An Efficient Agrobacterium-Mediated Genetic Transformation System for Persimmon (Diospyros kaki Thunb.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Plant Growth Regulator Combinations for Shoot Multiplication
2.3. Regeneration and Rooting of Adventitious Buds
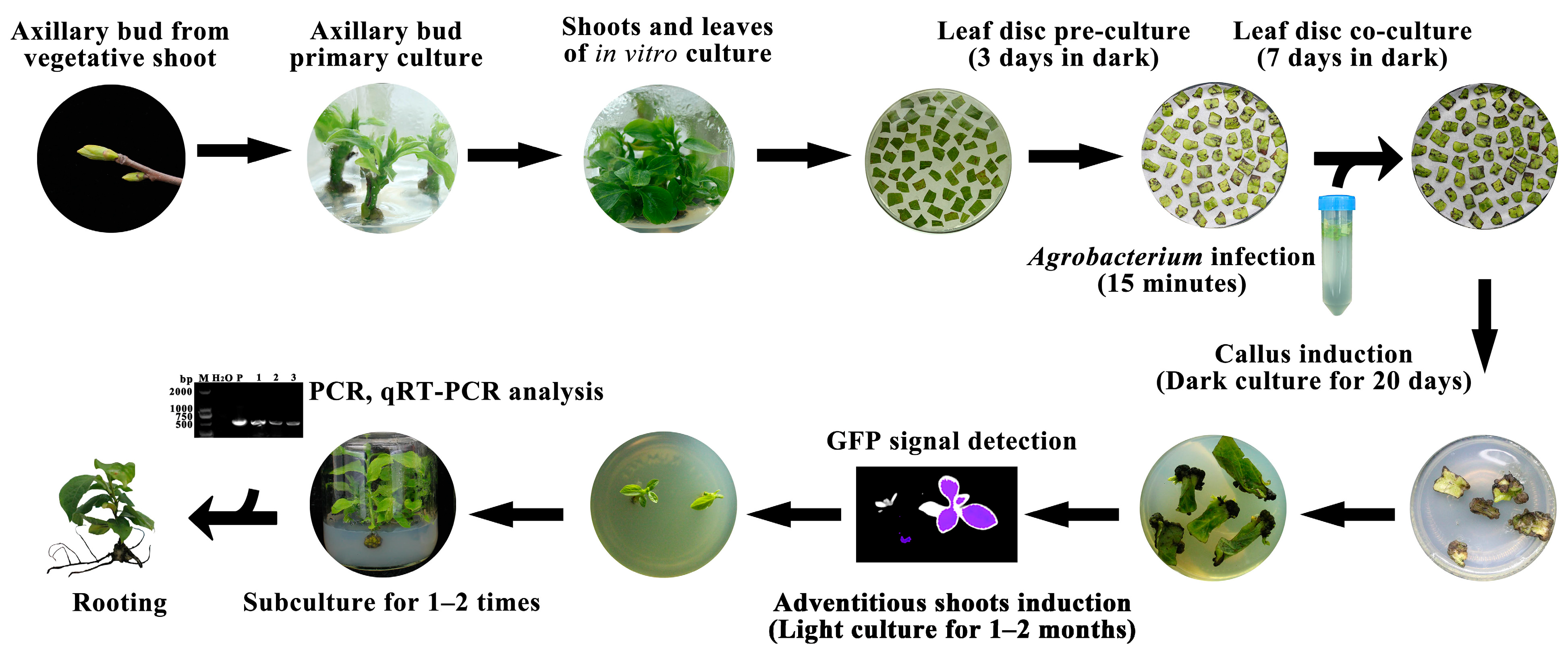
2.4. Agrobacterium tumefaciens-Mediated Gene Transformation
2.5. Green Fluorescent Protein Detection
2.6. Molecular Detection of Transformants
2.7. Southern Blotting Assay to Identify Positive Transgenic Plants
2.8. RNA Extraction and cDNA Synthesis
2.9. Quantitative Reverse Transcription PCR (qRT–PCR)
2.10. PAs Content Determination
3. Results and Discussion
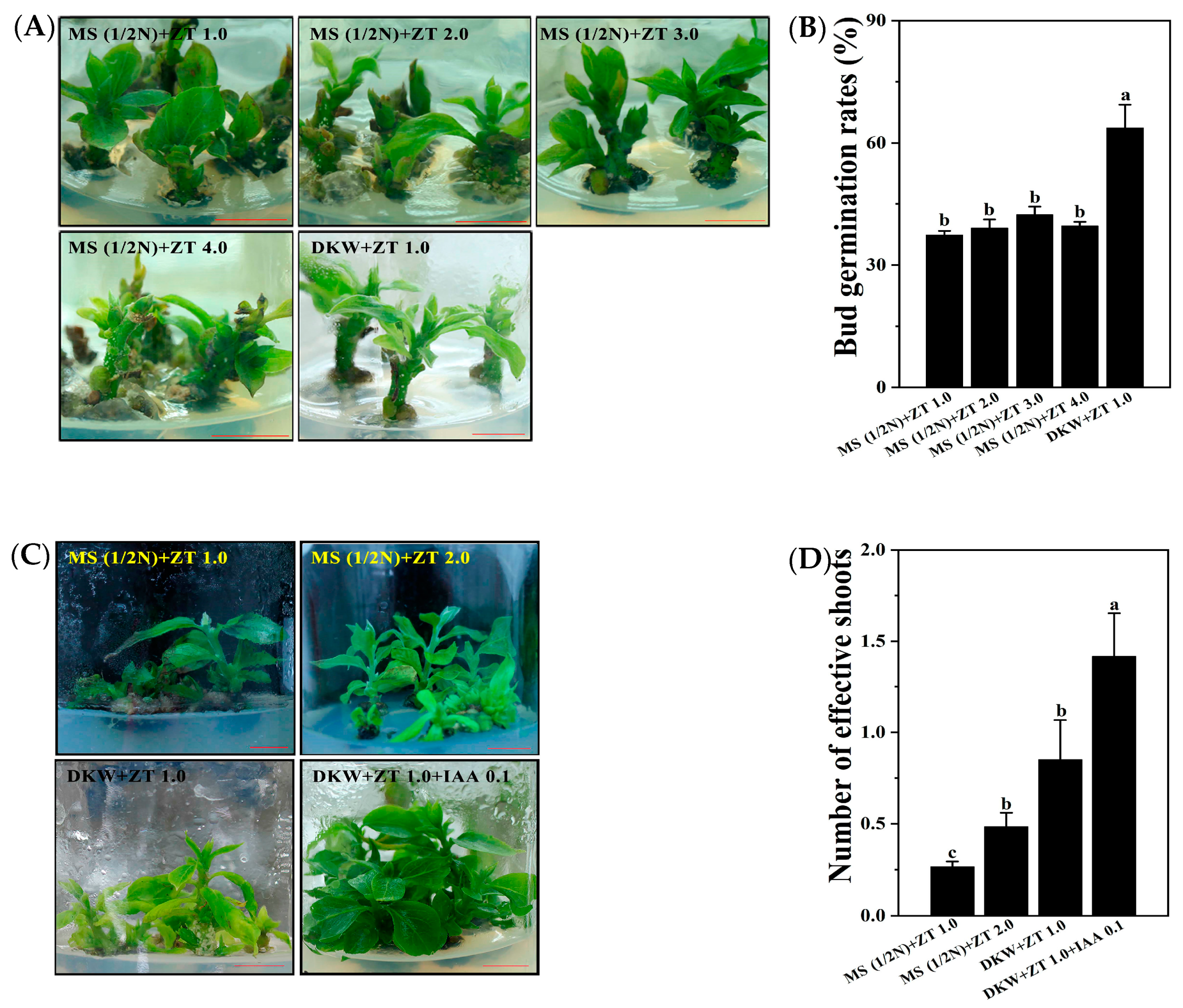
3.1. The Primary Culture and Subculture Proliferation of ‘Gongcheng Shuishi’
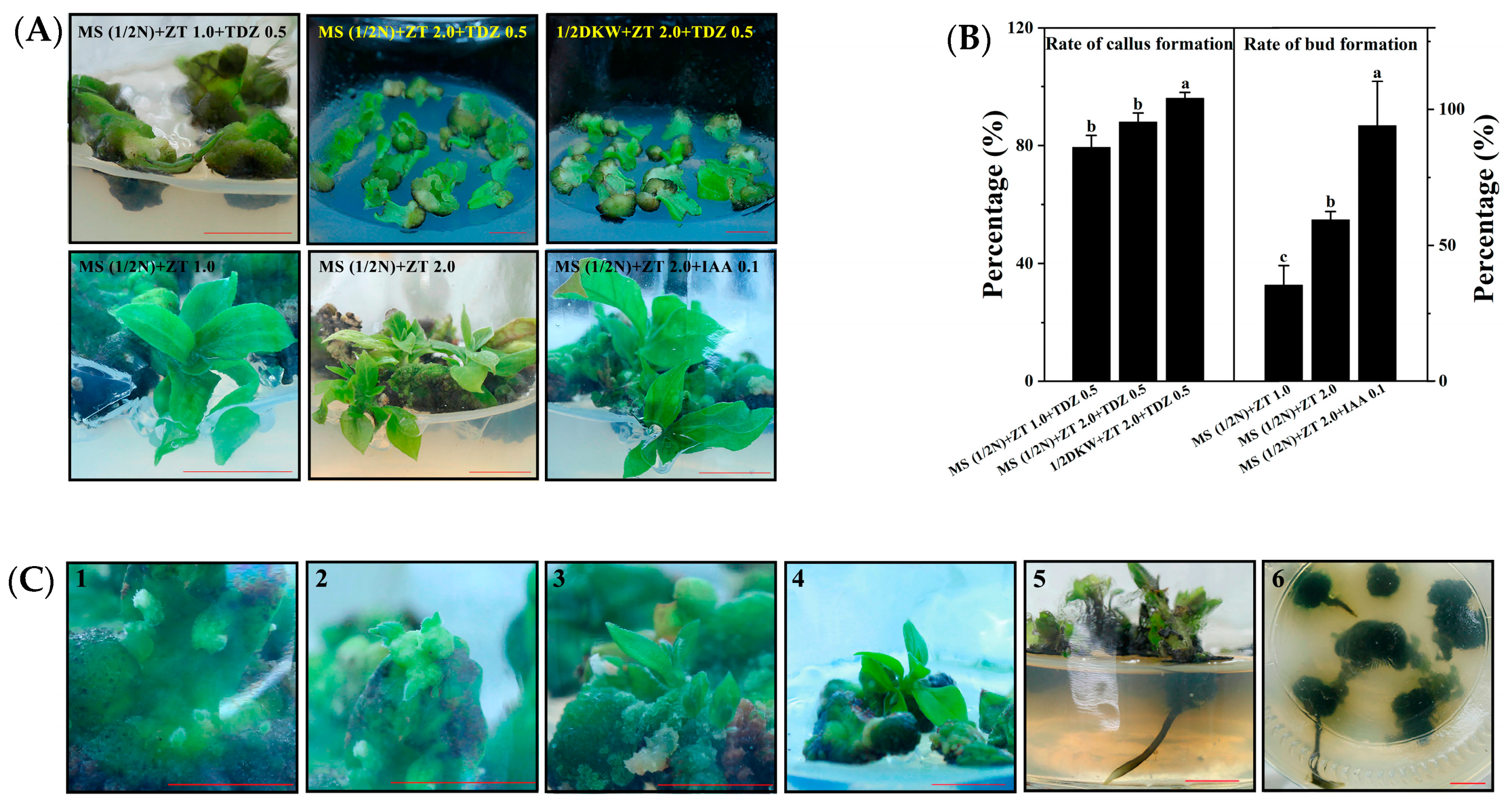
3.2. ‘Gongcheng Shuishi’ Persimmon Shoots Formation from Leaf Explants
3.3. Rooting of Regenerated Shoots
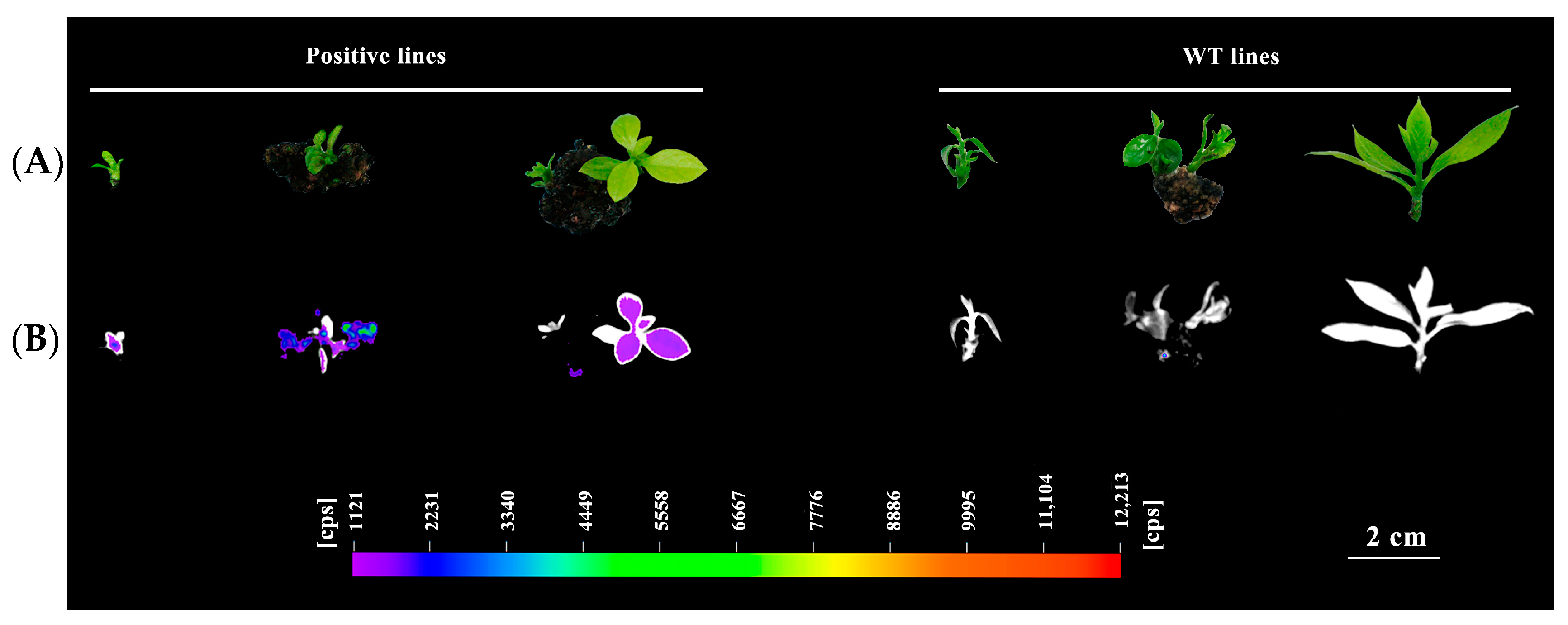
3.4. Transgenic Persimmon Plants with GFP
3.5. Identification of Stable Regenerated Transgenic Seedlings
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Tao, R.; Dandekar, A.M. Genetic Transformation of Diospyros kaki L. (Japanese Persimmon). In Transgenic Trees; Bajaj, Y.P.S., Ed.; Springer: Berlin/Heidelberg, Germany, 2000; Volume 44, pp. 77–87. [Google Scholar] [CrossRef]
