNA Viruses Tested in SE Experiments
3.2. Positive Sense RNA Viruses
3.3. Negative Sense RNA Viruses
3.4. Viroids
4. Genotype Effect
5. Genetic Variability of the Regenerated Plants
6. Embryogenesis Combined with other Treatments
7. Potential Transferability of Elimination Results by SE to Different Plant Species
8. Conclusions
9. Short Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jeger, M.J. The Impact of Climate Change on Disease in Wild Plant Populations and Communities. Plant Pathol. 2022, 71, 111–130. [Google Scholar] [CrossRef]
- Atlin, G.N.; Cairns, J.E.; Das, B. Rapid Breeding and Varietal Replacement Are Critical to Adaptation of Cropping Systems in the Developing World to Climate Change. Glob. Food Secur. 2017, 12, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Magyar-Tábori, K.; Mendler-Drienyovszki, N.; Hanász, A.; Zsombik, L.; Dobránszki, J. Phytotoxicity and Other Adverse Effects on the in Vitro Shoot Cultures Caused by Virus Elimination Treatments: Reasons and Solutions. Plants 2021, 10, 670. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, K.B.; Najar, A.; Jemai, N.; Jemmali, A. Advances in Sanitation Methods for Fruit Tree Species through in Vitro Technologies: Possibilities and Limits Scientific Review. Agric. Biotechnol. 2017, 45, 2483–2495. [Google Scholar]
- Panattoni, A.; Luvisi, A.; Triolo, E. Review. Elimination of Viruses in Plants: Twenty Years of Progress. Span. J. Agric. Res. 2013, 11, 173–188. [Google Scholar] [CrossRef]
- Halperin, W. In Vitro Embryogenesis: Some Historical Issues and Unresolved Problems. In In Vitro Embryogenesis in Plants; Springer: Berlin/Heidelberg, Germany, 1995; pp. 1–16. [Google Scholar]
- Oláh, R.; Zok, A.; Pedryc, A.; Howard, S.; Kovács, L.G. Somatic Embryogenesis in a Broad Spectrum of Grape Genotypes. Sci. Hortic. 2009, 120, 134–137. [Google Scholar] [CrossRef]
- Guan, Y.; Li, S.G.; Fan, X.F.; Su, Z.H. Application of Somatic Embryogenesis in Woody Plants. Front. Plant Sci. 2016, 7, 938. [Google Scholar] [CrossRef]
- Carabez, J.R.S.; Ortiz, D.T.; Pérez, M.R.V.; Peña, H.B. The Avocado Sunblotch Viroid: An Invisible Foe of Avocado. Viruses 2019, 11, 491. [Google Scholar] [CrossRef]
- Legg, J.P.; Lava Kumar, P.; Makeshkumar, T.; Tripathi, L.; Ferguson, M.; Kanju, E.; Ntawuruhunga, P.; Cuellar, W. Cassava Virus Diseases: Biology, Epidemiology, and Management. In Advances in Virus Research; Academic Press: Cambridge, MA, USA, 2015; Volume 91. [Google Scholar]
- Umer, M.; Liu, J.; You, H.; Xu, C.; Dong, K.; Luo, N.; Kong, L.; Li, X.; Hong, N.; Wang, G.; et al. Genomic, Morphological and Biological Traits of the Viruses Infecting Major Fruit Trees. Viruses 2019, 11, 515. [Google Scholar] [CrossRef]
- Afloukou, F.; Dossou, L.; Zinsou, V. Virus and Virus-like Diseases of Citrus in West-Africa: An Overview. J. Hortic. Postharvest Res. 2020, 3, 129–138. [Google Scholar]
- Bendix, C.; Lewis, J.D. The Enemy within: Phloem-Limited Pathogens. Mol. Plant Pathol. 2018, 19, 238–254. [Google Scholar] [CrossRef]
- Belabess, Z.; Sagouti, T.; Rhallabi, N.; Tahiri, A.; Massart, S.; Tahzima, R.; Lahlali, R.; Haissam Jijakli, M. Citrus Psorosis Virus: Current Insights on a Still Poorly Understood Ophiovirus. Microorganisms 2020, 8, 1197. [Google Scholar] [CrossRef]
- Zhou, C.; da Graça, J.V.; Freitas-Astúa, J.; Vidalakis, G.; Duran-Vila, N.; Lavagi, I. Citrus Viruses and Viroids. In The Genus Citrus; Elsevier: Amsterdam, The Netherlands, 2020. [Google Scholar]
- Muller, E. Cacao Swollen Shoot Virus (CSSV): History, Biology, and Genome. In Cacao Diseases: A History of Old Enemies and New Encounters; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Bhat, A.I.; Hohn, T.; Selvarajan, R. Badnaviruses: The Current Global Scenario. Viruses 2016, 8, 177. [Google Scholar] [CrossRef]
- Winiarczyk, K.; Solarska, E.; Sienkiewicz, W. Prevalence of Infections with Onion Yellow Dwarf Virus, Leek Yellow Stripe Virus and Garlic Common Latent Virus in Plants from the Genus Allium|Rozpowszechnienie Zakażeń Roślin z Rodzaju Allium Przez Wirusy Żółtej Karłowatości Cebuli (Oyd), Żółtej Smugo. Acta Sci. Pol. Hortorum Cultus 2014, 13, 123–133. [Google Scholar]
