Genome-Wide Analyses of Tea Plant Stress-Associated Proteins (SAPs) Reveal the Role of CsSAP12 in Increased Drought Tolerance in Transgenic Tomatoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of CsSAPs in Tea Plant
2.2. Evolutionary Analysis of Tea Plant CsSAPs
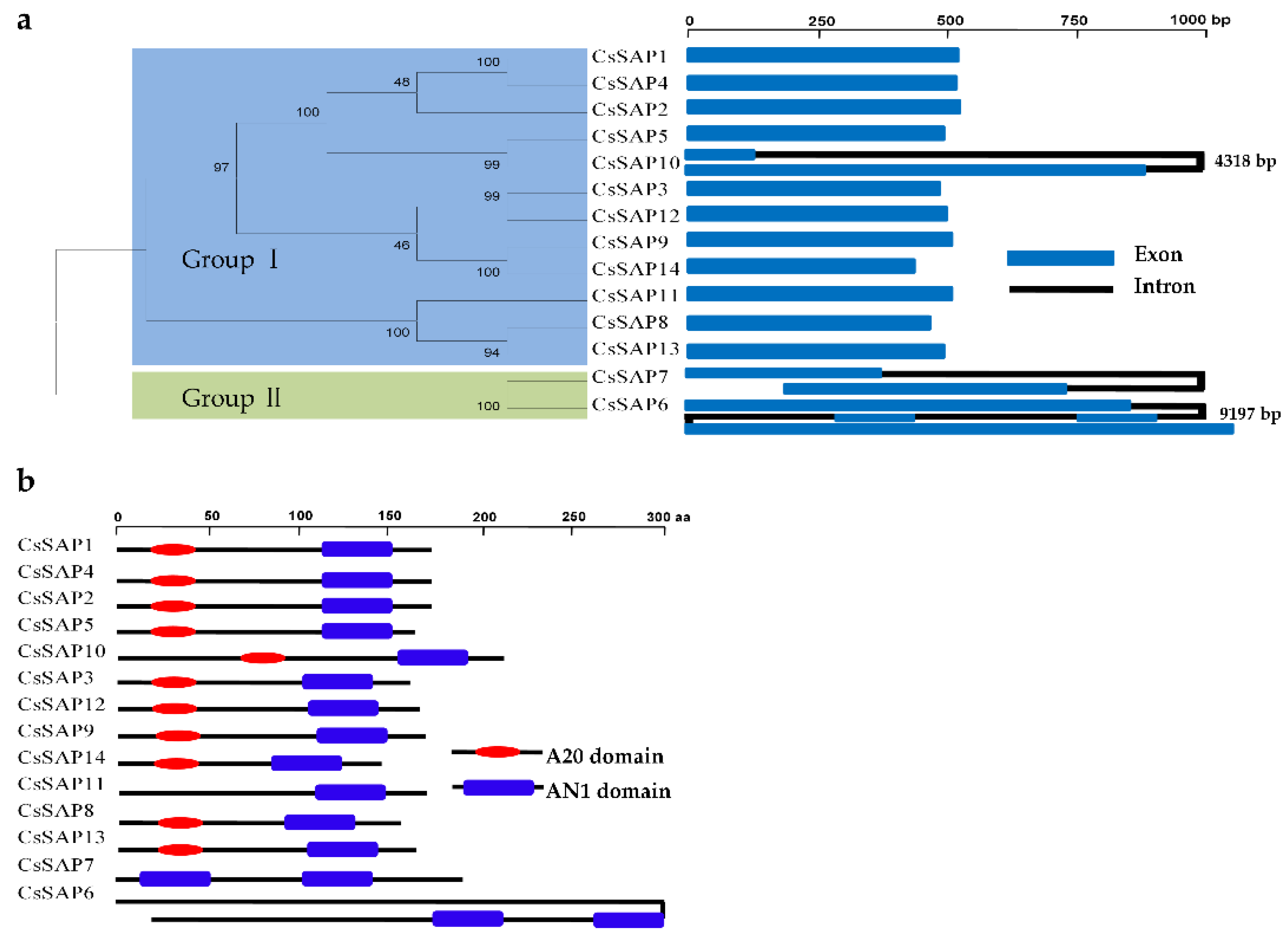
2.3. Gene Structure and Genome Localization of CsSAP Genes
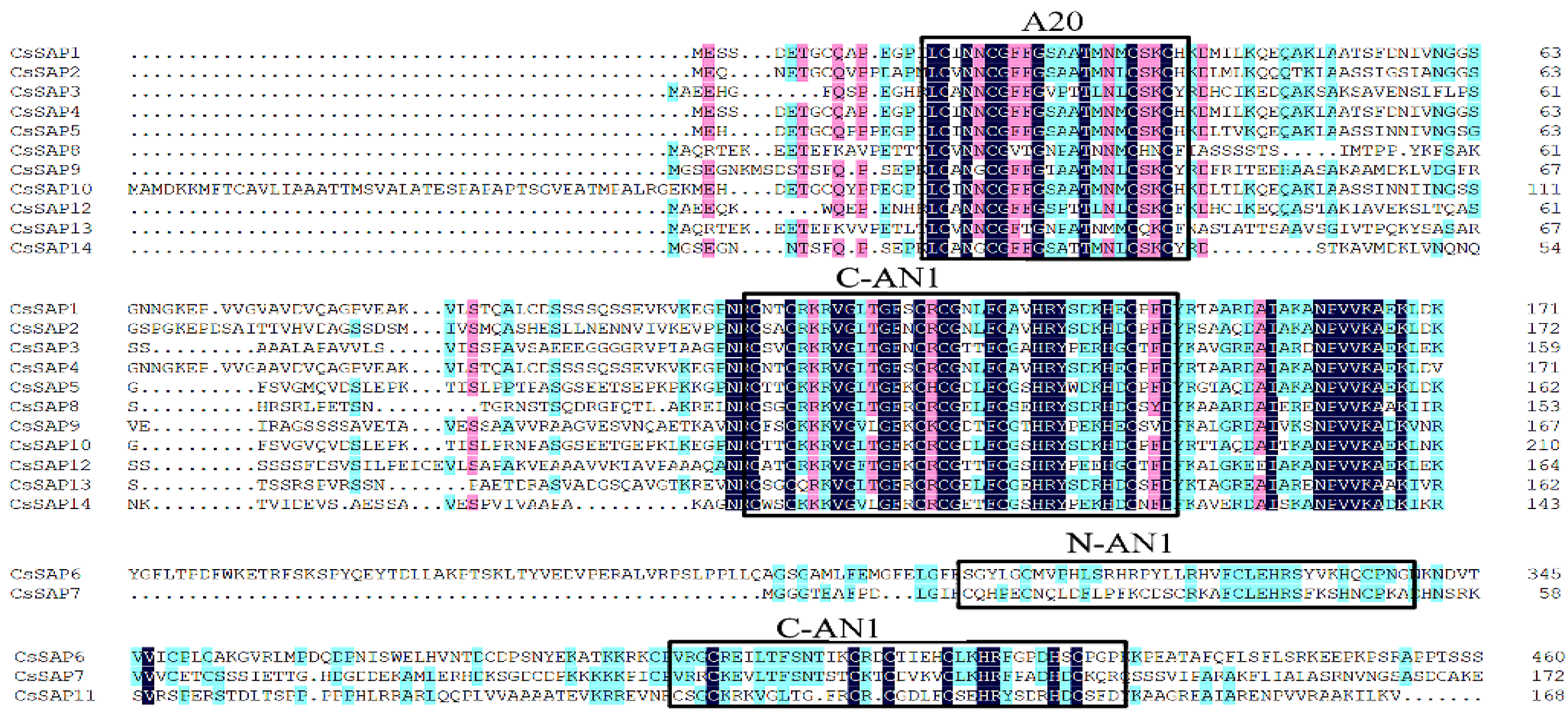
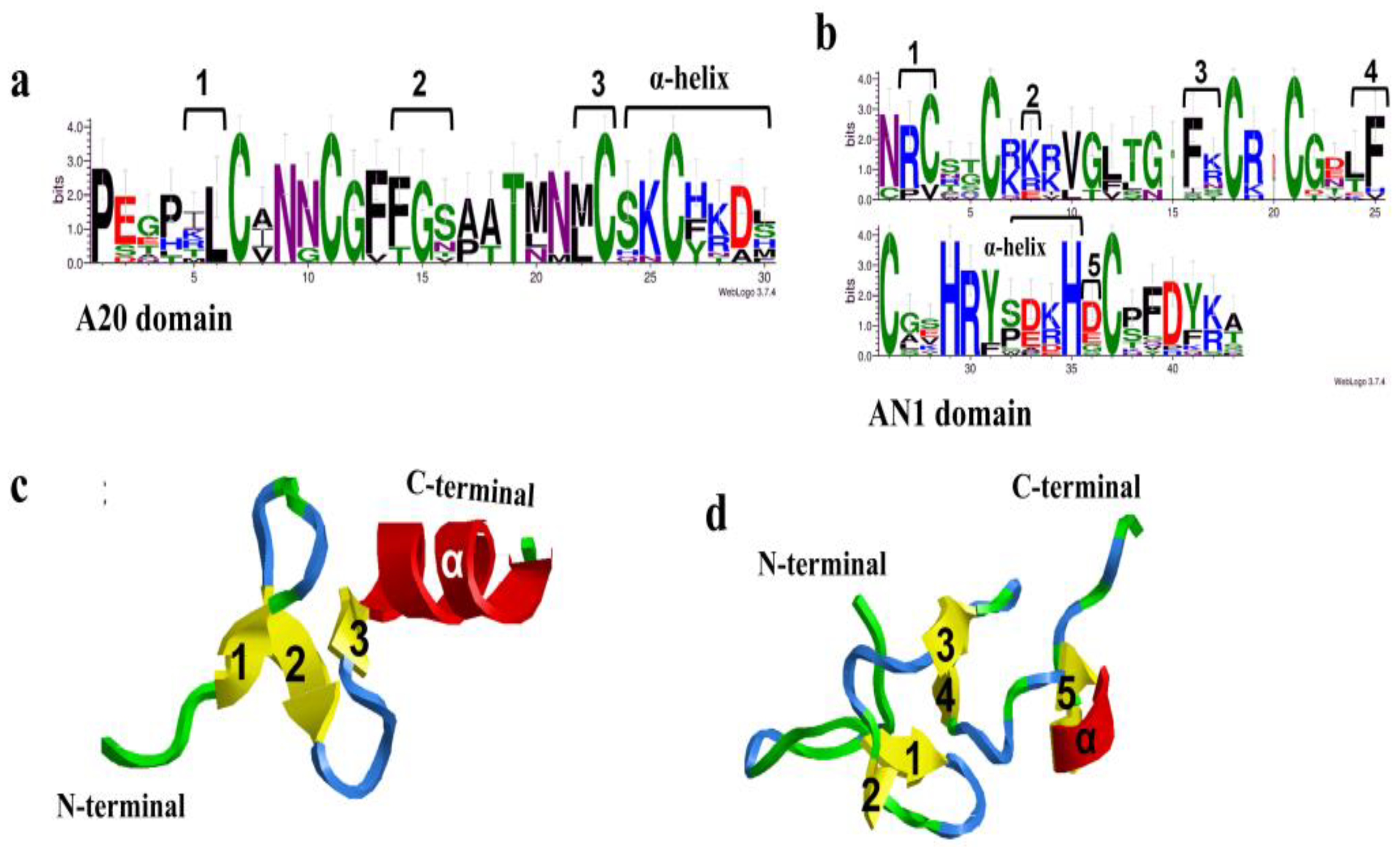
2.4. Amino Acid Sequences and Conserved Elements of CsSAPs
2.5. Cis-Acting Elements in Promoters
2.6. Plant Materials, Growth Conditions and Stress Treatments
2.7. The qRT-PCR Analysis of CsSAPs
2.8. Vector Construction, Plant Transformation and Drought Stress Treatment
2.9. Measurement of Physiological Indices
3. Results
3.1. Identification of CsSAPs in Tea Plant
3.2. Analysis of CsSAPs Structure and Conservative Elements
3.3. Phylogenetic Analysis of CsSAPs
3.4. Functional Prediction of CsSAPs
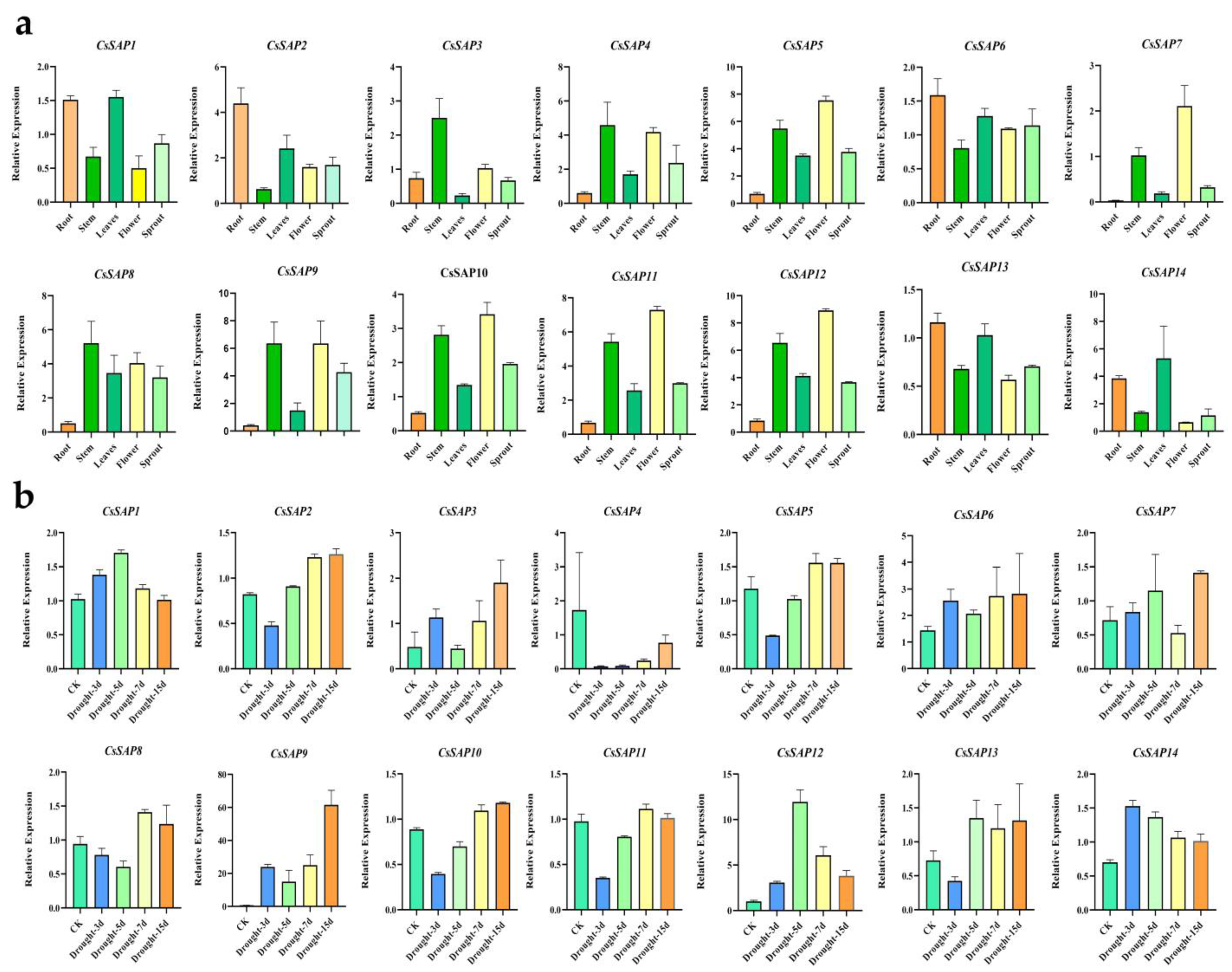
3.5. Expression Pattern of CsSAPs in Tea Plant
3.6. CsSAP12 Overexpression Enhances Drought Tolerance in Transgenic Tomato
3.7. Measurement of Physiological Indices of CsSAP12 Transgenic Plants
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gill, S.S.; Tuteja, N. Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 2010, 5, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Huang, C.; Zhang, X.; Zhang, L.; Wu, Z. Advances of plant zinc finger proteins involved in abiotic stress. Biotechnol. Bull. 2005, 6, 12–16. [Google Scholar]
- Gao, W.; Long, L.; Tian, X.; Jin, J.; Liu, H.; Zhang, H.; Xu, F.; Song, C. Genome-wide identification and expression analysis of stress-associated proteins (SAPs) containing A20/AN1 zinc finger in cotton. Mol. Genet. Genom. 2016, 291, 2199–2213. [Google Scholar] [CrossRef]
- Ghosh, S.; Tewari, R.; Dixit, D.; Sen, E. TNF alpha induced oxidative stress dependent Akt signaling affects actin cytoskeletal organization in glioma cells. Neurochem. Int. 2010, 56, 194–201. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Duan, D.; Zhao, S.; Xu, B.; Luo, J.; Wang, Q.; Huang, D.; Liu, C.; Li, C.; Gong, X.; et al. Genome-wide analysis and cloning of the apple stress-associated protein gene family reveals MdSAP15, which confers tolerance to drought and osmotic stresses in transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2478. [Google Scholar] [CrossRef]