- Akagi, T.; Katayama-Ikegami, A.; Yonemori, K. Proanthocyanidin biosynthesis of persimmon (Diospyros kaki Thunb.) fruit. Sci. Hort. 2011, 130, 373–380. [Google Scholar] [CrossRef]
- Akagi, T.; Shirasawa, K.; Nagasaki, H.; Hirakawa, H.; Tao, R.; Comai, L.; Henry, I.M. The persimmon genome reveals clues to the evolution of a lineage-specific sex determination system in plants. PLoS Genet. 2020, 16, e1008566. [Google Scholar] [CrossRef]
- Luo, C.; Zhang, Q.L.; Luo, Z.R. Genome-wide transcriptome analysis of Chinese pollination-constant nonastringent persimmon fruit treated with ethanol. BMC Genom. 2014, 15, 112. [Google Scholar] [CrossRef]
- Chen, W.X.; Xiong, Y.L.; Xu, L.Q.; Zhang, Q.L.; Luo, Z.R. An integrated analysis based on transcriptome and proteome reveals deastringency-related genes in CPCNA persimmon. Sci. Rep. 2017, 7, 44671. [Google Scholar] [CrossRef]
- Luo, Y.J.; Zhang, X.N.; Luo, Z.R.; Zhang, Q.L.; Liu, J.H. Identification and characterization of microRNAs from Chinese pollination constant non-astringent persimmon using high-throughput sequencing. BMC Plant Biol. 2015, 15, 11. [Google Scholar] [CrossRef]