- Lunello, P.; Ducasse, D.A.; Helguera, M.; Nome, S.F.; Conci, V.C. An Argentinean Isolate of Leek Yellow Stripe Virus from Leek Can Be Transmitted to Garlic. J. Plant Pathol. 2002, 84, 11–17. [Google Scholar]
- Verma, R.K.; Mishra, R.; Petrov, N.M.; Stoyanova, M.; Stoev, A.; Bakardjieva, N.V.; Gaur, R.K. Molecular Characterization and Recombination Analysis of an Indian Isolate of Onion Yellow Dwarf Virus. Eur. J. Plant Pathol. 2015, 143, 437–445. [Google Scholar] [CrossRef]
- Martelli, G.P. Directory of Virus and Virus-like Diseases of the Grapevine and Their Agents. J. Plant Pathol. 2014, 96, 1–136. [Google Scholar]
- Meng, B.; Martelli, G.P.; Golino, D.A.; Fuchs, M. Grapevine Viruses: Molecular Biology, Diagnostics and Management; Springer International Publishing AG: Cham, Switzerland, 2017. [Google Scholar]
- San Pedro, T.; Gammoudi, N.; Peiró, R.; Olmos, A.; Gisbert, C. Somatic Embryogenesis from Seeds in a Broad Range of Vitis vinifera L. Varieties: Rescue of True-to-Type Virus-Free Plants. BMC Plant Biol. 2017, 17, 226. [Google Scholar] [CrossRef]
- Jo, Y.; Song, M.K.; Choi, H.; Park, J.S.; Lee, J.W.; Lian, S.; Lee, B.C.; Cho, W.K. Genome Sequence of Grapevine Virus T, a Novel Foveavirus Infecting Grapevine. Genome Announc. 2017, 5, e00995-17. [Google Scholar] [CrossRef]
- Nuzzo, F.; Moine, A.; Nerva, L.; Pagliarani, C.; Perrone, I.; Boccacci, P.; Gribaudo, I.; Chitarra, W.; Gambino, G. Grapevine Virome and Production of Healthy Plants by Somatic Embryogenesis. Microb. Biotechnol. 2022, 15, 1357–1373. [Google Scholar] [CrossRef]
- ElSayed, A.I.; Komor, E.; Boulila, M.; Viswanathan, R.; Odero, D.C. Biology and Management of Sugarcane Yellow Leaf Virus: An Historical Overview. Arch. Virol. 2015, 160, 2921–2934. [Google Scholar] [CrossRef] [PubMed]
- Quainoo, A.K.; Wetten, A.C.; Allainguillaume, J. The Effectiveness of Somatic Embryogenesis in Eliminating the Cocoa Swollen Shoot Virus from Infected Cocoa Trees. J. Virol. Methods 2008, 149, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Quainoo, A.K. Age of Callus Tissues and Cotyledonary Materials on the Selection of Cocoa Swollen Shoot Virus-Free Somatic Embryos. Res. Biotechnol. 2011, 2, 78–81. [Google Scholar]
- Edward, R.; Wetten, A. Virus Detection and Elimination in Cocoa (Theobroma Cacao L.) Through Somatic Embryogenesis. J. Plant Sci. 2016, 4, 52–57. [Google Scholar]
- Sasi, S.; Bhat, A.I. In Vitro Elimination of Piper Yellow Mottle Virus from Infected Black Pepper through Somatic Embryogenesis and Meristem-Tip Culture. Crop Prot. 2018, 103, 39–45. [Google Scholar] [CrossRef]
- Damba, Y.; Quainoo, A.; Sowley, E. Effectiveness of Somatic Embryogenesis in Eliminating the Cassava Mosaic Virus from Infected Cassava (Manihot Esculenta Crantz) Plant Materials. Int. J. Sci. Technol. Res. 2013, 2, 282–287. [Google Scholar]
- Mutai, G.K.; Wagacha, J.M.; Nyaboga, E.N. Potential of Somatic Embryogenesis in Elimination of East Africa Cassava Mosaic Virus from Infected Cassava Cultivars in Kenya. Annu. Res. Rev. Biol. 2017, 15, 1–13. [Google Scholar] [CrossRef]
- Gambino, G.; Vallania, R.; Gribaudo, I. In Situ Localization of Grapevine Fanleaf Virus and Phloem-Restricted Viruses in Embryogenic Callus of Vitis Vinifera. Eur. J. Plant Pathol. 2010, 127, 557–570. [Google Scholar] [CrossRef]
- Gambino, G.; Bondaz, J.; Gribaudo, I. Detection and Elimination of Viruses in Callus, Somatic Embryos and Regenerated Plantlets of Grapevine. Eur. J. Plant Pathol. 2006, 114, 397–404. [Google Scholar] [CrossRef]
- Scagliusi, S.M.M.; Vega, J.; Kuniyuki, H. Cytopathology of Callus Cells Infected with Grapevine Leafroll-Associated Virus 3. Fitopatol. Bras. 2002, 27, 384–388. [Google Scholar] [CrossRef]