- Wang, Z.; Kuang, J.; Han, B.; Chen, S.; Liu, A. Genomic characterization and expression profiles of stress-associated proteins (SAPs) in castor bean (Ricinus communis). Plant Divers. 2021, 43, 152–162. [Google Scholar] [CrossRef]
- Linnen, J.M.; Bailey, C.P.; Weeks, D.L. 2 related localized messenger-RNAs form Xenopus laevis encode ubiquitin-like fusion proteins. Gene 1993, 128, 181–188. [Google Scholar] [CrossRef]
- DeValck, D.; Heyninck, K.; VanCriekinge, W.; Contreras, R.; Beyaert, R.; Fiers, W. A20, an inhibitor of cell death, self-associates by its zinc finger domain. FEBS Lett. 1996, 384, 61–64. [Google Scholar] [CrossRef]
- Jin, Y.; Wang, M.; Fu, J.J.; Xuan, N.; Zhu, Y.; Lian, Y.; Jia, Z.W.; Zheng, J.; Wang, G.Y. Phylogenetic and expression analysis of ZnF-AN1 genes in plants. Genomics 2007, 90, 265–275. [Google Scholar] [CrossRef]
- Lai, W.; Zhou, Y.; Pan, R.; Liao, L.; He, J.; Liu, H.; Yang, Y.; Liu, S. Identification and expression analysis of stress-associated Proteins (SAPs) containing A20/AN1 zinc finger in cucumber. Plants 2020, 9, 400. [Google Scholar] [CrossRef]
- Tyagi, H.; Jha, S.; Sharma, M.; Giri, P.; Tyagi, A.K. Rice SAPs are responsive to multiple biotic stresses and overexpression of OsSAP1, an A20/AN1 zinc-finger protein, enhances the basal resistance against pathogen infection in tobacco. Plant Sci. 2014, 225, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Jia, H.X.; Li, J.B.; Zhang, J.; Ren, Y.Q.; Hu, J.J.; Lu, M.Z. Genome-wide survey and expression analysis of the stress-associated protein gene family in desert poplar, Populus euphratica. Tree Genet. Genomes 2016, 12, 78. [Google Scholar] [CrossRef]
- Huang, J.; Wang, M.M.; Jiang, Y.; Bao, Y.M.; Huang, X.; Sun, H.; Xu, D.Q.; Lan, H.X.; Zhang, H.S. Expression analysis of rice A20/AN1-type zinc finger genes and characterization of ZFP177 that contributes to temperature stress tolerance. Gene 2008, 420, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.Y.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fu, X.; Xie, L.; Qin, W.; Li, L.; Sun, X.; Xing, S.; Tang, K. Stress associated protein 1 regulates the development of glandular trichomes in Artemisia annua. Plant Cell Tissue Organ Cult. 2019, 139, 249–259. [Google Scholar] [CrossRef]