- Tao, R.; Handa, T.; Tamura, M.; Sugiura, A. Genetic transformation of Japanese persimmon (Diospyros kaki L.) by Agrobacterium rhizogenes wild type strain A4. J. Jpn. Soc. Hort. Sci. 1994, 63, 283–289. [Google Scholar] [CrossRef][Green Version]
- Tao, R.; Dandekar, A.M.; Uratsu, S.L.; Vail, P.V.; Tebbts, J.S. Engineering genetic resistance against insects in Japanese persimmon using the cryIA(c) gene of Bacillus thuringiensis. J. Amer. Soc. Hort. Sci. 1997, 122, 764–771. [Google Scholar] [CrossRef]
- Nakamura, Y.; Kobayashi, S.; Nakajima, I. Agrobacterium-mediated transformation and plant regeneration from hypocotyl segments of Japanese persimmon (Diospyros kaki Thunb). Plant Cell Rep. 1998, 17, 435–440. [Google Scholar] [CrossRef]
- Li, X.H.; Jiang, Z.Y.; Shen, Y.Y.; Li, F.H.; Yu, X.Y.; Qu, S.C. In vitro regeneration and Agrobacterium tumefaciens-mediated genetic transformation of D. lotus (Diospyros lotus L.). Sci. Hort. 2018, 236, 229–237. [Google Scholar] [CrossRef]
- Koshita, Y.; Nakamura, Y.; Kobayashi, S.; Morinaga, K. Introduction of the rolC gene into the genome of the Japanese persimmon causes dwarfism. J. Jpn. Soc. Hort. Sci. 2002, 71, 529–531. [Google Scholar] [CrossRef]