- Popescu, C.F.; Buciumeanu, E.C.; Visoiu, E. Somatic Embryogenesis, a Reliable Method for Grapevine Fleck Virus Free Grapevine Regeneration. In Proceedings of the Extended Abstracts 14th Meeting of ICVG, Bari, Italy, 12–17 September 2003; p. 243. [Google Scholar]
- Goussard, P.G.; Wiid, J. The Elimination of Fanleaf Virus from Grapevines Using in Vitro Somatic Embryogenesis Combined with Heat Therapy. S. Afr. J. Enol. Vitic. 2017, 13, 81–83. [Google Scholar] [CrossRef][Green Version]
- D’Onghia, A.M.; Carimi, F.; De Pasquale, F.; Djelouah, K.; Martelli, G.P. Elimination of Citrus Psorosis Virus by Somatic Embryogenesis from Stigma and Style Cultures. Plant Pathol. 2001, 50, 266–269. [Google Scholar] [CrossRef]
- Suarez, I.E.; Schnell, R.A.; Kuhn, D.N.; Litz, R.E. Recovery and Indexing of Avocado Plants (Persea Americana) from Embryogenic Nucellar Cultures of an Avocado Sunblotch Viroid-Infected Tree. Plant Cell Tissue Organ Cult. 2005, 84, 27–37. [Google Scholar] [CrossRef]
- Gambino, G.; Navarro, B.; Vallania, R.; Gribaudo, I.; Di Serio, F. Somatic Embryogenesis Efficiently Eliminates Viroid Infections from Grapevines. Eur. J. Plant Pathol. 2011, 130, 511–519. [Google Scholar] [CrossRef]
- Meziane, M.; Frasheri, D.; Carra, A.; Boudjeniba, M.; D’Onghia, A.M.; Mercati, F.; Djelouah, K.; Carimi, F. Attempts to Eradicate Graft-Transmissible Infections through Somatic Embryogenesis in Citrus Ssp. and Analysis of Genetic Stability of Regenerated Plants. Eur. J. Plant Pathol. 2017, 148, 85–95. [Google Scholar] [CrossRef]
- Nkaa, F.A.; Ene-Obong, E.E.; Taylor, N.; Fauquet, C.; Mbanaso, E.N.A. Elimination of African Cassava Mosaic Virus (ACMV) and East African Cassava Mosaic Virus (EACMV) from Cassava (Manihot Esculenta Crantz) Cv. “Nwugo” via Somatic Embryogenesis. Am. J. Biotechnol. Mol. Sci. 2013, 3, 33–40. [Google Scholar]
- Borroto-Fernandez, E.G.; Sommerbauer, T.; Popowich, E.; Schartl, A.; Laimer, M. Somatic Embryogenesis from Anthers of the Autochthonous Vitis Vinifera Cv. Domina Leads to Arabis Mosaic Virus-Free Plants. Eur. J. Plant Pathol. 2009, 124, 171–174. [Google Scholar] [CrossRef]
- El-Sawy, A.; Gomaa, A.; Abd-El-Zaher, M.H.; Reda, A.; Danial, N. Production of Somatic Embryogenesis via in Vitro Culture of Stigma and Style for Elimination of Citrus Psorosisvirus (CpsV) from Some Citrus Genotypes. J. Hortic. Sci. Ornam. Plants 2013, 5, 110–117. [Google Scholar]
- Jin, S.B.; Park, J.H.; Park, S.M.; Lee, D.H.; Yun, S.H. Production of Citrus Plants from Ovule Cell Culture and Verification of CTV—Free Plants. Korean J. Hortic. Sci. Technol. 2017, 35, 121–130. [Google Scholar] [CrossRef]
- Kereša, S.; Kurtović, K.; Ban, S.G.; Vončina, D.; Jerčić, I.H.; Bolarić, S.; Lazarević, B.; Godena, S.; Ban, D.; Mihovilović, A.B. Production of Virus-Free Garlic Plants through Somatic Embryogenesis. Agronomy 2021, 11, 876. [Google Scholar] [CrossRef]
- Malenica, N.; Jagic, M.; Pavletic, B.; Bauer, N.; Vončina, D.; Zdunic, G.; Levanic, D.L. Somatic Embryogenesis as a Tool for Virus Elimination in Croatian Indigenous Grapevine Cultivars. Acta Bot. Croat. 2020, 79, 26–34. [Google Scholar] [CrossRef]
- Turcsan, M.; Demian, E.; Varga, T.; Jaksa-Czotter, N.; Szegedi, E.; Olah, R.; Varallyay, E. Hts-Based Monitoring of the Efficiency of Somatic Embryogenesis and Meristem Cultures Used for Virus Elimination in Grapevine. Plants 2020, 9, 1782. [Google Scholar] [CrossRef]
- Gambino, G.; Di Matteo, D.; Gribaudo, I. Elimination of Grapevine Fanleaf Virus from Three Vitis Vinifera Cultivars by Somatic Embryogenesis. Eur. J. Plant Pathol. 2009, 123, 57–60. [Google Scholar] [CrossRef]
- Goussard, P.G.; Wiid, J.; Kasdon, G.G.F. The Effectiveness of in Vitro Somatic Embryogenesis in Eliminating Fanleaf Virus and Leafroll Associated Viruses from Grapevines. S. Afr. J. Enol. Vitic. 1991, 12, 77–81. [Google Scholar] [CrossRef]
- Peiró, R.; Gammoudi, N.; Yuste, A.; Olmos, A.; Gisbert, C. Mature Seeds for in Vitro Sanitation of the Grapevine Leafroll Associated Virus (GLRaV-1 and GLRaV-3) from Grape (Vitis vinifera L.). Span. J. Agric. Res. 2015, 13, e1005. [Google Scholar] [CrossRef]
