Tomato Response to Fusarium spp. Infection under Field Conditions: Study of Potential Genes Involved
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sampling
2.2. Evaluation of Fusarium spp. Infection Level in Tomato Plants
2.2.1. gDNA Extraction
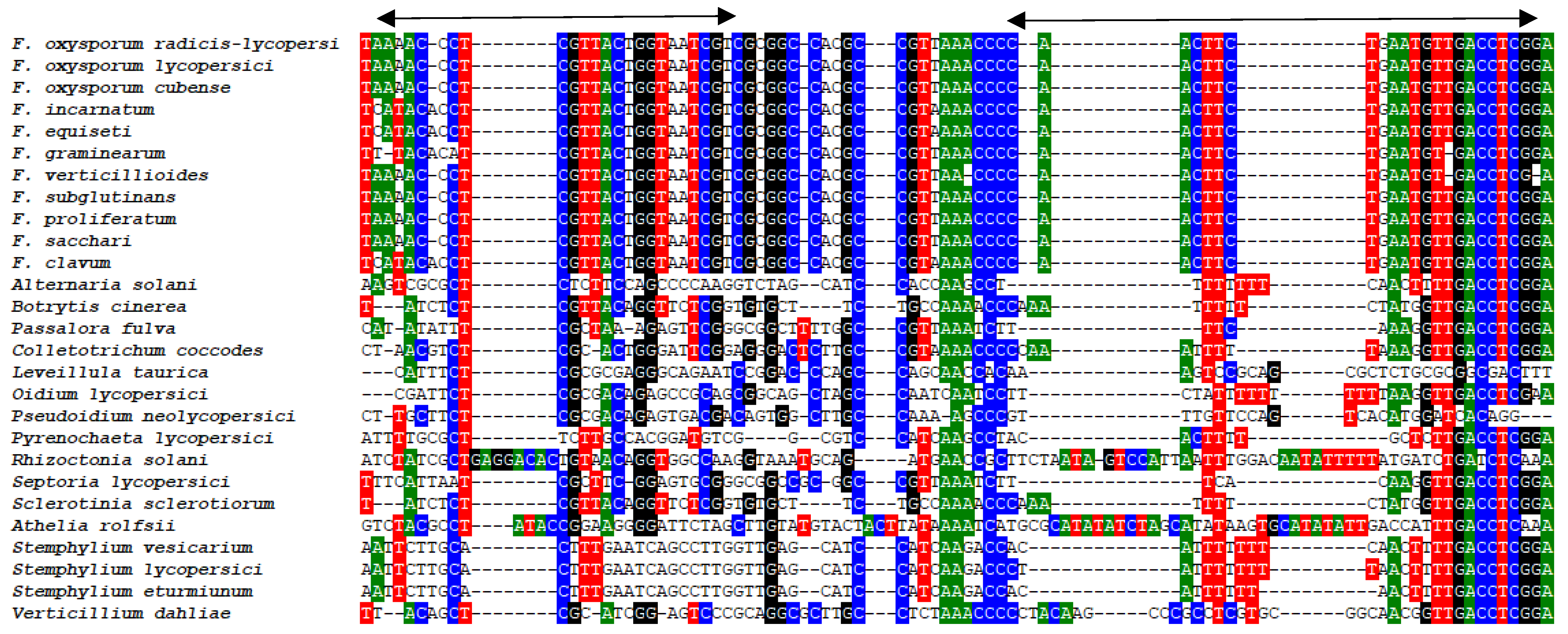
2.2.2. Real-Time Quantitative PCR (qPCR) Conditions for Fusarium spp. Detection and Quantification
2.3. Target Genes Expression
2.3.1. RNA Extraction and Complementary DNA (cDNA) Synthesis
2.3.2. qPCR Conditions for Gene Expression Analysis
2.3.3. Statistical Analysis
3. Results
3.1. Evaluation of Fusarium Infection Level in Tomato Plants
3.2. Target Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bawa, I. Management strategies of Fusarium wilt disease of tomato incited by Fusarium oxysporum f. sp. lycopersici (Sacc.): A review. Int. J. Adv. Acad. Res.|Sci. Technol. Eng. 2016, 2, 2488–9849. [Google Scholar]
- Pritesh, P.; Subramanian, R.B. PCR based method for testing Fusarium wilt resistance of tomato. Afr. J. Basic Appl. Sci. 2011, 3, 219–222. [Google Scholar]
- Almeida, D. Manual de Culturas Hortícolas, 2nd ed.; Editorial Presença: Lisboa, Portugal, 2014. [Google Scholar]
- Jones, J.B.; Stall, R.E.; Zitter, T.A. Compendium of Tomato Diseases and Pests, 2nd ed.; APS Press, The American Phytopathological Society: Saint Paul, MN, USA, 2014. [Google Scholar]
- Elshafie, H.S.; Sakr, S.; Bufo, S.A.; Camele, I. An attempt of biocontrol the tomato-wilt disease caused by Verticillium dahliae using Burkholderia gladioli pv. agaricicola and its bioactive secondary metabolites. Int. J. Plant Biol. 2017, 8, 57–60. [Google Scholar] [CrossRef][Green Version]
- Akbar, A.; Hussain, S.; Ali, G.S. Germplasm Evaluation of Tomato for Resistance to the Emerging Wilt Pathogen Fusarium equiseti. J. Agric. Stud. 2018, 5, 174. [Google Scholar] [CrossRef]
- Manikandan, R.; Harish, S.; Karthikeyan, G.; Raguchander, T. Comparative proteomic analysis of different isolates of Fusarium oxysporum f. sp. lycopersici to exploit the differentially expressed proteins responsible for virulence on tomato plants. Front. Microbiol. 2018, 9, 420. [Google Scholar] [CrossRef] [PubMed]
- Irzykowska, L.; Bocianowski, J.; Waśkiewicz, A.; Weber, Z.; Karolewski, Z.; Goliński, P.; Kostecki, M.; Irzykowski, W. Genetic variation of Fusarium oxysporum isolates forming fumonisin B 1 and moniliformin. J. Appl. Genet. 2012, 53, 237–247. [Google Scholar] [CrossRef]
- Jamiołkowska, A. Pathogenicity of some isolates of Colletotrichum coccodes and Fusarium spp. to sweet pepper (Capsicum annuum) seedlings. Phytopathol. Pol. 2008, 49, 65–71. [Google Scholar]
- Li, P.L.; Shi, Y.X.; Guo, M.Y.; Xie, X.W.; Chai, A.L.; Li, B.J. Fusarium wilt of cauliflower caused by Fusarium equiseti in China. Can. J. Plant Pathol. 2017, 39, 77–82. [Google Scholar] [CrossRef]
