Six First Reports of Pin Nematodes from Portugal, with an Update of the Systematics, Genetic Diversity, and Phylogeny of the Genus Paratylenchus (Nematoda: Tylenchulidae)
Abstract
1. Introduction
2. Materials and Methods
2.1. Nematode Population Sampling
2.2. Morphological and Morphometrical Study
2.3. DNA Extraction
2.4. PCR Amplification, DNA Purification and Sequencing
2.5. Phylogenetic Analyses
3. Results
3.1. Systematics
3.1.1. Morphological Features and Morphometric Measurements
3.1.2. Molecular Results and Phylogenetic Relationships of the Six Known Paratylenchus spp. and Other Members of Genus Paratylenchus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jairajpuri, M.S.; Ahmad, W. Dorylaimida: Free-Living, Predaceous and Plant-Parasitic Nematodes; Brill Publishers: Leiden, The Netherlands, 1992. [Google Scholar]
- Ahmed, M.; Holovachov, O. Twenty Years after De Ley and Blaxter—How Far Did We Progress in Understanding the Phylogeny of the Phylum Nematoda? Animals 2021, 11, 3479. [Google Scholar] [CrossRef] [PubMed]
- Decraemer, W.; Hunt, D.J. Structure and classification. In plant Nematology; Perry, R.N., Moens, M., Eds.; CABI Publishing: Wallingford, UK, 2006; pp. 3–32. [Google Scholar]
- Siddiqi, M.R. Tylenchida: Parasites of Plants and Insects, 2nd ed.; CABI Publishing: Wallingford, UK, 2000. [Google Scholar]
- Clavero-Camacho, I.; Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; León-Ropero, G.; Martín-Barbarroja, J.; Archidona-Yuste, A.; Castillo, P. Integrative taxonomy reveals hidden cryptic diversity within pin nematodes of the genus Paratylenchus (Nematoda: Tylenchulidae). Plants 2021, 10, 1454. [Google Scholar] [CrossRef] [PubMed]
- Clavero-Camacho, I.; Cantalapiedra-Navarrete, C.; Archidona-Yuste, A.; Castillo, P.; Palomares-Rius, J.E. Remarkable cryptic diversity of Paratylenchus spp. (Nematoda: Tylenchulidae) in Spain. Animals 2021, 11, 1161. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.R.; Karssen, G.; Couvreur, M.; Subbotin, S.A.; Bert, W. Integrative taxonomy and molecular phylogeny of the plant-parasitic nematode genus Paratylenchus (Nematoda: Paratylenchinae): Linking species with molecular barcodes. Plants 2021, 10, 408. [Google Scholar] [CrossRef]
- Kantor, M.R.; Handoo, Z.A.; Subbotin, S.A.; Bauchan, G.R.; Mowery, J.D. Morphological and molecular characterization of Paratylenchus beltsvillensis n. sp. (Tylenchida: Paratylenchidae) from the rhizosphere of pine tree (Pinus virginiana Mill) in Maryland, USA. J. Nematol. 2021, 53, 1–10. [Google Scholar] [CrossRef]
- Andrássy, I. Free-Living Nematodes of Hungary, II (Nematoda errantia); Hungarian Natural History Museum: Budapest, Hungary, 2007. [Google Scholar]
- Brzeski, M.W. Nematodes of Tylenchina in Poland and Temperate Europe; Muzeum i Instytut Zoologii, Polska Akademia Nauk: Warszawa, Poland, 1998. [Google Scholar]
- Esser, R.P. A diagnostic compendium to species included in Paratylenchinae Thorne, 1949 and Tylenchocriconematinae Raski & Siddiqi, 1975 (Nematoda: Criconematoidea). Nematologica 1992, 38, 146–163. [Google Scholar]
- Raski, D.J. Tylenchulidae of agricultural soils. In Manual of Agricultural Nematology; Nickle, W.R., Ed.; Marcel Dekker Inc.: New York, NY, USA, 1991; pp. 761–794. [Google Scholar]
- Ebsary, B.A. Catalog of the Order Tylenchida (Nematoda); Agriculture Canada, Research Branch: Ottawa, ON, Canada, 1991. [Google Scholar]