- Gao, M.; Sakamoto, A.; Miura, K.; Murata, N.; Sugiura, A.; Tao, R. Transformation of Japanese persimmon (Diospyros kaki Thunb.) with a bacterial gene for choline oxidase. Mol. Breed. 2000, 6, 501–510. [Google Scholar] [CrossRef]
- Gao, M.; Tao, R.; Miura, K.; Dandekar, A.M.; Sugiura, A. Transformation of Japanese persimmon (Diospyros kaki Thunb.) with apple cDNA encoding NADP-dependent sorbitol-6-phosphate dehydrogenase. Plant Sci. 2001, 160, 837–845. [Google Scholar] [CrossRef]
- Deguchi, M.; Koshita, Y.; Gao, M.; Tao, R.; Tetsumura, T.; Yamaki, S.; Kanayama, Y. Engineered sorbitol accumulation induces dwarfism in Japanese persimmon. J. Plant Physiol. 2004, 161, 1177–1184. [Google Scholar] [CrossRef]
- Tamura, M.; Gao, M.; Tao, R.; Labavitch, J.M.; Dandekar, A.M. Transformation of persimmon with a pear fruit polygalacturonase inhibiting protein (PGIP) gene. Sci. Hort. 2004, 103, 19–30. [Google Scholar] [CrossRef]
- Mo, R.L.; Huang, Y.M.; Yang, S.C.; Zhang, Q.L.; Luo, Z.R. Development of Agrobacterium-mediated transient transformation in persimmon (Diospyros kaki Thunb.). Sci. Hort. 2015, 192, 29–37. [Google Scholar] [CrossRef]