- Ramdial, H.; Hosein, F.; Rampersad, S.N. First Report of Fusarium incarnatum associated with fruit disease of bell peppers in Trinidad. Plant Dis. 2016, 100, 526. [Google Scholar] [CrossRef]
- Rowe, R. Comparative pathogenicity and host ranges of Fusarium oxysporum isolates causing crown and root rot of greenhouse and field-grown tomatoes in North America and Japan. Phytopathology 1980, 70, 1143–1148. [Google Scholar] [CrossRef]
- Nelson, P.; Toussoun, T.; Marasas, W. Fusarium Species: An Illustrated Manual for Identification; Park, U., Ed.; Pennsylvania State University Press: University Park, PA, USA, 1983. [Google Scholar]
- McGovern, R.J. Management of tomato diseases caused by Fusarium oxysporum. Crop Prot. 2015, 73, 78–92. [Google Scholar] [CrossRef]
- Paul, N.C.; Park, S.W.; Liu, H.; Choi, S.; Ma, J.; MacCready, J.S.; Chilvers, M.I.; Sang, H. Plant and Fungal Genome Editing to Enhance Plant Disease Resistance Using the CRISPR/Cas9 System. Front. Plant Sci. 2021, 12, 1534. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.D.; Félix, M.D.R.; Patanita, M.; Materatski, P.; Varanda, C. High throughput sequencing unravels tomato-pathogen interactions towards a sustainable plant breeding. Hortic. Res. 2021, 8, 171. [Google Scholar] [CrossRef] [PubMed]
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier-Academic Press: San Diego, CA, USA, 2005. [Google Scholar]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandi, A. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the mediterranean basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- López, W.R.; Garcia-Jaramillo, D.J.; Ceballos-Aguirre, N.; Castaño-Zapata, J.; Acuña-Zornosa, R.; Jovel, J. Transcriptional responses to Fusarium oxysporum f. sp. lycopersici (Sacc.) Snyder & Hansen infection in three Colombian tomato cultivars. BMC Plant Biol. 2021, 21, 412. [Google Scholar] [CrossRef]
- Galindo-González, L.; Deyholos, M.K. RNA-seq transcriptome response of flax (Linum usitatissimum L.) to the pathogenic fungus Fusarium oxysporum f. sp. lini. Front. Plant Sci. 2016, 7, 1766. [Google Scholar] [CrossRef]
- Campos, M.D.; Patanita, M.; Materatski, P.; Albuquerque, A.; Ribeiro, J.A.; Varanda, C. Defense strategies: The role of transcription factors in tomato–pathogen interaction. Biology 2022, 11, 235. [Google Scholar] [CrossRef]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef]
- Vallad, G.E.; Goodmanc, R.M. Review & Interpretation Systemic Acquired Resistance and Induced Systemic Resistance in Conventional Agriculture. Crop Sci. 2004, 44, 1920–1934. [Google Scholar] [CrossRef]