- Maggenti, A.R.; Luc, M.; Raski, D.J.; Fortuner, R.; Geraert, E. A reappraisal of Tylenchina (Nemata). 11. List of generic and supra-generic taxa, with their junior synonyms. Rev. Nématol. 1988, 11, 177–188. [Google Scholar]
- Raski, D.J.; Luc, M. A reappraisal of Tylenchina (Nemata). 10: The superfamily Criconematoidea Taylor, 1936. Rev. Nématol. 1987, 10, 409–444. [Google Scholar]
- Goodey, T. Soil and Freshwater Nematodes, 2nd ed.; Goodey, J.B., Ed.; Methuen: London, UK, 1963. [Google Scholar]
- Ghaderi, R. The damage potential of pin nematodes, Paratylenchus Micoletzky, 1922 sensu lato spp. (Nematoda: Tylenchulidae). J. Crop Prot. 2019, 8, 243–257. [Google Scholar]
- Subbotin, S.A.; Chitambar, J.J. Plant Parasitic Nematodes in Sustainable Agriculture of North America; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Upadhaya, A.; Yan, G.P.; Pasche, J. Reproduction ability and growth effect of pin nematode, Paratylenchus nanus, with selected field pea cultivars. Plant Dis. 2019, 103, 2520–2526. [Google Scholar] [CrossRef]
- Claerbout, J.; Vandevelde, I.; Venneman, S.; Kigozi, A.; de Sutter, N.; Neukermans, J.; Bleyaert, P.; Bert, W.; Hofte, M.; Viaene, N. A thorough study of a Paratylenchus sp. in glasshouse-grown lettuce: Characterisation, population dynamics, host plants and damage threshold as keys to its integrated management. Ann. Appl. Biol. 2020, 178, 62–79. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Xie, H.; Wu, W.J.; Xu, C.L. Pin nematode slow decline of Anthurium andraeanum, a new disease caused by the pin nematode Paratylenchus shenzhenensis. Plant Dis. 2016, 100, 940–945. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Braun, A.L.; Lownsbery, B.F. The pin nematode, Paratylenchus neoamblycephalus, on Myrobalan plum and other hosts. J. Nematol. 1975, 7, 336–343. [Google Scholar]
- Riga, E.; Porter, L.D.; Mojtahedi, H.; Erickson, D. Pratylenchus neglectus, P. thornei, and Paratylenchushamatus nematodes causing yield reduction to dryland peas and lentils in Idaho. Plant Dis. 2008, 92, 979. [Google Scholar] [CrossRef] [PubMed]
- Philis, J. Controlling parasitic nematodes in an established vineyard in Cyprus. Nematol. Medit. 2003, 31, 61–63. [Google Scholar]
- MacDonald, D.H. Effects of Paratylenchus hamatus on productivity of greenhouse roses. J. Nematol. 1976, 8, 294. [Google Scholar]
- Abivardi, C. Occurrence of Paratylenchus hamatus on citrus in Iran and its sensitivity to two nematicides under laboratory conditions. Plant Dis. Rep. 1970, 54, 1085–1088. [Google Scholar]
- French, A.M.; Lownsbery, B.F.; Ayoub, S.M.; Weiner, A.C.; Gholl, N.E. Pythiaceous fungi and plant-parasitic nematodes in California pear orchards. II. Incidence and distribution of parasitic nematodes in orchard soils. Hilgardia 1964, 35, 603–610. [Google Scholar] [CrossRef][Green Version]
- Faulkner, L.R. Pathogenicity and population dynamics of Paratylenchus hamatus on Mentha spp. Phytopathology 1964, 54, 344–348. [Google Scholar]
- Raski, D.J.; Lider, L.A. Nematodes in grape production. California Agriculture 1959, 13, 13–15. [Google Scholar]
- Weischcr, B. Neuere Gesichtspunktezur Frage der biologie und ökologie der w and erndenwurzel nematodes. Nematologica 1957, 2, 406–412. [Google Scholar]
- Rich, A.E. The occurrence and control of Paratylenchus hamatus on celery in New Hampshire. Plant Dis. Rep. 1955, 39, 307–308. [Google Scholar]