- Mo, R.L.; Yang, S.C.; Zhang, Q.L.; Xu, L.Q.; Luo, Z.R. Vacuum infiltration enhances the Agrobacterium-mediated transient transformation for gene functional analysis in persimmon (Diospyros kaki Thunb.). Sci. Hort. 2019, 251, 174–180. [Google Scholar] [CrossRef]
- Zhang, M.; Wang, S.D.; Xu, L.Q.; Luo, Z.R.; Zhang, Q.L. Development of Agrobacterium-mediated transient transformation for fruit discs in persimmon (Diospyros kaki Thunb.). Eur. J. Hort. Sci. 2021. [Google Scholar] [CrossRef]
- Sugiura, A.; Matsuda-Habu, Y.; Gao, M.; Esumi, T.; Tao, R. Somatic embryogenesis and plant regeneration from immature persimmon (Diospyros kaki Thunb.) embryos. HortScience 2008, 43, 211–214. [Google Scholar] [CrossRef]
- Tao, R.; Sugiura, A. Adventitious bud formation from callus cultures of Japanese persimmon. HortScience 1992, 27, 259–261. [Google Scholar] [CrossRef]
- Tetsumura, T.; Yukinaga, H. High-frequency shoot regeneration from roots of Japanese persimmon. HortScience 1996, 31, 463–464. [Google Scholar] [CrossRef]
- Yin, H.; Song, C.L.; Ma, J.L.; Zhang, Z.D.; Tang, X. Optimization of Agrobacterium tumefaciens mediated genetic transformation system of Mopanshi. Acta Agric. Boreali-Sin. 2007, 22, 56–59. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=HBNB200702014&DbName=CJFQ2007 (accessed on 1 March 2022).