- Zhao, M.; Ji, H.M.; Gao, Y.; Cao, X.X.; Mao, H.Y.; Ouyang, S.Q.; Liu, P. An integrated analysis of mRNA and srna transcriptional profiles in tomato root: Insights on tomato wilt disease. PLoS ONE 2018, 13, e0206765. [Google Scholar] [CrossRef]
- Greeff, C.; Roux, M.; Mundy, J.; Petersen, M. Receptor-like kinase complexes in plant innate immunity. Front. Plant Sci. 2012, 3, 209. [Google Scholar] [CrossRef] [PubMed]
- Akbudak, M.A.; Yildiz, S.; Filiz, E. Pathogenesis related protein-1 (PR-1) genes in tomato (Solanum lycopersicum L.): Bioinformatics analyses and expression profiles in response to drought stress. Genomics 2020, 112, 4089–4099. [Google Scholar] [CrossRef] [PubMed]
- Nair, A.; Kolet, S.P.; Thulasiram, H.V.; Bhargava, S. Role of methyl jasmonate in the expression of mycorrhizal induced resistance against Fusarium oxysporum in tomato plants. Physiol. Mol. Plant Pathol. 2015, 92, 139–145. [Google Scholar] [CrossRef]
- Cheval, C.; Aldon, D.; Galaud, J.P.; Ranty, B. Calcium/calmodulin-mediated regulation of plant immunity. Biochim. Biophys. Acta-Mol. Cell Res. 2013, 1833, 1766–1771. [Google Scholar] [CrossRef]
- Zhang, L.; Du, L.; Poovaiah, B.W. Calcium signaling and biotic defense responses in plants. Plant Signal. Behav. 2014, 9, 11. [Google Scholar] [CrossRef]
- Heinz. Heinz Seed International Brochure; H.J. Heinz Company Brands LLC: Stockton, CA, USA, 2019. [Google Scholar]
- Varanda, C.M.R.; Materatski, P.; Landum, M.; Campos, M.D.; Félix, M.d.R. Fungal Communities Associated with Peacock and Cercospora Leaf Spots in Olive. Plants 2019, 8, 169. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987, 19, 11–15. [Google Scholar]
- Campos, M.D.; Patanita, M.; Campos, C.; Materatski, P.; Varanda, C.M.R.; Brito, I.; Rosário Félix, M. do Detection and quantification of Fusarium spp. (F. oxysporum, F. verticillioides, F. graminearum) and Magnaporthiopsis maydis in maize using real-time PCR targeting the ITS region. Agronomy 2019, 9, 45. [Google Scholar] [CrossRef]
- Hall, T. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Ghareeb, H.; Bozsó, Z.; Ott, P.G.; Wydra, K. Silicon and Ralstonia solanacearum modulate expression stability of housekeeping genes in tomato. Physiol. Mol. Plant Pathol. 2011, 75, 176–179. [Google Scholar] [CrossRef]
- Müller, O.A.; Grau, J.; Thieme, S.; Prochaska, H.; Adlung, N.; Sorgatz, A.; Bonas, U. Genome-wide identification and validation of reference genes in infected tomato leaves for quantitative RT-PCR analyses. PLoS ONE 2015, 10, 8. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.W.; Hoshikawa, K.; Fujita, S.; Thi, D.P.; Mizoguchi, T.; Ezura, H.; Ito, E. Evaluation of internal control genes for quantitative realtime PCR analyses for studying fruit development of dwarf tomato cultivar ‘Micro-Tom’. Plant Biotechnol. 2018, 35, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Vandesompepe, J.; De Prete, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 7. [Google Scholar]