- Ghneim-Herrera, T.; Selvaraj, M.G.; Meynard, D.; Fabre, D.; Pena, A.; Ben Romdhane, W.; Ben Saad, R.; Ogawa, S.; Rebolledo, M.C.; Ishitani, M.; et al. Expression of the Aeluropus littoralis AlSAP gene enhances rice yield under field drought at the reproductive stage. Front. Plant Sci. 2017, 8, 994. [Google Scholar] [CrossRef]
- Xing, W.; Xu, B.; Wang, Z.; Jia, C.; Liu, J.; Jin, Z. Cloning and Expression of stress associated protein fene from banana (Musa acuminata L.AAA group,cv.Brazilian). Acta Bot. Boreali-Occident. Sin. 2014, 34, 225–230. [Google Scholar]
- Ben Saad, R.; Safi, H.; Ben Hsouna, A.; Brini, F.; Ben Romdhane, W. Functional domain analysis of LmSAP protein reveals the crucial role of the zinc-finger A20 domain in abiotic stress tolerance. Protoplasma 2019, 256, 1333–1344. [Google Scholar] [CrossRef]
- Zhang, F.; Lu, X.; Chen, H.; Wang, C.; Li, Y.; Zheng, J.; Cai, X. New Maize Stress-Associated Protein Kinase ZmSAPK3 Gene Useful for Improving Plant Osmotic Stress Resistance and Cultivating High-Yielding Transgenic Crops. Patent CN107312784-A, 27 April 2016. [Google Scholar]
- Liu, S.X.; Wang, J.L.; Jiang, S.Y.; Wang, H.; Gao, Y.Z.; Zhang, H.J.; Li, D.Y.; Song, F.M. Tomato SlSAP3, a member of the stress-associated protein family, is a positive regulator of immunity against Pseudomonas syringae pv. tomato DC3000. Mol. Plant Pathol. 2019, 20, 815–830. [Google Scholar] [CrossRef]
- Zhang, Y.; Lan, H.X.; Shao, Q.L.; Wang, R.Q.; Chen, H.; Tang, H.J.; Zhang, H.S.; Huang, J. An A20/AN1-type zinc finger protein modulates gibberellins and abscisic acid contents and increases sensitivity to abiotic stress in rice (Oryza sativa). J. Exp. Bot. 2016, 67, 315–326. [Google Scholar] [CrossRef]
- Zhang, N.; Xu, J.; Liu, X.Y.; Liang, W.X.; Xin, M.M.; Du, J.K.; Hu, Z.R.; Peng, H.R.; Guo, W.L.; Ni, Z.F.; et al. Identification of HSP90C as a substrate of E3 ligase TaSAP5 through ubiquitylome profiling. Plant Sci. 2019, 287, 110170. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Huang, C.; Xie, Y.; Song, F.; Zhou, X. A tomato glutaredoxin gene SlGRX1 regulates plant responses to oxidative, drought and salt stresses. Planta 2010, 232, 1499–1509. [Google Scholar] [CrossRef] [PubMed]