- Thorne, G.; Allen, M.W. Paratylenchus hamatus n.sp, and Xiphinema index n. sp., two nematodes associated with fig roots, with a note on Paratylenchus anceps Cobb. Proc. Helm. Soc. Wash. 1950, 17, 27–35. [Google Scholar]
- Lowensbry, B.F.; Stoddars, E.M.; Lownsbery, J.W. Paratylenchus hamatus pathogenic on celery. Phytopathology 1952, 42, 651–653. [Google Scholar]
- Fisher, J.M. Effect of temperature and host on Paratylenchus neoamblycephalus and effect of the nematode on the host. Aust. J. Agric. Res. 1967, 18, 921–929. [Google Scholar] [CrossRef]
- Viketoft, M.; Palmborg, C.; Sohlenius, B.; Huss-Danell, K.; Bengtsson, J. Plant species effects on soil nematode communities in experimental grasslands. Appl. Soil Ecol. 2005, 30, 90–103. [Google Scholar] [CrossRef]
- Viketoft, M. Effects of six grassland plant species on soil nematodes: A glasshouse experiment. Soil Biol. Biochem. 2008, 40, 906–915. [Google Scholar] [CrossRef]
- Bell, N.L. The biology of the plant parasitic nematodes Paratylenchus nanus and Paratrichodorus minor in soil under pasture. Ph.D. Thesis, Massey University, Palmerston North, New Zealand, 1999. [Google Scholar]
- Archidona-Yuste, A.; Wiegand, T.; Castillo, P.; Navas-Cortés, J.A. Dataset on the diversity of plant-parasitic nematodes in cultivated olive trees in southern Spain. Data Brief. 2019, 27, 104658. [Google Scholar] [CrossRef]
- Ali, N.; Chapuis, E.; Tavoillot, J.; Mateille, T. Plant-parasitic nematodes associated with olive tree (Olea europaea L.) with a focus on the Mediterranean Basin: A review. Comptes Rendus Biologies 2014, 337, 423–442. [Google Scholar] [CrossRef]
- Téliz, D.; Landa, B.B.; Rapoport, H.F.; Pérez Camacho, F.; Jiménez Díaz, R.M.; Castillo, P. Plant-parasitic nematodes infecting grapevine in Southern Spain and susceptible reaction to root-knot nematodes of rootstocks reported as moderately resistant. Plant Dis. 2007, 91, 1147–1154. [Google Scholar] [CrossRef]
- Peña-Santiago, R.; Castillo, P.; Escuer, M.; Guerrero, P.; Talavera, M.; Vieira, P. Tylenchid Species (Nematoda, Tylenchida) Recorded in the Iberian Peninsula and the Balearic Islands: A Compendium; Universidad de Jaén: Jaén, Spain, 2004; pp. 1–127. [Google Scholar]
- Munawar, M.; Yevtushenko, D.P.; Palomares-Rius, J.E.; Castillo, P. Species diversity of pin nematodes (Paratylenchus spp.) from potato growing regions of southern Alberta, Canada. Plants 2021, 10, 188. [Google Scholar] [CrossRef] [PubMed]
- Lima, M.B. First record of the genera Hemicycliophora, Paralongidorus and Paratylenchus in Portugal. Agronomía lush. 1966, 28, 143–144. [Google Scholar]
- Macara, A.M. Nematodos asociados a plantas forestales en Portugal. Bol San. Veg. Plagas 1988, 14, 185–225. [Google Scholar]
- Ghaderi, R.; Kashi, L.; Karegar, A. Contribution to the study of the genus Paratylenchus Micoletzky, 1922 sensu lato (Nematoda: Tylenchulidae). Zootaxa 2014, 3841, 151–187. [Google Scholar] [CrossRef]
- Munawar, M.; Miao, W.; Castillo, P.; Zheng, J.-W. A new pin nematode, Paratylenchus sinensis n. sp. (Nematoda: Paratylenchinae) in the rhizosphere of white mulberry from Zhejiang Province, China. Eur. J. Plant Pathol. 2020, 156, 1023–1029. [Google Scholar] [CrossRef]