- Clarke, K.R.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 2nd ed.; Plymouth Marine Laboratory: Plymouth, UK, 2001. [Google Scholar]
- Anderson, M.J.; Gorley, R.N.; Clarke, K.R. PERMANOVA+ for PRIMER: Guide to Software and Statistical Methods; PRIMER-E: Plymouth, UK, 2008. [Google Scholar]
- Clarke, K.R.; Green, R.H. Statistical Design and Analysis for a “Biological Effects” Study. Mar. Ecol. Prog. Ser. 1988, 46, 213–226. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Gomes, L.; Nobre, T.; Rei, F.; Félix, M.d.R. Effect of Long-Term Fungicide Applications on Virulence and Diversity of Colletotrichum spp. Associated to Olive Anthracnose. Plants 2019, 8, 311. [Google Scholar] [CrossRef]
- Materatski, P.; Varanda, C.; Carvalho, T.; Dias, A.B.; Campos, M.D.; Rei, F.; Félix, M.d.R. Diversity of Colletotrichum Species Associated with Olive Anthracnose and New Perspectives on Controlling the Disease in Portugal. Agronomy 2018, 8, 301. [Google Scholar] [CrossRef]
- López, M.M.; Llop, P.; Olmos, A.; Marco-Noales, E.; Cambra, M.; Bertolini, E. Are molecular tools solving the challenges posed by detection of plant pathogenic bacteria and viruses? Curr. Issues Mol. Biol. 2009, 11, 13–46. [Google Scholar] [CrossRef]
- Bryan, G.T.; Daniels, M.J.; Osbourn, A.E. Comparison of fungi within the Gaeumannomyces-Phialophora complex by analysis of ribosomal DNA sequences. Appl. Environ. Microbiol. 1995, 61, 681–689. [Google Scholar] [CrossRef]
- Abd-Elsalam, K.A.; Aly, I.N.; Abdel-Satar, M.A.; Khalil, M.S.; Verreet, J.A. PCR identification of Fusarium genus based on nuclear ribosomal-DNA sequence data. Afr. J. Biotechnol. 2003, 2, 96–103. [Google Scholar] [CrossRef][Green Version]
- Singha, I.M.; Kakoty, Y.; Unni, B.G.; Das, J.; Kalita, M.C. Identification and characterization of Fusarium sp. using ITS and RAPD causing fusarium wilt of tomato isolated from Assam, North East India. J. Genet. Eng. Biotechnol. 2016, 14, 99–105. [Google Scholar] [CrossRef]
- Zeng, X.; Kong, F.; Halliday, C.; Chen, S.; Lau, A.; Playford, G.; Sorrell, T.C. Reverse line blot hybridization assay for identification of medically important fungi from culture and clinical specimens. J. Clin. Microbiol. 2007, 45, 2872–2880. [Google Scholar] [CrossRef] [PubMed]
- Lanubile, A.; Pasini, L.; Marocco, A. Differential gene expression in kernels and silks of maize lines with contrasting levels of ear rot resistance after Fusarium verticillioides infection. J. Plant Physiol. 2011, 168, 298. [Google Scholar] [CrossRef]