- Narancio, R.; John, U.; Mason, J.; Giraldo, P.; Spangenberg, G. Digital PCR (dPCR) and qPCR mediated determination of transgene copy number in the forage legume white clover (Trifolium repens). Mol. Biol. Rep. 2021, 48, 3069–3077. [Google Scholar] [CrossRef] [PubMed]
- Ellul, P.; Garcia-Sogo, B.; Pineda, B.; Rios, G.; Roig, L.; Moreno, V. The ploidy level of transgenic plants in Agrobacterium-mediated transformation of tomato cotyledons (Lycopersicon esculentum Mill) is genotype and procedure dependent [corrected]. Theor. Und Angew. Genet. 2003, 106, 231–238. [Google Scholar] [CrossRef]
- Liu, S.; Yuan, X.; Wang, Y.; Wang, H.; Wang, J.; Shen, Z.; Gao, Y.; Cai, J.; Li, D.; Song, F. Tomato stress-associated protein 4 contributes positively to immunity against necrotrophic fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 2019, 32, 566–582. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Jeyasri, R.; Selvaraj, A.; Kalaiyarasi, D.; Aruni, W.; Pandian, S.T.K.; Ramesh, M. Global transcriptome analysis of novel stress associated protein (SAP) genes expression dynamism of combined abiotic stresses in Oryza sativa (L.). J. Biomol. Struct. Dyn. 2021, 39, 2106–2117. [Google Scholar] [CrossRef]
- Richards, S.; Murali, S.C. Best practices in insect genome sequencing: What works and what doesn’t. Curr. Opin. Insect Sci. 2015, 7, 1–7. [Google Scholar] [CrossRef]
- Scalbert, A.; Williamson, G. Dietary intake and bioavailability of polyphenols. J. Nutr. 2000, 130, 2073S–2085S. [Google Scholar] [CrossRef]
- RiceEvans, C.A.; Miller, J.; Paganga, G. Antioxidant properties of phenolic compounds. Trends Plant Sci. 1997, 2, 152–159. [Google Scholar] [CrossRef]
- Dos Santos, R.A.C.; Berretta, A.A.; Barud, H.d.S.; Ribeiro, S.J.L.; Gonzalez-Garcia, L.N.; Zucchi, T.D.; Goldman, G.H.; Riano-Pachon, D.M. Draft genome sequence of Komagataeibacter rhaeticus strain AF1, a high producer of cellulose, isolated from Kombucha tea. Genome Announc. 2014, 2, e00731-14. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef]