- Bouamama-Gzara, B.; Selmi, I.; Chebil, S.; Melki, I.; Mliki, A.; Ghorbel, A.; Carra, A.; Carimi, F.; Mahfoudhi, N. Elimination of Grapevine Leafroll Associated Virus-3, Grapevine Rupestris Stem Pitting Associated Virus and Grapevine Virus A from a Tunisian Cultivar by Somatic Embryogenesis and Characterization of the Somaclones Using Ampelographic Descriptors. Plant Pathol. J. 2017, 33, 561–571. [Google Scholar] [CrossRef]
- Gribaudo, I.; Gambino, G.; Cuozzo, D.; Mannini, F. Attempts to Eliminate Grapevine Rupestris Stem Pitting-Associated Virus from Grapevine Clones. J. Plant Pathol. 2006, 88, 293–298. [Google Scholar]
- Parmessur, Y.; Aljanabi, S.; Saumtally, S.; Dookun-Saumtally, A. Sugarcane Yellow Leaf Virus and Sugarcane Yellows Phytoplasma: Elimination by Tissue Culture. Plant Pathol. 2002, 51, 561–566. [Google Scholar] [CrossRef]
- Mahmoud, K.B.; Najar, A.; JedidI, E.; HamdI, I.; Jemmali, A. Detection of Two Viroids in the Tunisian Sweet Orange (Citrus Sinensis L) Cv. Maltese and Sanitation via Somatic Embryogenesis. J. Chem. Pharm. Res. 2017, 9, 154–159. [Google Scholar]
- D’Onghia, A.M.; De Pasquale, F.; Carimi, F.; Savino, V.; Crescimanno, F.G. Somatic Embryogenesis from Style Culture as a Possible Means for Virus Elimination in Cityus. J. Phytopathol. 1997, 145, 77–79. [Google Scholar] [CrossRef]
- Hervé, E.; Romain, G.; Thierry, B.; Jean-Christophe, B.; Estelle, J. Plant Fidelity in Somatic Embryogenesis-Regenerated Plants. In Somatic Embryogenesis: Fundamental Aspects and Applications; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Jahnke, G.; Májer, J.; Varga, P.; SzÖke, B. Analysis of Clones of Pinots Grown in Hungary by SSR Markers. Sci. Hortic. 2011, 129, 32–37. [Google Scholar] [CrossRef]
- Aleza, P.; Juárez, J.; Ollitrault, P.; Navarro, L. Polyembryony in Non-Apomictic Citrus Genotypes. Ann. Bot. 2010, 106, 533–545. [Google Scholar] [CrossRef] [PubMed]
| Disease | Name | Family/Genus | Properties | References | ||
|---|---|---|---|---|---|---|
| avocado | sunblotch | ASBVd | Avocado sunblotch viroid | Avsunviroidae Avsunviroid | nonprotein-coding ssRNA, accumulating in the chloroplast, seed/graft transmissible | [9] |
| cassava | mosaic disease | ACMV | African cassava mosaic virus | Geminiviridae Begomovirus | ssDNA genome, vector transmissible | [10] |
| EACMV | East African cassava mosaic virus | Geminiviridae Begomovirus | ssDNA genome, vector transmissible | [10] | ||
| citrus | tristeza | CTV | Citrus tristeza virus | Closteroviridae Closterovirus | +ssRNA genome, graft/vector transmissible, phloem limited | [11,12,13] |
| psorosis complex | CPsV | Citrus psorosis ophiovirus | Aspiviridae Ophiovirus | -ssRNA genome, seems to be graft transmissible, seedborne | [11,14] | |
| exocortis | CEVd | Citrus exocortis viroid | Pospiviroideae Pospiviroid | nonprotein-coding, small, circular ssRNA, graft transmitted | [12,15] | |
| cachexia | HSVd | Hop stunt viroid | Pospiviroideae Hostuviroid | nonprotein-coding, small, circular ssRNA, accumulates within the nucleus, graft transmitted | [12,15] | |
| infectious variegation | CVV | Citrus variegation virus | Bromoviridae Ilarvirus | +ssRNA genome, seed/graft transmissible | [11,15] | |
| dwarfing symptoms | CDVd | Citrus dwarfing viroid | Pospiviroidae Apscaviroid | nonprotein-coding, small, circular ssRNA, graft transmissible | [15] | |
| bark cracking symptoms | CBCVd | Citrus bark cracking viroid | Pospiviroidae Cocadviroid | nonprotein-coding, small, circular ssRNA, graft transmissible | [15] | |
| cocoa | swollen shoot disease | CSSV | Cocoa swollen shoot virus | Caulimoviridae Badnavirus | dsDNA-RT genome, replicating through an RNA intermediate, seed/graft/vector transmissible | [16,17] |
| garlic | symptomless | GCLV | Garlic common latent virus | Betaflexiviridae Carlavirus | +ssRNA genome, not seed borne, mechanically/vector transmissible | [18] |
| mosaic | LYSV | Leek yellow stripe virus | Potyviridae Potyvirus | +ssRNA genome, not seed borne, mechanically/vector transmissible | [19] | |