- Kim, B.R.; Nam, H.Y.; Kim, S.U.; Kim, S.I.; Chang, Y.J. Normalization of reverse transcription quantitative-PCR with housekeeping genes in rice. Biotechnol. Lett. 2003, 25, 1869–1872. [Google Scholar] [CrossRef] [PubMed]
- Volkov, R.A.; Panchuk, I.I.; Schöffl, F. Heat-stress-dependency and developmental modulation of gene expression: The potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR. J. Exp. Bot. 2003, 54, 2343–2349. [Google Scholar] [CrossRef] [PubMed]
- Nicot, N.; Hausman, J.F.; Hoffmann, L.; Evers, D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005, 56, 2907–2914. [Google Scholar] [CrossRef] [PubMed]
- Salava, H.; Thula, S.; Mohan, V.; Kumar, R.; Maghuly, F. Application of genome editing in tomato breeding: Mechanisms, advances, and prospects. Int. J. Mol. Sci. 2021, 22, 682. [Google Scholar] [CrossRef] [PubMed]
- Lian, Q.; He, X.; Zhang, B.; Wang, Y.; Ma, Q. Identification and Characterization of WRKY41, a Gene Conferring Resistance to Powdery Mildew in Wild Tomato (Solanum habrochaites) LA1777. Int. J. Mol. Sci. 2022, 23, 1267. [Google Scholar] [CrossRef]
- Karkute, S.G.; Gujjar, R.S.; Rai, A.; Akhtar, M.; Singh, M.; Singh, B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 2018, 13, 8–17. [Google Scholar] [CrossRef]
- Chinnapandi, B.; Bucki, P.; Braun Miyara, S. SlWRKY45, nematode-responsive tomato WRKY gene, enhances susceptibility to the root knot nematode M. javanica infection. Plant Signal. Behav. 2017, 12, 12. [Google Scholar] [CrossRef]
- Huang, S.; Gao, Y.; Liu, J.; Peng, X.; Niu, X.; Fei, Z.; Cao, S.; Liu, Y. Genome-wide analysis of WRKY transcription factors in Solanum lycopersicum. Mol. Genet. Genom. 2012, 287, 495–513. [Google Scholar] [CrossRef]
- Phukan, U.J.; Jeena, G.S.; Shukla, R.K. WRKY transcription factors: Molecular regulation and stress responses in plants. Front. Plant Sci. 2016, 7, 760. [Google Scholar] [CrossRef] [PubMed]

| Gene | Gene ID | Primer Sequence (5′ → 3′) | AS (bp) | R2 | E | Ref. |
|---|---|---|---|---|---|---|
| TUB | TC170178 a | F: CCTGGTGGTGACCTTGCTAAG R: CTCACCGACATACCAATGCAC | 143 | 0.995 | 103.9 | [35] |
| ACT | U60480 b | F: GGAATCCACGAGACTACATAC R: GGGAAGCCAAGATAGAGC | 228 | 0.990 | 94.4 | [35] |
| PGK | TC181003 a | F: TCTACAAGGCCCAAGGTTATG R: GCAGCAAACTTGTCCGCAATC | 148 | 0.982 | 61.8 | [35] |
| UBI | TC193502 a | F: GGACGGACGTACTCTAGCTGAT R: AGCTTTCGACCTCAAGGGTA | 134 | 0.995 | 90.7 | [36] |
| GAPDH | TC198136 a | F: CTGCTCTCTCAGTAGCCAACAC R: CTTCCTCCAATAGCAGAGGTTT | 156 | 0.998 | 92.3 | [36] |
| PHD | Solyc06g051420.2.1 c | F: GGGATGGGATGGAGCGTAGAGA R: CATCACTCTCCTCTTGCAGCCT | 279 | 0.944 | 71.8 | [36] |
| LmS7 | Solyc09g009640.2.1 c | F: GGTGGAAGACAAGTGGTTGGAACAC R: CGTCTGGCTGAACAAAAGGATTGG | 220 | 0.997 | 99.9 | [36] |
| EXPRESSED | Solyc07g025390.2.1 c | F: GCTAAGAACGCTGGACCTAATG R: TGGGTGTGCCTTTCTGAATG | 183 | 0.999 | 95.0 | [37] |