- Wang, Y.-X.; Liu, Z.-W.; Wu, Z.-J.; Li, H.; Wang, W.-L.; Cui, X.; Zhuang, J. Genome-wide identification and expression analysis of GRAS family transcription factors in tea plant (Camellia sinensis). Sci. Rep. 2018, 8, 3949. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gao, T.; Chen, J.; Yang, J.; Huang, H.; Yu, Y. The late embryogenesis abundant gene family in tea plant (Camellia sinensis): Genome-wide characterization and expression analysis in response to cold and dehydration stress. Plant Physiol. Biochem. 2019, 135, 277–286. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-wide identification, classification and expression analysis of the HSP gene superfamily in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Guo, Y.; Chen, X.; Zheng, Y.; Sun, Y.; Yang, J.; Ye, N. Genome-wide identification of WOX genes and their expression patterns under different hormone and abiotic stress treatments in tea plant (Camellia sinensis). Trees-Struct. Funct. 2019, 33, 1129–1142. [Google Scholar] [CrossRef]
- Wang, P.; Yue, C.; Chen, D.; Zheng, Y.; Zhang, Q.; Yang, J.; Ye, N. Genome-wide identification of WRKY family genes and their response to abiotic stresses in tea plant (Camellia sinensis). Genes Genom. 2019, 41, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wu, A.; He, Y.; Li, F.; Wei, C. Genome-wide characterization of the basic leucine zipper transcription factors in Camellia sinensis. Tree Genet. Genomes 2018, 14, 27. [Google Scholar] [CrossRef]
- Teng, R.-M.; Wang, Y.-X.; Wang, W.-L.; Li, H.; Shen, W.; Zhuang, J. Genome-wide identification, classification and expression pattern of LBD gene family in Camellia sinensis. Biotechnol. Biotechnol. Equip. 2018, 32, 1387–1397. [Google Scholar] [CrossRef]
- Vij, S.; Tyagi, A.K. Genome-wide analysis of the stress associated protein (SAP) gene family containing A20/AN1 zinc-finger(s) in rice and their phylogenetic relationship with Arabidopsis. Mol. Genet. Genom. 2006, 276, 565–575. [Google Scholar] [CrossRef]
- Solanke, A.U.; Sharma, M.K.; Tyagi, A.K.; Sharma, A.K. Characterization and phylogenetic analysis of environmental stress-responsive SAP gene family encoding A20/AN1 zinc finger proteins in tomato. Mol. Genet. Genom. 2009, 282, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Roberts, N.D.; Wala, J.A.; Shapira, O.; Schumacher, S.E.; Kumar, K.; Khurana, E.; Waszak, S.; Korbel, J.O.; Haber, J.E.; et al. Patterns of somatic structural variation in human cancer genomes. Nature 2020, 578, 112–121. [Google Scholar] [CrossRef] [PubMed]