| OYDV | Onion yellow dwarf virus | Potyviridae Potyvirus | +ssRNA genome, mechanically/vector transmissible | [20] | ||
| grapevine | infectious degeneration | ArMV | Arabis mosaic virus | Secoviridae Nepovirus | +ssRNA genome, non phloem limited, seed/sap/vector transmissible | [21,22] |
| GFLV | Grapevine fanleaf virus | Secoviridae, Nepovirus | +ssRNA genome, non phloem limited, seed/graft/vector transmitted | [22,23] | ||
| fleck complex | GFkV | Grapevine fleck virus | Tymoviridae, Maculavirus | +ssRNA genome, phloem limited, not seed transmitted, graft transmitted | [21,22] | |
| GRVFV | Grapevine rupestris vein feathering virus | Tymoviridae Marafivirus | +ssRNA genome, phloem limited | [21,22] | ||
| leafroll disease | GLRaV-1 | Grapevine leafroll-associated virus-1 | Closteroviridae Ampelovirus | +ssRNA genome, phloem limited, graft/vector transmissible | [11,21] | |
| GLRaV-2 | Grapevine leafroll-associated virus-2 | Closteroviridae Closterovirus | +ssRNA genome, phloem limited, graft transmissible | [11,22] | ||
| GLRaV-3 | Grapevine leafroll-associated virus-3 | Closteroviridae Ampelovirus | +ssRNA genome, phloem limited, seed/graft/vector transmissible | [11,23] | ||
| GLRaV-4 | Grapevine leafroll-associated virus-4 | Closteroviridae Ampelovirus | +ssRNA genome, phloem limited, graft/vector transmissible | [22] | ||
| rugose wood complex | GRSPaV | Grapevine rupestris stem pitting-associated virus | Betaflexiviridae Foveavirus | +ssRNA genome, maybe not phloem limited, may not be seed transmitted, graft transmissible | [21,22] | |
| GVA | Grapevine virus A | Betaflexiviridae Vitivirus | +ssRNA genome, phloem limited, graft/vector transmissible | [11,22] | ||
| GVD | Grapevine virus D | Betaflexiviridae Vitivirus | +ssRNA genome, graft transmissible | [22] | ||
| GVT | Grapevine virus T | Betaflexiviridae Foveavirus | +ssRNA genome | [24,25] | ||
| leaf mottling and deformation | GPGV | Grapevine Pinot gris virus | Betaflexiviridae Tichovirus | +ssRNA genome, graft transmissible, vector supposed | [21,22] | |
| yellow speckle | GYSVd-1 | Grapevine yellow speckle viroid-1 | Pospiviroideae Apscaviroid | nonprotein-coding, small, circular ssRNA, accumulating within the nucleus, seed transmitted | [20,21] | |
| symptomless or unknown symptomes | GSyV-1 | Grapevine Syrah virus-1 | Tymoviridae Marafivirus | +ssRNA genome, phloem limited | [21,22] | |
| HSVd | Hop stunt viroid | Pospiviroideae Hostuviroid | nonprotein-coding, small, circular ssRNA, accumulating within the nucleus | [21,22] | ||
| black pepper | yellow mottle | PYMoV | Piper yellow mottle virus | Caulimoviridae Badnavirus | dsDNA-RT genome, replicating through an RNA intermediate, seed/vector transmission | [17] |
| sugarcane | yellow leaf | SCYLV | Sugarcane yellow leaf virus | Luteoviridae Polerovirus | +ssRNA genome, phloem restricted, vector transmissible | [26] |
| Species/Cultivar | Elimination Efficiency of Regenerated Plants | Key Points of the Protocol 1 | Virus Diagnostic Method | References | ||
|---|---|---|---|---|---|---|
| viruses | ACMV | Manihot esculenta ‘Nwugo’ | 100% | indirect embryogenesis, immature leaf lobes (picloram) | PCR | [42] |
| ArMV | Vitis vinifera ‘Domina’ | 100% | direct and indirect embryogenesis, anthers (2,4-D+BAP) | ELISA, immuno capture RT-PCR | [43] | |
| CPsV | Citrus limon ‘Béni’, ‘Abbès’, ‘Sans pépins’, Citrus sinensis ‘Mitidja navel’, ‘Shamouti de station’, ‘Washington navel 251’ | 100% | indirect embryogenesis, stigmas, and styles (BAP) | DAS– ELISA | [41] | |
| Citrus reticulata, Citrus sinensis ‘Navelina’ Citrus sinenesis x C. reticulata ‘Dweet tangor’ | 100% | indirect embryogenesis, stigmas, and styles (BAP) | DAS– ELISA | [38] | ||
| Citrus sinensis ‘Washington navel’ | 100% | indirect embryogenesis, stigma (BAP) | RT-PCR | [44] | ||
| CSSV | Theobroma cacao ‘Amolenado’ | 100% | indirect embryogenesis, staminode explants (2,4-D, TDZ) | PCR, qPCR | [27] | |