- Munawar, M.; Miao, W.; Ye, W.; Zheng, J. Updated description of Paratylenchus lepidus Raski 1975 and P. minor Sharma, Sharma and Khan, 1986 by integrating molecular and ultra-structural observations. J. Nematol. 2019, 51, 1–13. [Google Scholar] [CrossRef]
- Munawar, M.; Cai, R.; Ye, W.; Powers, T.O.; Zheng, J. Description of Gracilacus paralatescens n. sp. (Nematoda: Paratylenchinae) found from the rhizosphere of Bamboo in Zhejiang, China. J. Nematol. 2018, 50, 611–622. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Yan, G.; Kantor, M.; Handoo, Z. On the molecular identity of Paratylenchus nanus Cobb, 1923 (Nematoda: Tylenchida). J. Nematol. 2020, 52, 1–7. [Google Scholar] [CrossRef]
- Le, T.M.L.; Nguyen, H.T.; Nguyen, T.D.; Trinh, Q.P. First report of Paratylenchus lepidus Raski, 1975 associated with green tea (Camellia sinensis (L.) Kuntze) in Vietnam. J. Nematol. 2020, 52, 1–4. [Google Scholar] [CrossRef]
- Li, Y.; Wang, K.; Xie, H.; Xu, C.L. Morphology and molecular analysis of Paratylenchus chongqingensis n. sp. (Nematoda: Paratylenchinae) from soil associated with Ophiopogon japonicas in China. Eur. J. Plant Pathol. 2019, 154, 597–605. [Google Scholar] [CrossRef]
- Hesar, A.M.; Karegar, A.; Ghaderi, R. Phylogenetic relationships of Cacopaurus pestis Thorne, 1943 within representatives of the Tylenchulidae Skarbilovich, 1947 as inferred from ITS and D2–D3 expansion segments of 28S-rRNA sequences. Nematology 2019, 21, 971–994. [Google Scholar] [CrossRef]
- Etongwe, C.M.; Singh, P.R.; Bert, W.; Subbotin, S.A. Molecular characterisation of some plant-parasitic nematodes (Nematoda: Tylenchida) from Belgium. Russ. J. Nematol. 2020, 28, 1–28. [Google Scholar]
- Phani, V.; Somvanshi, V.S.; Rao, U.; Khan, M.R. Paratylenchus jasmineae sp. n. (Nematoda: Paratylenchinae) from rhizosphere of Jasminum sambac in India. Nematology 2019, 21, 469–478. [Google Scholar] [CrossRef]
- Zhuo, K.; Liu, X.; Tao, Y.; Wang, H.; Lin, B.; Liao, J. Morphological and molecular characterisation of three species of Paratylenchus Micoletzky, 1922 (Tylenchida: Paratylenchidae) from China, with a first description of the male P. rostrocaudatus. Nematology 2018, 20, 837–850. [Google Scholar] [CrossRef]
- Wang, K.; Xie, H.; Li, Y.; Wu, W.J.; Xu, C.L. Morphology and molecular analysis of Paratylenchus nanjingensis n. sp. (Nematoda: Paratylenchinae) from the rhizosphere soil of Pinus massoniana in China. J. Helminthol. 2016, 90, 166–173. [Google Scholar] [CrossRef]
- Wang, K.; Li, Y.; Xie, H.; Xu, C.L.; Wu, W.J. Morphology and molecular analysis of Paratylenchus guangzhouensis n. sp. (Nematoda: Paratylenchinae) from the soil associated with Bambusa multiplex in China. Eur. J. Plant Pathol. 2016, 145, 255–264. [Google Scholar] [CrossRef]
- Yu, Q.; Ye, W.; Powers, T. Morphological and molecular characterization of Gracilacus wuae n. sp. (Nematoda: Criconematoidea) associated with cow parsnip (Heracleum maximum) in Ontario, Canada. J. Nematol. 2016, 48, 203–213. [Google Scholar] [CrossRef]
- Esmaeili, M.; Heydari, R.; Castillo, P.; Bidhendi, M.Z.; Palomares-Rius, J.E. Molecular characterisation of two known species of Paratylenchus Micoletzky, 1922 from Iran with notes on the validity of Paratylenchus audriellus Brown, 1959. Nematology 2016, 18, 591–604. [Google Scholar] [CrossRef]
- Van den Berg, E.; Tiedt, L.R.; Subbotin, S.A. Morphological and molecular characterisation of several Paratylenchus Micoletzky, 1922 (Tylenchida: Paratylenchidae) species from South Africa and USA, together with some taxonomic notes. Nematology 2014, 16, 323–358. [Google Scholar] [CrossRef]