| Gene | Gene ID | Primer Sequence (5′ → 3′) | AS (bp) | R2 | E | Ref. |
|---|---|---|---|---|---|---|
| WRKY41 | Solyc01g095630 a | F: TCCTCATTTGGTGGAGAAGG R: TAGCTTAGGATCAATTAGGC | 171 | 0.997 | 102.0 | [24] |
| WRKY40 | Solyc06g068460 a | F: GAGTTGGCTAGATTGAGACTG R: TTGATGCCACAAAAGAGTTG | 144 | 0.999 | 97.0 | [24] |
| RLK | Solyc03g059080 a | F: GCAGTGTGTAGATCCTAAGC R: CAGTGCCTTGACGACAATTG | 210 | 0.995 | 103.2 | [24] |
| PR1b | Solyc00g174340 a | F: ATACTCAAGTAGTCTGGCGC R: GTAAGGACGTTGTCCGATCC | 106 | 0.972 | 98.2 | [24] |
| CBEF | Solyc10g006660 a | F: ATTAAGTCCTGAGTTGATGG R: GATAACAGTGCATCAGAAGGG | 107 | 0.951 | 110.8 | [24] |
| CNGC | Solyc05g050350 a | F: CACAAATGCATCAAGTCTTGG R: CTAAAATCTGGTTCAGCTGG | 141 | 0.991 | 103.3 | [24] |
| OPR3 | NM_001246944 b | F: CCTCTTTCAAACAACAATGGCG R: TGAACTTGCCCATCTTGTAAGG | 115 | 0.996 | 101.4 | [27] |
| PR1 | EU589238 b | F: GCAGCTCGTAGACAAGTTGGAGTCG R: TGTTGCATCCTGCAGTCCCC | 107 | 0.995 | 100.8 | [27] |
| Sample | Ct Mean | ±SE |
|---|---|---|
| 1 | ND | - |
| 2 | 21.34 | ±0.095 |
| 3 | 26.38 | ±0.022 |
| 4 | 23.16 | ±0.015 |
| 5 | 23.77 | ±0.150 |
| 6 | 24.62 | ±0.060 |
| 7 | ND | - |
| 8 | 25.01 | ±0.425 |
| 9 | 23.36 | ±0.000 |
| 10 | 21.24 | ±0.005 |
| 11 | 22.37 | ±0.200 |
| 12 | 21.47 | ±0.055 |
| 13 | 24.11 | ±0.085 |
| 14 | 21.98 | ±0.145 |
| 15 | ND | - |
| 16 | 23.38 | ±0.170 |
| 17 | ND | - |
| 18 | 22.28 | ±0.010 |
| 19 | 17.8 | ±0.090 |
| 20 | 24.04 | ±0.031 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, J.A.; Albuquerque, A.; Materatski, P.; Patanita, M.; Varanda, C.M.R.; Félix, M.d.R.; Campos, M.D. Tomato Response to Fusarium spp. Infection under Field Conditions: Study of Potential Genes Involved. Horticulturae 2022, 8, 433. https://doi.org/10.3390/horticulturae8050433
Ribeiro JA, Albuquerque A, Materatski P, Patanita M, Varanda CMR, Félix MdR, Campos MD. Tomato Response to Fusarium spp. Infection under Field Conditions: Study of Potential Genes Involved. Horticulturae. 2022; 8(5):433. https://doi.org/10.3390/horticulturae8050433
Chicago/Turabian StyleRibeiro, Joana A., André Albuquerque, Patrick Materatski, Mariana Patanita, Carla M. R. Varanda, Maria do Rosário Félix, and Maria Doroteia Campos. 2022. "Tomato Response to Fusarium spp. Infection under Field Conditions: Study of Potential Genes Involved" Horticulturae 8, no. 5: 433. https://doi.org/10.3390/horticulturae8050433
APA StyleRibeiro, J. A., Albuquerque, A., Materatski, P., Patanita, M., Varanda, C. M. R., Félix, M. d. R., & Campos, M. D. (2022). Tomato Response to Fusarium spp. Infection under Field Conditions: Study of Potential Genes Involved. Horticulturae, 8(5), 433. https://doi.org/10.3390/horticulturae8050433