| Theobroma cacao ‘CL 19/10’, ‘ICS 68’ | 100% | indirect embryogenesis, staminode explants (2,4-D, TDZ) | PCR | [29] | ||
| CTV | Citrus limon ‘Dellys’, ‘Villafranca’ Citrus sinensis ‘Mitidja navel’, ‘Shamouti de station’, ‘Washington navel 251’ | 100% | indirect embryogenesis, stigmas, and styles (BAP) | DAS– ELISA | [41] | |
| Citrus erythrosa ‘Dongjeongkyool’ Citrus nippokoreana ‘Cheongkyool’ Citrus aurantium ‘Jikak’ Citrus unshiu ‘Miyagawa wase’, ‘Haryejosaeng’ | 100% | direct and indirect embrygenesis, ovules from immature fruits (Kin) | RT-PCR, immune-Strip test | [45] | ||
| CVV | Citrus limon ‘Bornèo’, ‘Eureka 4’, ‘Sans pépins’, Citrus sinensis ‘Washington navel 251’ | 100% | indirect embryogenesis, stigmas, and styles (BAP) | TAS-ELISA | [41] | |
| EACMV | Manihot esculenta ‘Nwugo’ | 100% | indirect embryogenesis, immature leaf lobes (picloram) | PCR | [42] | |
| Manihot esculenta ‘TME 14’, ‘Ex-Mariakani’, ‘Sagalato’, ‘Kibandameno’, ‘TMS 60444’ | 100%, 71.4–91.7%, 100%, 100%, 66.7–75% | embryogenesis, nodal cuttings (picloram) | PCR | [32] | ||
| GCLV | Allium sativum ‘Istarski crveni’, ‘IPT012’ | 18.2–50%, 0–20% | indirect embryogenesis, cutted cloves (2,4-D/2,4-D+Kin) | ELISA, RT-PCR | [46] | |
| GFkV | Vitis vinifera ‘Mission’, ‘Coarna negra’, ‘Ranai Magaraci’ | 100% | direct or indirect embryogenesis, anthers (IAA+BAP), ovules (IBA+BAP) | ELISA | [36] | |
| Vitis vinifera ‘Pinot blanc’, ‘Cabernet franc’, ‘Valenci blanc’ | 100% | direct or indirect embryogenesis, immature cut seeds (TDZ) | RT-qPCR | [23] | ||
| Vitis vinifera ‘Babica’ | 100% | immature anthers, (BAP+NOA+2,4-D) | RT-PCR | [47] | ||
| Vitis vinifera ‘Muscat Ottonel H-7-3’, ‘Muscat Ottonel H-14-1’ | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | ||
| Vitis vinifera ‘Glória’, ‘Muscat Ottonel H-13-4’, ‘Müller-Thurgau’, Vitis sp. ‘9/143’ complex hybrid | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR | this paper | ||
| Vitis vinifera ‘Brachetto’, ‘Cabernet Sauvignon’, ‘Nebbiolo’, ‘Sangiovese’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GFLV | Vitis vinifera ‘Cari’, ‘Proviné’, ‘Roussan’ | 100%, 97%, 100% | anthers and ovaries (2,4-D+BAP) | RT-PCR | [49] | |
| Vitis rupestris ‘Rupestris du Lot’ | 0% | indirect embryogenesis, anthers (2,4-D+BAP), and ovaries (2,4-D+NOA+BAP) | ELISA, immuno-sorbent electron microscopy | [50] | ||
| Vitis vinifera ‘Gewürztraminer’, Vitis rupestris ‘Rupestris du lot’ | 100% | indirect embryogenesis combined with heat therapy (35 °C), anthers (2,4-D+BAP), and ovaries (2,4-D+NOA+BAP) | ELISA, immuno-sorbent electron microscopy | [37] | ||
| Vitis vinifera ‘Pinot blanc’, ‘Tempranillo’, ‘Godello’, ‘Merlot’ | 88%, 69%, 79%, 90% | direct or indirect embryogenesis, immature cut seeds (TDZ) | RT-qPCR | [23] | ||
| Vitis vinifera ‘Babica’, ‘Plavac mali’ | 66%, 33% | immature anthers, (BAP+NOA+2,4-D) | RT-PCR | [47] | ||
| Vitis sp. ‘9/143’ hybrid, Vitis vinifera ‘Muscat Ottonel H-13-4’ | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR | this paper | ||
| Vitis vinifera ‘Cabernet Sauvignon’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GLRaV-1 | Vitis vinifera ‘Grignolino’ | 100% | indirect embryogenesis, stamens, and pistils (2,4-D+BAP) | ELISA, RT-PCR, tissue blot immuno-assay | [34] | |
| Vitis vinifera ‘Grumet Negre’ | 100% | mature cut seeds (TDZ) | RT-qPCR | [51] | ||
| Vitis vinifera ‘Plavac mali’ | 100% | immature anthers, (BAP+NOA+2,4-D) | RT-PCR | [47] | ||
| Vitis vinifera ‘Müller-Thurgau’ | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR | this paper | ||
| GLRaV-2 | Vitis vinifera ‘Roobernet’ | 100% | indirect embryogenesis, anthers (2,4-D+BAP), and ovaries (2,4-D+NOA+BAP) | ELISA, immuno-sorbent electron microscopy | [50] | |
| Vitis vinifera ‘Cabernet Sauvignon’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GLRaV-3 | Vitis vinifera ‘Roobernet’ | 100% | indirect embryogenesis, anthers (2,4-D+BAP), and ovaries (2,4-D+NOA+BAP) | ELISA, immunosorbent electron microscopy | [50] | |