- Messer, P.W.; Arndt, P.F. The majority of recent short DNA insertions in the human genome are tandem duplications. Mol. Biol. Evol. 2007, 24, 1190–1197. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xie, S.; Xie, P.; Yao, M.; Liu, W.; Qin, L.W.; Liu, Z.S.; Zheng, M.; Liu, H.F.; Guan, M.; et al. Genome-wide identification of stress-associated proteins (SAP) with A20/AN1 zinc finger domains associated with abiotic stresses responses in Brassica napus. Environ. Exp. Bot. 2019, 165, 108–119. [Google Scholar] [CrossRef]
- Mukhopadhyay, A.; Vij, S.; Tyagi, A.K. Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. USA 2004, 101, 6309–6314. [Google Scholar] [CrossRef]
- Ben-Saad, R.; Meynard, D.; Ben-Romdhane, W.; Mieulet, D.; Verdeil, J.-L.; Al-Doss, A.; Guiderdoni, E.; Hassairi, A. The promoter of the AlSAP gene from the halophyte grass Aeluropus littoralis directs a stress-inducible expression pattern in transgenic rice plants. Plant Cell Rep. 2015, 34, 1791–1806. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008, 66, 445–462. [Google Scholar] [CrossRef]
- Ben-Saad, R.; Ben-Ramdhan, W.; Zouari, N.; Azaza, J.; Mieulet, D.; Guiderdoni, E.; Ellouz, R.; Hassairi, A. Marker-free transgenic durum wheat cv. Karim expressing the AlSAP gene exhibits a high level of tolerance to salinity and dehydration stresses. Mol. Breed. 2012, 30, 521–533. [Google Scholar] [CrossRef]
- Sreedharan, S.; Shekhawat, U.K.S.; Ganapathi, T.R. MusaSAP1, a A20/AN1 zinc finger gene from banana functions as a positive regulator in different stress responses. Plant Mol. Biol. 2012, 80, 503–517. [Google Scholar] [CrossRef]
- Kim, G.-D.; Cho, Y.-H.; Yoo, S.-D. Regulatory functions of evolutionarily conserved AN1/A20-like Zinc finger family proteins in Arabidopsis stress responses under high temperature. Biochem. Biophys. Res. Commun. 2015, 457, 213–220. [Google Scholar] [CrossRef]
- Wu, L.; Yu, M.; Holowachuk, J.; Sharpe, A.; Lydiate, D.; Hegedus, D.; Gruber, M. Overexpression of Brassica napus myrosinase-associated protein 1 improved Sclerotinia sclerotiorum tolerance in Arabidopsis thaliana. Can. J. Plant Sci. 2017, 97, 842–851. [Google Scholar] [CrossRef][Green Version]