- Powers, T.; Harris, T.S.; Higgins, R.S.; Mullin, P.G.; Powers, K.S. Nematode biodiversity assessments need vouchered databases: A BOLD reference library for plant-parasitic nematodes in the superfamily Criconematoidea. Genome 2020, 5, 1–10. [Google Scholar] [CrossRef]
- Whitehead, A.G.; Hemming, J.R. A comparison of some quantitative methods of extracting small vermiform nematodes from soil. Ann. Appl. Biol. 1965, 55, 25–38. [Google Scholar] [CrossRef]
- Jenkins, W.R.B. A rapid centrifugal-flotation technique for separating nematodes from soil. Plant Dis. Rep. 1964, 48, 692. [Google Scholar]
- Seinhorst, J.W. Killing nematodes for taxonomic study with hot F.A. 4:1. Nematologica 1966, 12, 178. [Google Scholar] [CrossRef]
- Fawzia H Abdel Rahman. Chemical drying of nematodes for Scanning Electron Microscopy observations. World J. Agr. Soil Sci. 2021, 7, 1–5. [CrossRef]
- Gutiérrez-Gutiérrez, C.; Mota, M.; Castillo, P.; Teixeira-Santos, M.; Palomares-Rius, J.E. Description and molecular phylogeny of one new and one known needle nematode of the genus Paralongidorus (Nematoda: Longidoridae) from grapevine in Portugal using integrative approach. Eur. J. Plant Pathol. 2018, 151, 155–172. [Google Scholar] [CrossRef]
- Gutiérrez-Gutiérrez, C.; Teixeira Santos, M.; Inácio, M.L.; Eisenback, J.D.; Mota, M. Description of Longidorus bordonensis sp. nov. from Portugal, with systematics and molecular phylogeny of the genus (Nematoda, Longidoridae). Zoosyst. Evol. 2020, 96, 175–193. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Castresana, J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef]
- Tanabe, A.S. Kakusan4 and Aminosan: Two programs for comparing nonpartitioned, proportional, and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Notes 2011, 11, 914–921. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Suchard, M.A.; Xie, D.; Drummond, A.J. Tracer v1.6. Available online: http://beast.bio.ed.ac.uk/Tracer (accessed on 2 December 2021).
- Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 2 December 2021).
- Oostenbrink, M. A note on Paratylenchus in the Netherlands with the description of P. goodeyi n. sp. (Nematoda, Criconematidae). Tijdschr. Plantenziekten 1953, 59, 207–216. [Google Scholar] [CrossRef]
- Castillo, P.; González-País, M.A.; Gómez-Barcina, A. El género Gracilacus Raski, 1962 en España (Paratylenchinae: Tylenchida). Rev. Ibér. Parasitol. 1989, 49, 321–328. [Google Scholar]
- Sturhan, D. Plant-parasitic nematodes in Germany—an annotated checklist. Soil Org. 2014, 83, 22. [Google Scholar]
- Háněl, L.; Čerevková, A. Diversity of soil nematodes in meadows of the White Carpathians. Helminthologia 2006, 43, 109–116. [Google Scholar] [CrossRef]
- Brzeski, M.W. Paratylenchinae: Morphology of some known species and descriptions of Gracilacus bilineata sp. n. and G. vera sp. n. (Nematoda: Tylenchulidae). Nematologica 1995, 41, 535–565. [Google Scholar] [CrossRef]
- Szczygiel, A. Plant parasitic nematodes associated with strawberry plants in Poland. Zesz. Probl. Postepów Nauk. Rol. 1974, 154, 9–132. [Google Scholar]
- Geraert, E. The Genus Paratylenchus. Nematologica 1965, 11, 301–334. [Google Scholar] [CrossRef]