| Vitis vinifera ‘Grumet Negre’ | 100% | mature cut seeds (TDZ) | RT-qPCR | [51] | ||
| Vitis vinifera ‘Cabernet franc’, ‘Godello’, ‘Merlot’, ‘Valencí blanc’ | 100%, 100%, 90%, 100% | direct or indirect embryogenesis, immature cut seeds (TDZ) | RT-qPCR | [23] | ||
| Vitis vinifera ‘Müller-Thurgau’ | 100% | indirect embryogenesis, stamens, and pistils (2,4-D+BAP) | ELISA, RT-PCR, tissue blot immunoassay | [34] | ||
| Vitis vinifera ‘Hencha’ | 100% | direct embryogenesis, stamen (2,4-D+TDZ) | ELISA, RT-PCR | [52] | ||
| Vitis vinifera ‘Babica’, ‘Plavac mali’ | 100% | immature anthers, (BAP+NOA+2,4-D) | RT-PCR | [47] | ||
| Vitis vinifera ‘Cabernet Sauvignon’, ‘Sangiovese’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GLRaV-4 | Vitis vinifera ‘Sangiovese’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | |
| GPGV | Vitis vinifera ‘Trilla’, ‘Szirén’, ‘Muscat Ottonel H-14-1’ | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | |
| Vitis rupestris Vitis sp. ‘Richter 110’ Vitis vinifera ‘Brachetto’, ‘Cabernet Sauvignon’, ‘Chardonnay’, ‘Nebbiolo’, ‘Sangiovese’ | 100% 100% 100%, 100%, 100%, 91%, 100% | indirect embryogenesis, anthers and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GRSPaV | Vitis vinifera ‘Grignolino’, ‘Müller-Thurgau’, ‘Bosco’ | 100% | indirect embryogenesis, stamens, and pistils (2,4-D+BAP) | ELISA, RT-PCR, tissue blot immunoassay | [34] | |
| Vitis vinifera ‘Albarola’, ‘Bosco’, ‘Brachetto’, ‘Grignolino’, ‘Müller Thurgau’, ‘Rossese’, ‘Vermentino’ | 100% | indirect embryogenesis, immature anthers, and ovules (2,4-D+BAP) | RT-PCR | [53] | ||
| Vitis vinifera ‘Hencha’ | 100% | direct embryogenesis, stamen (2,4-D+TDZ) | ELISA, RT-PCR | [52] | ||
| Vitis vinifera ‘Trilla’, ‘Szirén’, ‘Muscat Ottonel H-7-3’, ‘Muscat Ottonel H-14-1’ | 100%, 54%, 100%, 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | ||
| Vitis sp. ‘Pamerzs’, ‘Abigél’, ‘Borsmenta’, ‘9/143’ hybrid Vitis vinifera ‘Glória’, ‘Muscat Ottonel’, ‘Müller-Thurgau’ | 100%, 88%, 100%, 100%, 60%, 100%, 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR | this paper | ||
| Vitis rupestris Vitis sp. ‘Richter 110’ Vitis vinifera ‘Brachetto’, ‘Cabernet Sauvignon’, ‘Chardonnay’, ‘Nebbiolo’, ‘Sangiovese’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GRVFV | Vitis vinifera ‘Muscat Ottonel H-7-3’, ‘Muscat Ottonel H-14-1’ | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | |
| GSyV-1 | Vitis vinifera ‘Trilla’, ‘Szirén’, ‘Muscat Ottonel H-7-3’, ‘Muscat Ottonel H-14-1’ | 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | |
| GVA | Vitis vinifera ‘Grignolino’ | 100% | indirect embryogenesis, stamens, and pistils (2,4-D+BAP) | ELISA, RT-PCR, tissue blot immunoassay | [34] | |
| Vitis vinifera ‘Hencha’ | 100% | direct embryogenesis, stamen (2,4-D+TDZ) | ELISA, RT-PCR | [52] | ||
| Vitis vinifera ‘Cabernet Sauvignon’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GVD | Vitis vinifera ‘Sangiovese’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | |
| GVT | Vitis vinifera ‘Trilla’, ‘Szirén’, ‘Muscat Ottonel H-14-1’ | 100%, 100%, 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | |
| Vitis vinifera ‘Chardonnay’ | 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| LYSV | Allium sativum ‘Istarski crveni’, ‘IPT012’ | 30.4–62.5%, 0–20% | indirect embryogenesiscut cloves (2,4-D/2,4-D+Kin) | ELISA, RT-PCR | [46] | |
| OYDV | Allium sativum ‘Istarski crveni’, ‘IPT012’ | 19.6–50% 0–13.3% | indirect embryogenesiscut cloves (2,4-D/2,4-D+Kin) | ELISA, RT-PCR | [46] | |
| PYMoV | Piper nigrum ‘IISR Malabar Excel’, ‘IISR Shakthi’, ‘IISR Thevam’, ‘Panniyur-1’, ‘Sreekara’, ‘Subhakara’ | 55%, 65%, 100%, 70%, 72%, 72% | embryogenesis, micropylar region of matured seeds (PGR-free) | PCR | [30] | |
| SCYLV | Saccharum sp. ‘CC8527’, ‘CC8215’, ‘R830288’, ‘R831592’, ‘R830395’, ‘R832065’, ‘R840653’, ‘R832276’, ‘G75368’, ‘N27’, ‘Q159’, ‘Q135’, ‘Q155’, ‘Q127’, ‘SP80185’, ‘ROC14’, ‘ROC13’, ‘SP803390’, ‘SP792233’ | 100% | indirect embryogenesis, young leaf rolls (2,4-D) | RT-PCR | [54] | |