- Liu, H.; Wang, Y.; Zhou, X.; Wang, C.; Wang, C.; Fu, J.; Wei, T. Overexpression of a harpin-encoding gene popW from Ralstonia solanacearum primed antioxidant defenses with enhanced drought tolerance in tobacco plants. Plant Cell Rep. 2016, 35, 1333–1344. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | Zinc-Finger Domain | CDS Length (bp) | Protein Length (aa) | Molecular Weight (kDa) | No. of Introns | Theoretical pI | Scaffold Location |
|---|---|---|---|---|---|---|---|---|
| CsSAP1 | TEA003271.1 | A20-AN1 | 519 | 173 | 18.31 | 0 | 7.72 | Scaffold622: 1698779:1699297:+ |
| CsSAP2 | TEA007661.1 | A20-AN1 | 522 | 174 | 17.45 | 0 | 7.51 | Scaffold560: 754898:755419:+ |
| CsSAP3 | TEA007758.1 | A20-AN1 | 483 | 161 | 17.06 | 0 | 8.14 | Scaffold390: 736618:737100:+ |
| CsSAP4 | TEA008252.1 | A20-AN1 | 516 | 172 | 18.14 | 0 | 7.44 | Scaffold2348: 1266811:1267326:+ |
| CsSAP5 | TEA009516.1 | A20-AN1 | 492 | 164 | 17.53 | 0 | 7.76 | Scaffold2009: 46003:46494:− |
| CsSAP6 | TEA013656.1 | AN1-AN1 | 1737 | 579 | 63.76 | 4 | 9.66 | Scaffold942: 1448206:1457402:− |
| CsSAP7 | TEA013686.1 | AN1-AN1 | 573 | 191 | 20.92 | 1 | 8.52 | Scaffold4677: 68010:69805:+ |
| CsSAP8 | TEA014231.1 | A20-AN1 | 465 | 155 | 17.24 | 0 | 8.91 | Scaffold3002: 9737:10201:+ |
| CsSAP9 | TEA016255.1 | A20-AN1 | 507 | 169 | 17.96 | 0 | 8.46 | Scaffold435: 1131711:1132217:+ |
| CsSAP10 | TEA016540.1 | A20-AN1 | 636 | 212 | 22.52 | 1 | 7.92 | Scaffold1761: 717585:722550:− |
| CsSAP11 | TEA016572.1 | AN1 | 507 | 169 | 18.75 | 0 | 9.11 | Scaffold1761: 663031:663537:− |
| CsSAP12 | TEA021384.1 | A20-AN1 | 498 | 166 | 17.85 | 0 | 8.08 | Scaffold4125: 306796:307293:+ |
| CsSAP13 | TEA023579.1 | A20-AN1 | 492 | 164 | 17.64 | 0 | 8.7 | Scaffold5358: 143051:143542:+ |
| CsSAP14 | TEA025409.1 | A20-AN1 | 435 | 145 | 15.69 | 0 | 8.6 | Scaffold2300: 157553:157987:+ |
| Cis-Acting Elements | ABRE | ARE | CGTCA | ERE | LTR | MBS | TCA | TC-Rich Repeat | W-Box |
|---|---|---|---|---|---|---|---|---|---|
| Stress to Response | ABA | Hypoxia | MeJA | Ethylene | Chilling | Drought | SA | Defence | Pathogen |
| CsSAP1 | 0/5 | 1/0 | 1/0 | 0/1 | 1/0 | ||||
| CsSAP2 | 0/2 | 1/1 | 1/0 | ||||||
| CsSAP3 | 1/1 | 3/0 | 1/0 | 0/1 | 2/0 | ||||
| CsSAP4 | 2/0 | 2/2 | 1/0 | 1/0 | 1/0 | ||||
| CsSAP5 | 1/0 | 1/1 | 0/1 | 1/0 | |||||
| CsSAP6 | 0/1 | 1/0 | 1/2 | 0/1 | |||||
| CsSAP7 | 1/1 | 1/0 | 1/1 | 0/1 | 1/0 | 1/0 | |||
| CsSAP8 | 1/1 | 0/1 | 0/1 | 1/0 | 0/1 | ||||
| CsSAP9 | 2/1 | 1/2 | 1/0 | ||||||