- Solov’eva, G.I. Parasitic Nematodes of Woody and Herbaceous Plants. A Review of the Genus Paratylenchus Micoletzky, 1922 (Nematoda: Criconematidae); Amerind Publishing Co.: New Delhi, India, 1975. [Google Scholar]
- Lisetskaya, L.F. Nematode fauna of ethereal oil plants of Moldavia. Mater. Nats. Konf. Po Parazitol. Sofia 1968, 13, 47–48. [Google Scholar]
- Izatullaeva, R.I. Nematodes of the flowering plants of Kazakhstan; Abstract Kandidat’s Dissertation: Alma-Ata, Kazakhstan, 1967; pp. 1–18. [Google Scholar]
- Wu, L.Y. Paratylenchus tenuicaudatus n. sp. (Nematoda: Criconematidae). Can. J. Zool. 1961, 39, 163–165. [Google Scholar] [CrossRef]
- Subbotin, S.A.; Vovlas, N.; Crozzoli, R.; Sturhan, D.; Lamberti, F.; Moens, M.; Baldwin, J.G. Phylogeny of Criconematina Siddiqi, 1980 (Nematoda: Tylenchida) based on morphology and D2–D3 expansion segments of the 28S-rRNA gene sequences with application of a secondary structure model. Nematology 2005, 7, 927–944. [Google Scholar] [CrossRef]
- Myers, R.F. Plant parasitic nematodes in New Jersey. J. Nematol. 1986, 18, 272–274. [Google Scholar] [PubMed]
- Hernández, M.; Mateo, M.D.; Jordana, R. Estudio comparativo entre grupos tróficos de nematodos del suelo de cinco bosques de Navarra (tres naturales y dos de repoblación). Actas II Congr. Mund. Vasco. Sec. Biol. Ambient. 1988, 2, 323–335. [Google Scholar]
- Raski, D.J. Revision of the genus Paratylenchus Micoletzky, 1922 and descriptions of new species. Part II of three parts. J. Nematol. 1975, 7, 274–295. [Google Scholar]
- Wu, L.Y. Paratylenchus veruculatus n. sp. (Criconematidae: Nematoda) from Scotland. Can. J. Zool. 1962, 40, 773–775. [Google Scholar] [CrossRef]
- Duyck, P.F.; Dortel, E.; Tixier, P.; Vinatier, F.; Loubana, P.M.; Chabrier, C.; Quénéhervé, P. Niche partitioning based on soil type and climate at the landscape scale in a community of plant-feeding nematodes. Soil Biol. Biochem. 2012, 44, 49–55. [Google Scholar] [CrossRef]
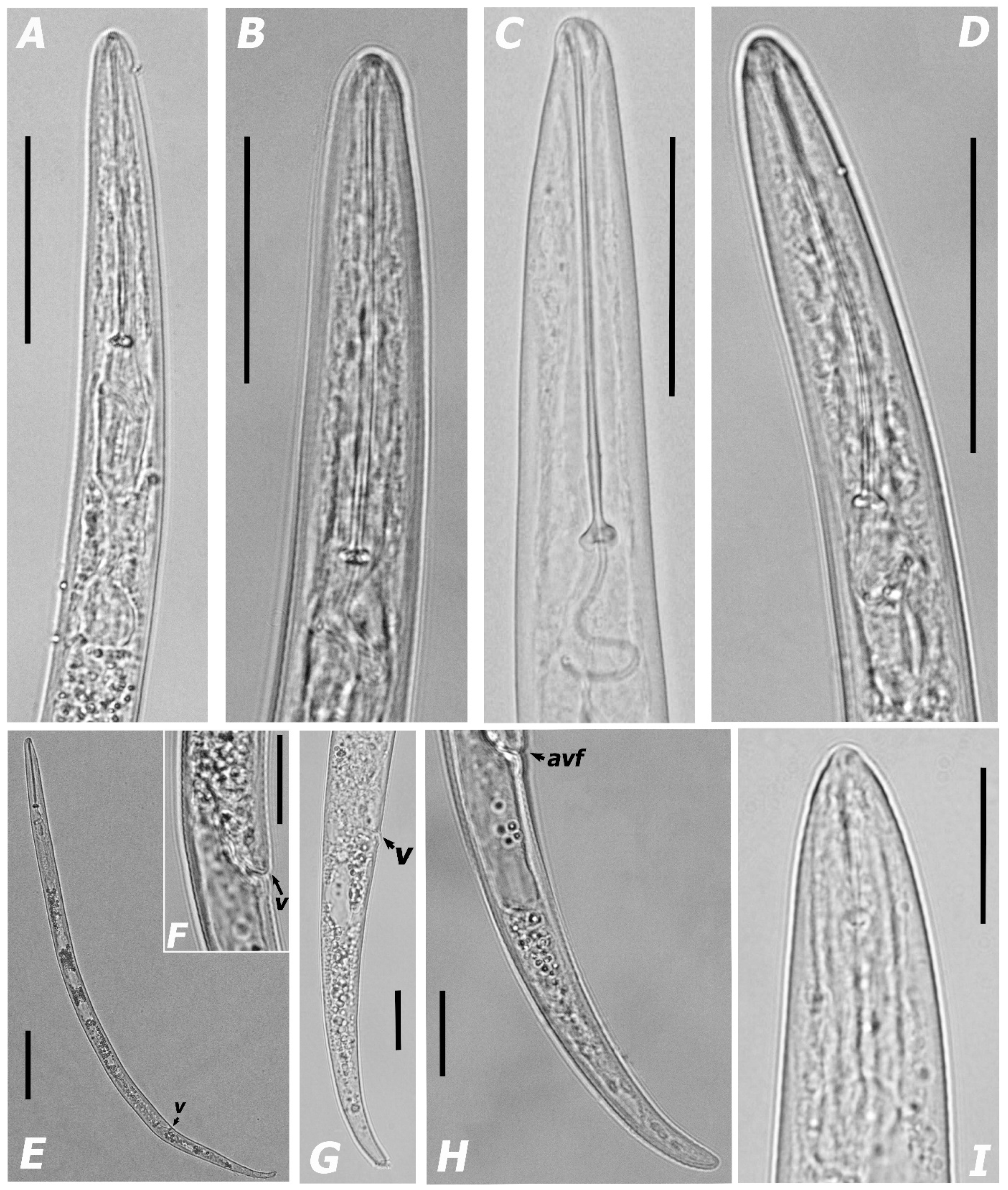
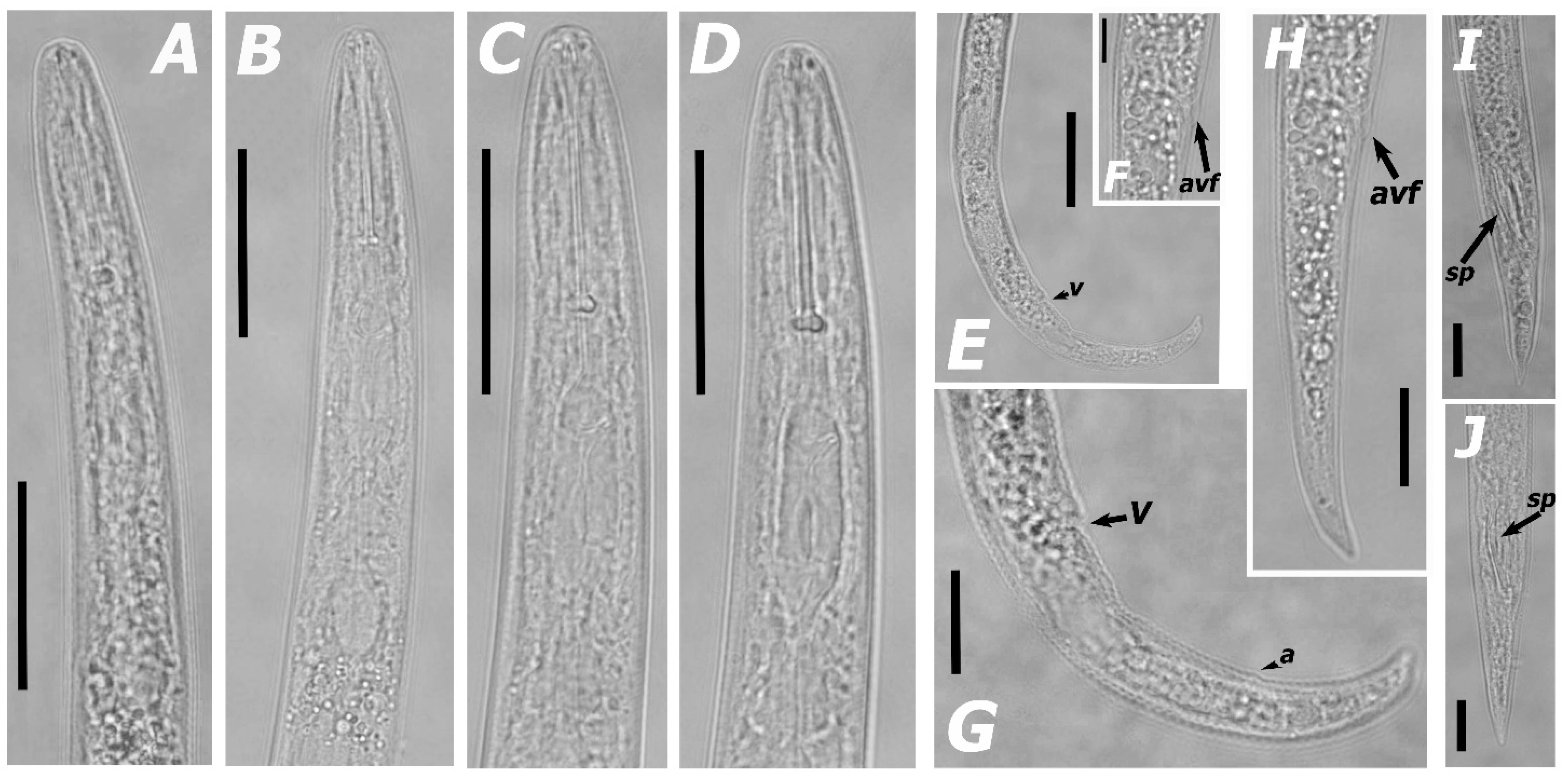
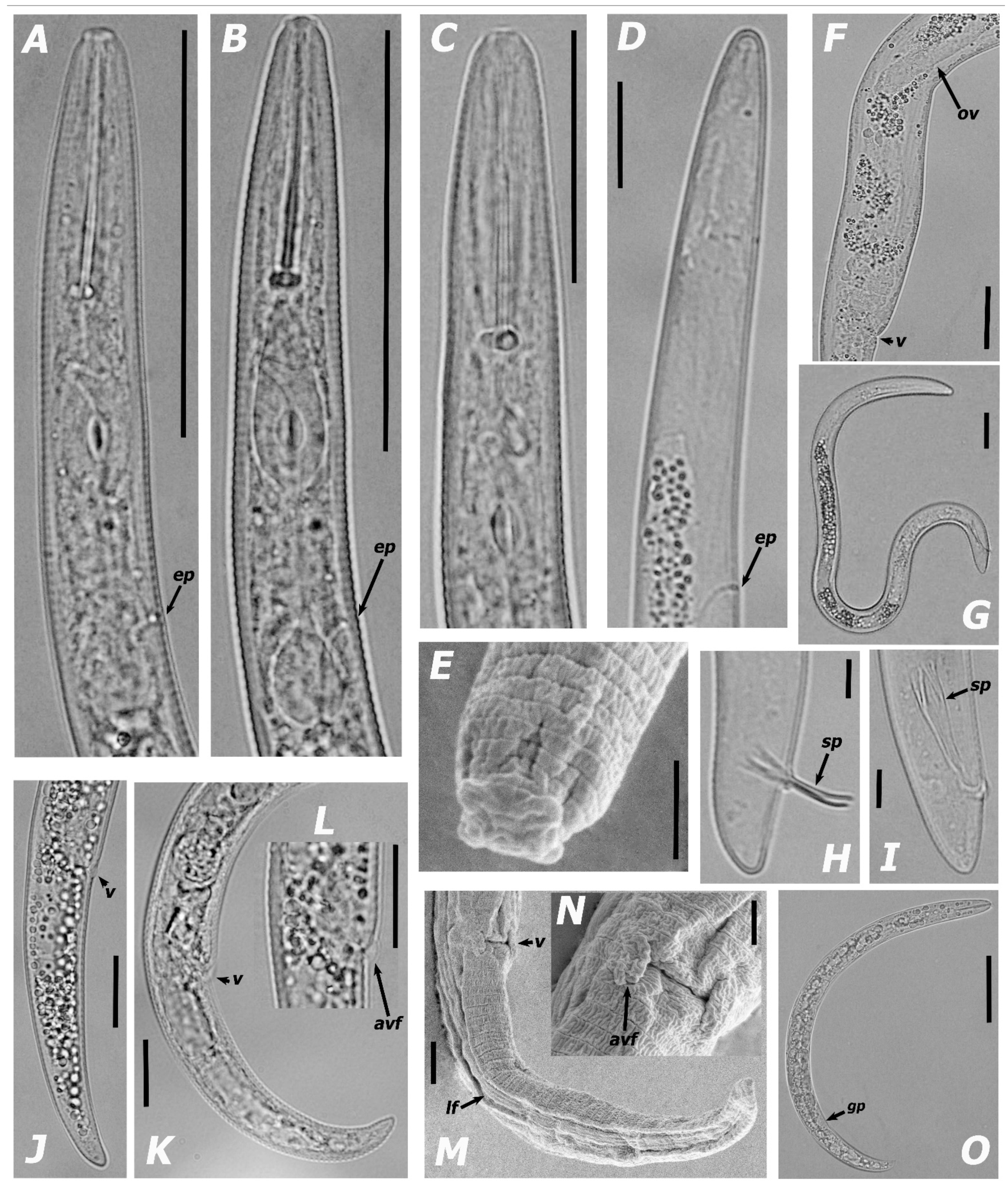
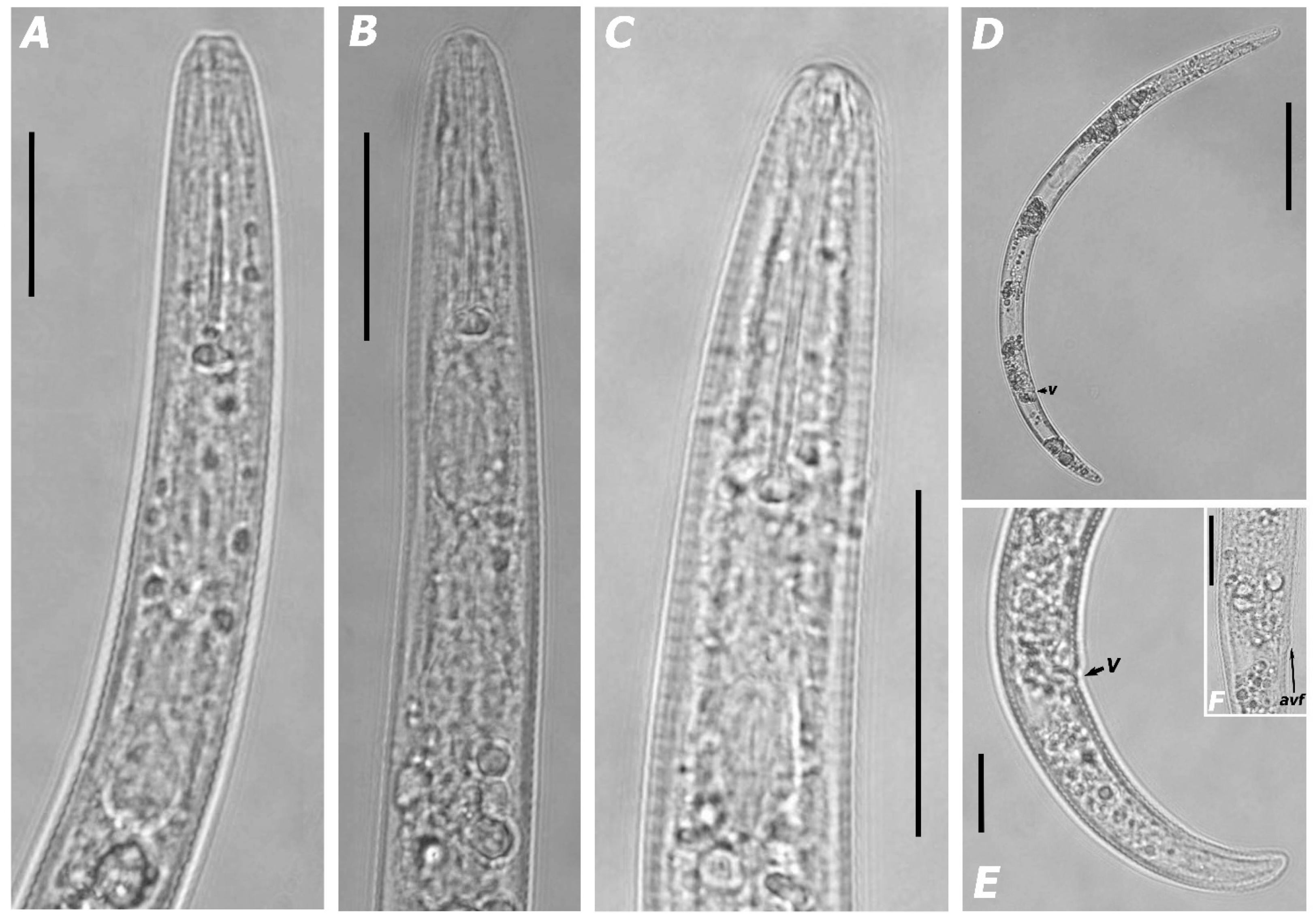

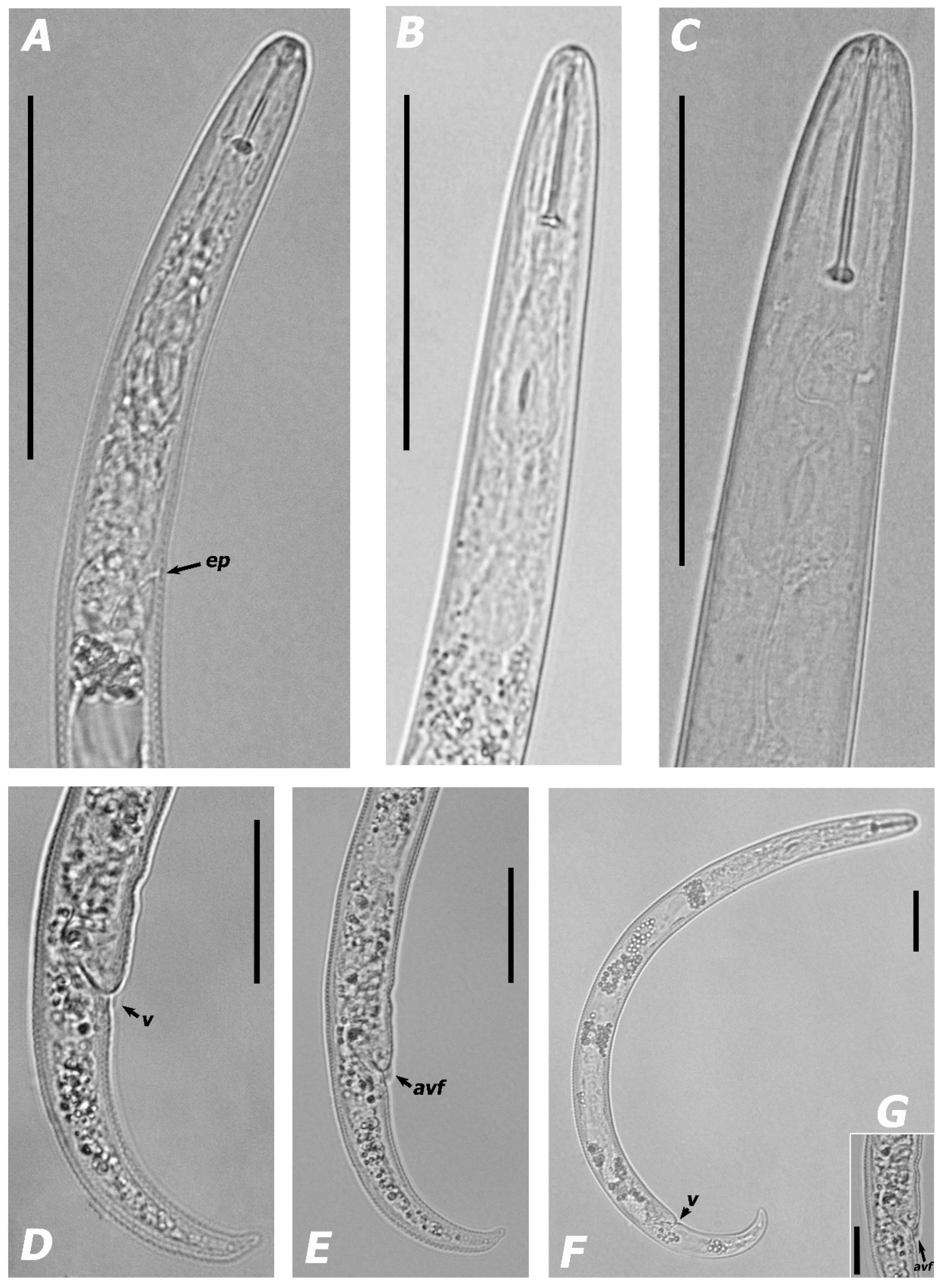

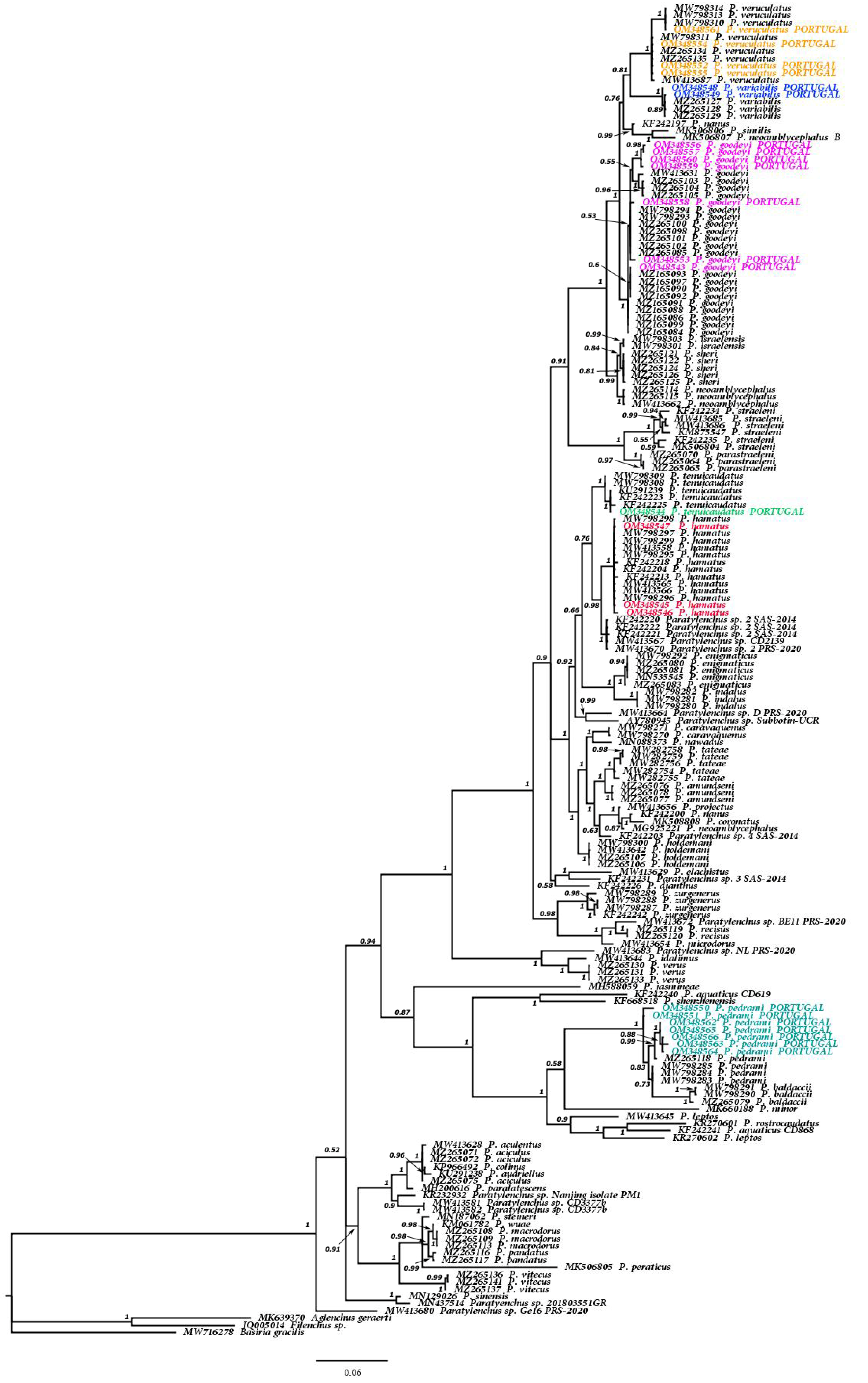
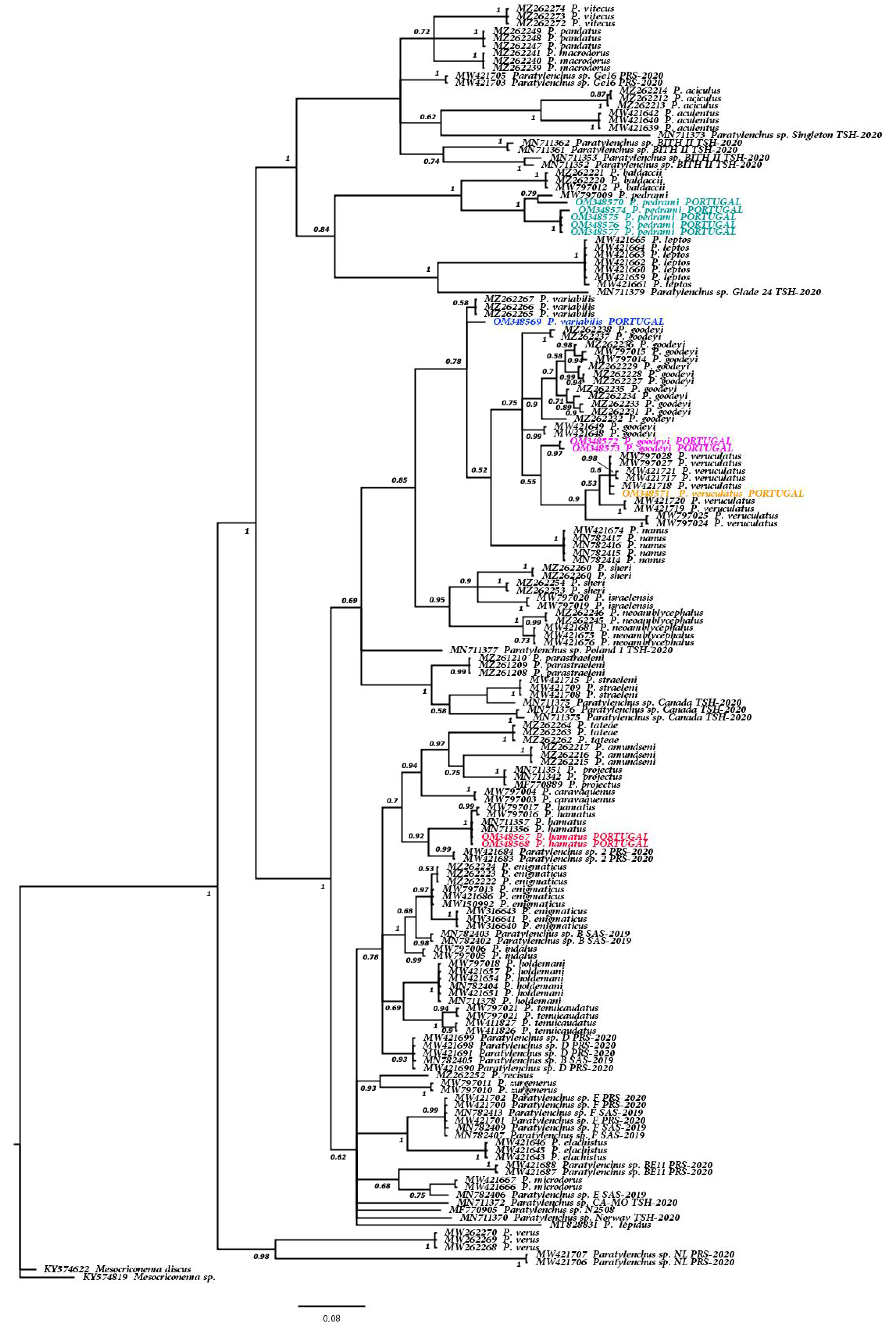
| Species | Sample Code | Locality | Host | Genbank Accessions | ||
|---|---|---|---|---|---|---|
| 18S | 28S | COI | ||||
| P. goodeyi | T1-3 | Monte da Ribeira, São Manços | grapevine | OM345189- OM345190 | OM348556- OM348560; OM348553 | OM348572- OM348573 |
| 09-02-20 | Carvalhal, Bombarral | grapevine | - | OM348543 | - | |
| P. hamatus | 45-007-20 | Roliça, Bombarral | grapevine | OM345185 | OM348545- OM348547 | OM348567- OM348568 |
| P. pedrami | AL-V4 | Santa Catarina de Sítimos, Alcácer do Sal | grapevine | OM345191- OM345192 | OM348562- OM348566- | OM348574- OM348577 |
| CF-1 | Aldeia Galega da Merceana, Alenquer | grapevine | - | OM348550- OM348551 | OM348570 | |
| P. tenuicaudatus | 198-33-19 | Carvalhal, Bombarral | grapevine | OM345184 | OM348544 | - |
| P. variabilis | 197-32-19 | São Domingos de Carmões, Torres Vedras | grapevine | OM345186 | OM348548- OM348549 | OM348569 |
| P. veruculatus | T1-3 | Monte da Ribeira, São Manços | grapevine | OM345187- OM345188 | OM348552; OM348554- OM348555; OM348561 | OM348571 |
| Measurements and Ratios | P. goodeyi | P. tenuicaudatus | ||
|---|---|---|---|---|
| Sample Code | T1-3 | 09-02-20 | 198-33-19 | |