| Species/Cultivar | Elimination Efficiency of Regenerated Plants | Key Points of the Protocol 1 | Viroid Diagnostic Method | Reference | ||
|---|---|---|---|---|---|---|
| viroids | ASBVd | Persea americana ‘Vero Beach’ | 0% | embryogenesis, nucellus of immature avocado seeds (picloram) | RT-PCR, fragments were cloned and sequenced | [39] |
| CBCVd (CVd-IV) | Citrus sinensis ‘Maltese’ | 100% | indirect embryogenesis, stigmas, and styles (BAP) | RT-PCR | [55] | |
| CDVd (CVd-III) | Citrus limon ‘Dellys’, ‘Eureka Maroc’, ‘Lunario’, ‘Villafranca’, ‘Sécile’ Citrus sinensis ‘Mitidja navel’, ‘Shamouti de station’ | 100%, 100%, 100%, 100%, 64%, 100%, 100% | indirect embryogenesis, stigmas, and styles (BAP) | RT-PCR | [41] | |
| Citrus sinensis ‘Maltese’ | 100% | indirect embryogenesis, stigmas, and styles (BAP) | RT-PCR | [55] | ||
| CEVd | Citrus limon ‘Béni Abbès’, ‘Bornèo’, ‘Dellys’, ‘Eureka 4’, ‘Femminello’, ‘Lunario’, ‘Sans pépins’, ‘Villafranca’, ‘Sécile’ Citrus sinensis ‘Mitidja navel’, ‘Shamouti de station’ | 100%, 100%, 100%, 100%, 100%, 100%, 100%, 100%, 82%, 100%, 100% | indirect embryogenesis, stigmas, and styles (BAP) | RT-PCR | [41] | |
| Citrus limon ‘Lunario’, ‘Femminello Zagara Bianca’, ‘Femminello Santa Teresa’ | 100% | indirect embryogenesis, styles (BAP) | woody indicator | [56] | ||
| HSVd | Citrus limon ‘Béni Abbès’, ‘Bornèo’, ‘Dellys’, ‘Eureka 4’, ‘Femminello’, ‘Lunario’, ‘Sans pépins’, ‘Villafranca’, ‘Sécile’ Citrus sinensis ‘Mitidja navel’, ‘Shamouti de station’ | 60%, 71%, 75%, 75%, 67%, 100%, 100%, 83%, 64%, 100%, 77% | indirect embryogenesis, stigmas, and styles (BAP) | RT-PCR | [41] | |
| Vitis vinifera ‘Cari’, ‘Proviné’, ‘Roussan’, ‘Nebbiolo’ | 100% | indirect embryogenesis, stamens, and pistils (2,4-D+BAP) | RT-PCR | [40] | ||
| Vitis vinifera ‘Trilla’, ‘Szirén’, ‘Muscat Ottonel H-7-3’, ‘Muscat Ottonel H-14-1’ | 83%, 81%, 100%, 100% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | ||
| Vitis rupestris Vitis sp. ‘Richter 110’ Vitis vinifera ‘Brachetto’, ‘Cabernet Sauvignon’, ‘Chardonnay’, ‘Nebbiolo’, ‘Sangiovese’ | 0% 60% 100%, 100%, 95%, 100%, 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
| GYSVd-1 | Vitis vinifera ‘Cari’, ‘Proviné’, ‘Roussan’, ‘Nebbiolo’ | 100% | indirect embryogenesis, stamens, and pistils (2,4-D+BAP) | RT-PCR | [40] | |
| Vitis vinifera ‘Trilla’, ‘Szirén’, ‘Muscat Ottonel H-7-3’, ‘Muscat Ottonel H-14-1’ | 8.3%, 82%, 86%, 75% | indirect embryogenesis, anthers (2,4-D+TDZ) | RT-PCR, sRNA HTS | [48] | ||
| Vitis rupestris Vitis vinifera ‘Brachetto’, ‘Chardonnay’, ‘Nebbiolo’, ‘Sangiovese’ | 100% 100%, 100%, 73%, 100% | indirect embryogenesis, anthers, and ovaries (2,4-D+BAP) | RNA HTS, RT-qPCR | [25] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olah, R.; Turcsan, M.; Olah, K.; Farkas, E.; Deak, T.; Jahnke, G.; Sardy, D.A.N. Somatic Embryogenesis: A Tool for Fast and Reliable Virus and Viroid Elimination for Grapevine and other Plant Species. Horticulturae 2022, 8, 508. https://doi.org/10.3390/horticulturae8060508
Olah R, Turcsan M, Olah K, Farkas E, Deak T, Jahnke G, Sardy DAN. Somatic Embryogenesis: A Tool for Fast and Reliable Virus and Viroid Elimination for Grapevine and other Plant Species. Horticulturae. 2022; 8(6):508. https://doi.org/10.3390/horticulturae8060508
Chicago/Turabian StyleOlah, Robert, Mihaly Turcsan, Krisztina Olah, Eszter Farkas, Tamas Deak, Gizella Jahnke, and Diana Agnes Nyitraine Sardy. 2022. "Somatic Embryogenesis: A Tool for Fast and Reliable Virus and Viroid Elimination for Grapevine and other Plant Species" Horticulturae 8, no. 6: 508. https://doi.org/10.3390/horticulturae8060508
APA StyleOlah, R., Turcsan, M., Olah, K., Farkas, E., Deak, T., Jahnke, G., & Sardy, D. A. N. (2022). Somatic Embryogenesis: A Tool for Fast and Reliable Virus and Viroid Elimination for Grapevine and other Plant Species. Horticulturae, 8(6), 508. https://doi.org/10.3390/horticulturae8060508