- Ikegami, A.; Eguchi, S.; Kitajima, A.; Inoue, K.; Yonemori, K. Identification of genes involved in proanthocyanidin biosynthesis of persimmon (Diospyros kaki) fruit. Plant Sci. 2007, 172, 1037–1047. [Google Scholar] [CrossRef]
- Akagi, T.; Ikegami, A.; Tsujimoto, T.; Kobayashi, S.; Sato, A.; Kono, A.; Yonemori, K. DkMyb4 is a Myb transcription factor involved in proanthocyanidin biosynthesis in persimmon fruit. Plant Physiol. 2009, 151, 2028–2045. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.P.; Vimolmangkang, S.; Soria-Guerra, R.E.; Korban, S.S. Introduction of apple ANR genes into tobacco inhibits expression of both CHI and DFR genes in flowers, leading to loss of anthocyanin. J. Exp. Bot. 2012, 63, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.X.; Du, G.Q.; Ma, J.L.; Gao, Y.; Wang, L. Callus formation and adventitious bud regeneration from leaves in vitro in persimmon (Diospyros kaki). J. Fruit Sci. 2004, 21, 376–378. [Google Scholar] [CrossRef]
- Doyle, J.I.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. Available online: https://www.scienceopen.com/document?vid=9f54eaa3-acbe-4864-a21c-7ab44385e409 (accessed on 1 March 2022).