| Locality | Monte da Ribeira, São Manços | Carvalhal, Bombarral | Carvalhal, Bombarral | |
| n | 5 females | 1 female | 5 females | 1 male |
| L | 449.2 ± 33.5 (399.5–481.5) | 414.5 | 296.8 ± 20.7 (276.7–331.3) | 354.4 |
| a | 31.1 ± 1.8 (28.4–32.7) | 28.1 | 24.4 ± 1.1 (22.8–25.5) | 26.4 |
| b | 3.6 ± 0.2 (3.5–3.9) | 3.4 | 4.1 ± 0.3 (3.7–4.4) | 4.6 |
| c | 8.7 ± 1.3 (7.3–10.0) | 9.1 | 11.1 ± 1.9 (8.4–13.3) | 13.1 |
| c’ | 5.0 ± 0.5 (4.5–5.5) | 4.2 | 3.0 ± 0.3 (2.6–3.5) | 2.7 |
| V or T | 79.8 ± 1.2 (78.2–81.0) | 80.8 | 81.2 ± 1.2 (79.2–81.9) | 63.6 |
| G1(%) | 31.9 ± 2.2 (29.6–34.8) | 41.7 | 22.8 ± 2.8 (20.9–26.0) | - |
| Stylet length | 57.9 ± 2.2 (54.8–60.6) | 49.6 | 30.2 ± 1.1 (29.0–31.6) | - |
| Cone length | 47.7 ± 1.7 (46.5–48.9) | 39.7 | 25.5 ± 0.3 (25.3–25.7) | - |
| (Stylet length/body length) × 100 | 13.0 ± 0.6 (12.3–14.0) | 12.0 | 10.2 ± 0.5 (9.5–10.8) | - |
| m | 84.6 ± 0.3 (84.4–84.9) | 80.1 | 81.6 ± 2.1 (80.1–83.0) | - |
| DGO | 5.2 ± 0.5 (4.8–5.6) | 5.5 | 5.1 ± 0.6 (4.7–5.6) | - |
| O | 9.2 ± 0.6 (8.8–9.6) | 11.0 | 16.5 ± 2.3 (14.9–18.1) | - |
| Lip width | 2.9 ± 0.5 (2.4–3.5) | 2.7 | 3.3 ± 0.7 (2.7–4.2) | 2.9 |
| Median bulb length | 22.8 ± 1.6 (20.7–25.0) | 35.5 | 14.3 ± 1.6 (12.0–16.2) | 16.5 |
| Median bulb width | 8.5 ± 0.5 (7.8–9.3) | 9.2 | 5.9 ± 0.9 (5.0–7.3) | 4.7 |
| Anterior end to center median bulb | 76.0 ± 4.2 (70.5–81.0) | 69.7 | 41.1 ± 2.1 (37.6–43.1) | 40.9 |
| MB | 61.8 ± 1.7 (59.9–63.6) | 57.3 | 56.2 ± 4.1 (49.5–59.5) | 49.1 |
| Excretory pore to anterior end | 93.9 ± 12.2 (84.0–115.0) | 86.1 | 63.3 ± 3.7 (58.0–68.4) | 75.5 |
| Pharynx length | 123.0 ± 4.7 (115.0–127.3) | 121.6 | 73.2 ± 2.7 (70.1–76.0) | 83.3 |
| Maximum body diam. | 14.4 ± 0.5 (13.8–14.9) | 14.7 | 12.2 ± 1.3 (11.5–14.5) | 13.4 |
| Tail length | 52.4 ± 8.5 (40.0–60.7) | 45.5 | 27.2 ± 4.3 (22.4–33.0) | 27.1 |
| Anal body diam. | 10.5 ± 1.0 (9.0–11.4) | 10.8 | 9.2 ± 1.1 (7.7–10.6) | 10.2 |
| Spicules | - | - | - | 19 |
| Gubernaculum | - | - | - | 6.5 |
| Measurements and Ratios | P. hamatus | P. variabilis | P. veruculatus | |
|---|---|---|---|---|
| Sample Code | 045-007-20 | 197-32-19 | T1-3 | |
| Locality | Roliça, Bombarral | São Domingos de Carmões, Torres Vedras | Monte da Ribeira, São Manços | |
| n | 4 females | 5 males | 13 females | 2 females |
| L | 320.1 ± 16.4 (298.3–337.8) | 442.9 ± 19.3 (321.9–362.7) | 296.1 ± 23.2 (247.7–336.2) | 306.8 ± 22.0 (291.3–322.4) |
| a | 22.8 ± 1.9 (20.4–25.0) | 27.1 ± 2.1 (24.7–29.6) | 22.8 ± 1.4 (20.5–25.0) | 21.9 ± 3.0 (19.8–24.0) |
| b | 4.1 ± 0.1 (4.0–4.1) | 4.6 ± 0.3 (4.1–4.8) | 4.2 ± 0.4 (3.8–4.9) | 3.8 ± 0.3 (3.6–4.0) |
| c | 10.6 ± 0.9 (9.4–11.6) | 13.3 ± 1.4 (11.7–14.8) | 12.9 ± 1.6 (9.1–15.0) | 11.7 ± 0.6 (11.3–12.1) |
| c’ | 3.2 ± 0.4 (2.8–3.6) | 3.1 ± 0.4 (2.5–3.5) | 2.6 ± 0.4 (2.0–3.4) | 2.7 ± 0.4 (2.4–2.9) |
| V or T | 81.8 ± 1.7 (80.6–84.1) | 68.2 ± 1.5 (66.4–69.7) | 83.9 ± 1.0 (82.4–85.4) | 83.8 ± 1.1 (83.0–84.6) |
| G1(%) | 35.7 ± 0.7 (34.9–36.4) | - | 30.1 ± 6.4 (18.0–38.4) | 19.8 ± 0.3 (19.5–20.0) |
| Stylet length | 30.6 ± 1.3 (28.8–31.9) | 19.0 ± 1.9 (16.0–20.7) | 17.6 ± 0.8 (16.3–19.0) | 14.0 ± 1.3 (13.1–14.9) |
| Cone length | 19.5 ± 1.9 (18.1–20.9) | 14.3 ± 0.5 (14.0–14.7) | 13.0 ± 1.0 (12.0–14.0) | 8.3 ± 1.1 (7.5–9.0) |
| (Stylet length/body length) × 100 | 9.6 ± 0.4 (9.0–10.0) | 5.6 ± 0.7 (4.4–6.1) | 6.0 ± 0.7 (5.1–7.7) | 5.2 ± 0.6 (4.8–5.6) |