| CsSAP10 | 1/1 | 2/1 | 0/1 | 2/1 | |||||
| CsSAP11 | 0/3 | 0/1 | 0/2 | 0/1 | 2/0 | 1/0 | |||
| CsSAP12 | 0/1 | 1/0 | 1/0 | 0/1 | |||||
| CsSAP13 | 1/0 | 0/1 | 3/2 | ||||||
| CsSAP14 | 0/1 | 1/0 | 1/0 | 1/2 |
| Plant Species | A20- | A20- | A20 | AN1 | AN1- | AN1- | AN1- | Total Number |
|---|---|---|---|---|---|---|---|---|
| AN1 | A20- | AN1 | AN1- | AN1- | ||||
| AN1 | C2H2 | C2H2- | ||||||
| C2H2 | ||||||||
| Camellia sinensis | 11 | 0 | 0 | 1 | 2 | 0 | 0 | 14 |
| Arabidopsis thaliana | 10 | 0 | 0 | 1 | 1 | 1 | 1 | 14 |
| Brassica napus | 32 | 0 | 7 | 9 | 5 | 4 | 0 | 57 |
| Gossypium hirsutum | 28 | 0 | 0 | 3 | 4 | 0 | 2 | 37 |
| Malus domestica | 23 | 0 | 3 | 0 | 3 | 0 | 1 | 30 |
| Medicago truncatula | 11 | 0 | 0 | 2 | 1 | 0 | 2 | 16 |
| Oryza sativa | 11 | 1 | 1 | 3 | 1 | 0 | 1 | 18 |
| Populus euphratica | 15 | 0 | 0 | 0 | 2 | 0 | 1 | 18 |
| Solanum lycopersicum | 9 | 0 | 0 | 1 | 2 | 0 | 1 | 13 |
| Zea mays | 8 | 0 | 0 | 1 | 1 | 0 | 1 | 11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.-C.; Li, C.; Li, S.-H.; Tang, J.; Shi, H.-D.; Yang, T.-M.; Liang, M.-Z.; Liu, D.-D. Genome-Wide Analyses of Tea Plant Stress-Associated Proteins (SAPs) Reveal the Role of CsSAP12 in Increased Drought Tolerance in Transgenic Tomatoes. Horticulturae 2022, 8, 363. https://doi.org/10.3390/horticulturae8050363
Fan S-C, Li C, Li S-H, Tang J, Shi H-D, Yang T-M, Liang M-Z, Liu D-D. Genome-Wide Analyses of Tea Plant Stress-Associated Proteins (SAPs) Reveal the Role of CsSAP12 in Increased Drought Tolerance in Transgenic Tomatoes. Horticulturae. 2022; 8(5):363. https://doi.org/10.3390/horticulturae8050363
Chicago/Turabian StyleFan, Shu-Chen, Chun Li, Shao-Hua Li, Jie Tang, Hong-Di Shi, Tian-Ming Yang, Ming-Zhi Liang, and Dan-Dan Liu. 2022. "Genome-Wide Analyses of Tea Plant Stress-Associated Proteins (SAPs) Reveal the Role of CsSAP12 in Increased Drought Tolerance in Transgenic Tomatoes" Horticulturae 8, no. 5: 363. https://doi.org/10.3390/horticulturae8050363
APA StyleFan, S.-C., Li, C., Li, S.-H., Tang, J., Shi, H.-D., Yang, T.-M., Liang, M.-Z., & Liu, D.-D. (2022). Genome-Wide Analyses of Tea Plant Stress-Associated Proteins (SAPs) Reveal the Role of CsSAP12 in Increased Drought Tolerance in Transgenic Tomatoes. Horticulturae, 8(5), 363. https://doi.org/10.3390/horticulturae8050363