- Oshida, M.; Yonemori, K.; Sugiura, A. On the nature of coagulated tannins in astringent-type persimmon fruit after an artificial treatment of astringency removal. Postharvest Biol. Technol. 1996, 8, 317–327. [Google Scholar] [CrossRef]
- Li, Y.G.; Tanner, G.; Larkin, P. The DMACA-HCl protocol and the threshold proanthocyanidin content for bloat safety in forage legumes. J. Sci. Food Agric. 1996, 70, 89–101. [Google Scholar] [CrossRef]
- Giordani, E.; Naval, M.; Benelli, C. In vitro propagation of persimmon (Diospyros kaki Thunb.). In Protocols for Micropropagation of Selected Economically-Important Horticultural Plants, Methods in Molecular Biology; Lambardi, M., Ozudogru, E.A., Jain, S.M., Eds.; Springer: New York, NY, USA, 2013; Volume 994, pp. 89–98. [Google Scholar] [CrossRef]
- Witte, C.P.; Tiller, S.A.; Taylor, M.A.; Davies, H.V. Addition of nickel to Murashige and Skoog medium in plant tissue culture activates urease and may reduce metabolic stress. Plant Cell Tiss. Org. 2002, 68, 103–104. [Google Scholar] [CrossRef]
- Li, J.; Luo, Y.J.; Zhang, Q.L.; Luo, Z.R.; Liu, J.H. In vivo culture system optimization and regeneration of date plum (Diospyros lotus Linn.) dormant buds and leaves. J. Huazhong Agric. Univ. 2016, 35, 14–19. [Google Scholar] [CrossRef]
- Fukui, H.; Sugiyama, M.; Nakamura, M. Shoot tip culture of Japanese persimmon (Diospyros kaki Thunb.). J. Jpn. Soc. Hort. Sci. 1989, 58, 43–47. [Google Scholar] [CrossRef]
- Fellman, C.D.; Read, P.E.; Hosier, M.A. Effects of thidiazuron and CPPU on meristem formation and shoot proliferation. HortScience. 1987, 22, 1197–1200. Available online: http://pascal-francis.inist.fr/vibad/index.php?action=getRecordDetail&idt=7678477 (accessed on 1 March 2022).
- Yokoyama, T.; Moriyasu, Y.; Sugawara, Y. Adventitious bud formation through nodule induction by thidiazuron in cultured leaf segments of the Japanese persimmon (Diospyros kaki Thunb.). Plant Biotechnol. 2011, 28, 339–344. [Google Scholar] [CrossRef][Green Version]
- Delia, G.; Farina, L.; Buccheri, M.; Rosato, S.; Insero, O.; Rega, P.; Damiano, C. Proteomic analysis of in vitro rooted persimmon (cv. ‘Kaki Tipo’) plants in response to various auxins and light conditions. Acta Hort. 2009, 833, 171–176. [Google Scholar] [CrossRef]
- Ma, J.L.; Liu, X.N.; Zhang, Z.D.; Yuan, S.Q. Rooting and adventitious shoot regeneration from root of Uenishiwase persimmon. Acta Agric. Boreali-Sin. 2004, 19, 115. Available online: https://kns.cnki.net/kcms/detail/detail.aspx?FileName=HBNB200404033&DbName=CJFQ2004 (accessed on 1 March 2022).
- Gambino, G.; Gribaudo, I. Genetic transformation of fruit trees: Current status and remaining challenges. Transgenic Res. 2012, 21, 1163–1181. [Google Scholar] [CrossRef]
- Tao, R. Agrobacterium-mediated genetic transformation in Japanese persimmon. Acta Hort. 2003, 601, 57–63. [Google Scholar] [CrossRef]
- Chen, W.X.; Zheng, Q.Y.; Li, J.W.; Liu, Y.; Xu, L.Q.; Zhang, Q.L.; Luo, Z.R. DkMYB14 is a bifunctional transcription factor that regulates the accumulation of proanthocyanidin in persimmon fruit. Plant J. 2021, 106, 1708–1727. [Google Scholar] [CrossRef]
- Zhu, Q.G.; Gong, Z.Y.; Wang, M.M.; Li, X.; Grierson, D.; Yin, X.R.; Chen, K.S. A transcription factor network responsive to high CO2/hypoxia is involved in deastringency in persimmon fruit. J. Exp. Bot. 2018, 69, 2061–2070. [Google Scholar] [CrossRef]
- Cui, M.L.; Liu, C.; Piao, C.L.; Liu, C.L. A stable Agrobacterium rhizogenes-mediated transformation of cotton (Gossypium hirsutum L.) and plant regeneration from transformed hairy root via embryogenesis. Front. Plant Sci. 2020, 11, 604255. [Google Scholar] [CrossRef]
- Yang, S.C.; Zhang, M.; Xu, L.Q.; Luo, Z.R.; Zhang, Q.L. MiR858b inhibits proanthocyanidin accumulation by the repression of DkMYB19 and DkMYB20 in persimmon. Front. Plant Sci. 2020, 11, 576378. [Google Scholar] [CrossRef] [PubMed]

| Donor Gene | Acceptor Genotype | Explants | Total Time of Callus Formation | Regeneration Rate (Positive)% | References |
|---|---|---|---|---|---|
| cryIA(c) | J-PCNA ‘Jiro’ | Leaf disc | 2 months | 2.1 (1.5) | [8] |
| - | J-PCNA ‘Saijo’ | Hypocotyl segments | 4–5 months | 27.0 (11.1) | [9] |
| codA | J-PCNA ‘Jiro’ | Leaf disc | - | 20.0 | [12] |
| S6PDH | J-PCNA ‘Jiro’ | Leaf disc | - | 3.5 (2.5) | [13] |
| PGIP | J-PCNA ‘Jiro’ | Leaf disc | 1 month | 7.8 (5.5) | [15] |
| LAC | C-PCNA‘Baogai Tianshi’, ‘Eshi 1’, non-PCNA ‘Mopanshi’ | Leaves in vivo | [16] | ||
| LAR | C-PCNA ‘Xiaoguo Tianshi’, non-PCNA ‘Mopanshi’ | Leaves in vivo | [17] | ||
| ANR | Non-PCNA ‘Gongheng Shuishi’ | Fruit discs | [18] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, M.; Yang, S.; Chen, W.; Xu, L.; Guo, D.; Luo, Z.; Zhang, Q. An Efficient Agrobacterium-Mediated Genetic Transformation System for Persimmon (Diospyros kaki Thunb.). Horticulturae 2022, 8, 422. https://doi.org/10.3390/horticulturae8050422
Zhang M, Yang S, Chen W, Xu L, Guo D, Luo Z, Zhang Q. An Efficient Agrobacterium-Mediated Genetic Transformation System for Persimmon (Diospyros kaki Thunb.). Horticulturae. 2022; 8(5):422. https://doi.org/10.3390/horticulturae8050422
Chicago/Turabian StyleZhang, Meng, Sichao Yang, Wenxing Chen, Liqing Xu, Dayong Guo, Zhengrong Luo, and Qinglin Zhang. 2022. "An Efficient Agrobacterium-Mediated Genetic Transformation System for Persimmon (Diospyros kaki Thunb.)" Horticulturae 8, no. 5: 422. https://doi.org/10.3390/horticulturae8050422
APA StyleZhang, M., Yang, S., Chen, W., Xu, L., Guo, D., Luo, Z., & Zhang, Q. (2022). An Efficient Agrobacterium-Mediated Genetic Transformation System for Persimmon (Diospyros kaki Thunb.). Horticulturae, 8(5), 422. https://doi.org/10.3390/horticulturae8050422