| m | 65.7 ± 4.0 (62.8–68.5) | 73.6 ± 3.8 (70.9–76.3) | 72.1 ± 8.1 (63.2–80.5) | 68.8 ± 8.8 (62.5–75.0) |
| DGO | 6.6 ± 1.0 (5.9–7.3) | - | 5.4 ± 0.2 (5.2–5.6) | - |
| O | 22.3 ± 4.2 (19.4–25.3) | - | 29.9 ± 2.2 (27.8–32.4) | - |
| Lip width | 3.4 ± 0.4 (3.0–3.9) | 3.1 ± 0.2 (2.9–3.4) | 3.5 ± 0.5 (2.7–4.3) | 3.8 ± 0.4 (3.5–4.1) |
| Median bulb length | 19.8 ± 2.7 (16.9–22.4) | 17.4 ± 3.1 (15.2–22.0) | 17.1 ± 1.5 (15.2–20.5) | 20.3 ± 1.4 (19.3–21.3) |
| Median bulb width | 6.8 ± 0.6 (5.8–7.2) | 4.6 ± 0.6 (4.0–5.2) | 7.4 ± 1.0 (5.7–9.9) | 7.4 ± 0.1 (7.4–7.3) |
| Anterior end to center median bulb | 41.5 ± 2.8 (37.5–44.0) | 39.9 ± 0.2 (38.8–40.0) | 36.4 ± 2.2 (32.1–40.5) | 41.9 ± 0.3 (51.7–42.2) |
| MB | 53.8 ± 3.3 (50.5–56.8) | 51.5 ± 1.6 (50.3–52.6) | 51.9 ± 1.8 (49.1–54.7) | 51.6 ± 0.3 (51.4–51.9) |
| Excretory pore to anterior end | 69.1 ± 3.5 (66.7–74.1) | 68.2 ± 2.0 (65.3–70.0) | 64.4 ± 3.1 (57.4–68.4) | 73.1 ± 0.3 (72.9–73.3) |
| Pharynx length | 77.1 ± 2.7 (74.3–80.9) | 75.1 ± 2.9 (71.9–79.0) | 70.2 ± 4.2 (62.1–75.0) | 81.2 ± 0.1 (81.1–81.3) |
| Maximum body diam. | 14.1 ± 0.5 (13.5–14.7) | 12.8 ± 1.6 (10.9–14.6) | 13.0 ± 1.0 (11.9–15.4) | 14.1 ± 0.9 (13.4–14.7) |
| Tail length | 30.4 ± 3.7 (27.7–35.9) | 26.0 ± 2.9 (21.8–28.2) | 23.3 ± 3.3 (16.5–31.7) | 26.3 ± 3.1 (24.2–28.5) |
| Anal body diam. | 9.6 ± 1.0 (8.1–10.4) | 8.5 ± 0.7 (8.1–9.7) | 9.1 ± 0.9 (6.9–1.3) | 9.8 ± 0.1 (9.8–9.9) |
| Spicules | - | 21.8 ± 1.1 (20.1–22.6) | - | - |
| Gubernaculum | - | 11.7 ± 2.0 (9.0–13.9) | - | - |
| Character/Sample Code | AL-V4 | CF-1 | |
|---|---|---|---|
| Locality | Santa Catarina de Sítimos, Alcácer do Sal | Aldeia Galega da Merceana | |
| n | 20 females | 3 males | 12 females |
| L | 298.5 ± 22.8 (239.3–337.4) | 255.2 ± 32.2 (227.3–290.5) | 294.3 ± 16.9 (270.0–330.6) |
| a | 24.4 2.7 (19.7–28.9) | 23.2 ± 4.3 (20.7–28.1) | 24.4 ± 1.9 (20.4–27.1) |
| b | 4.0 ± 0.3 (3.6–4.7) | 3.8 ± 0.1 (3.8–3.9) | 4.2 ± 0.5 (3.2–5.2) |
| c | 13.0 ± 2.4 (9.7–17.7) | 24.1 ± 4.5 (19.4–28.3) | 14.3 ± 1.5 (11.2–16.4) |
| c’ | 2.9 ± 0.5 (2.0–3.8) | 1.4 ± 0.3 (1.1–1.6) | 2.4 ± 0.3 (2.0–3.0) |
| V or T | 81.2 ± 1.4 (79.2–85.4) | 28.7 ± 1.9 (27.4–30.1) | 80.5 ± 1.3 (78.9–82.9) |
| G1 | 29.3 ± 9.7 (17.8–53.3) | - | 25.3 ± 3.2 (19.9–28.0) |
| Stylet length | 29.8 ± 1.2 (27.5–32.0) | - | 29.2 ± 1.3 (26.2–30.5) |
| Cone length | 20.1 ± 1.2 (18.3–22.4) | - | 20.6 ± 0.5 (20.0–21.0) |
| (Stylet length/body length) × 100 | 10.0 ± 0.7 (9.1–11.9) | - | 10.0 ± 0.3 (9.7–10.7) |
| m | 67.6 ± 2.6 (61.4–72.6) | - | 69.7 ± 1.6 (67.8–71.5) |
| DGO | 5.1 ± 0.6 (4.0–6.1) | - | 4.3 ± 0.4 (3.8–4.5) |
| O | 17.1 ± 1.9 (13.7–19.9) | - | 14.4 ± 1.3 (12.9–15.2) |
| Lip width | 3.0 ± 0.2 (2.5–3.4) | 1.8 ± 0.1 (1.6–1.9) | 3.3 ± 0.4 (2.3–3.8) |
| Median bulb length | 18.7 ± 1.6 (16.4–21.7) | - | 16.5 ± 1.1 (14.5–18.1) |
| Median bulb width | 7.1 ± 0.8 (6.2–8.3) | - | 5.9 ± 0.6 (4.9–7.2) |
| Anterior end to center median bulb | 44.2 ± 1.5 (41.5–46.6) | - | 38.7 ± 3.7 (32.0–42.6) |
| MB | 58.7 ± 2.9 (53.6–65.6) | - | 55.9 ± 4.7 (44.5–61.3) |
| Excretory pore to anterior end | 65.2 ± 3.7 (57.7–70.5) | 55.1 ± 3.0 (528–58.5) | 60.0 ± 3.2 (52.0–63.1) |
| Pharynx length | 74.3 ± 5.5 (64.0–83.2) | 65.2 ± 2.5 (60.4–64.0) | 69.5 ± 6.5 (59.6–83.2) |
| Maximum body diam. | 12.4 ± 1.4 (9.5–14.4) | 11.1 ± 0.8 (10.3–12.0) | 12.0 ± 1.0 (10.8–14.3) |
| Tail length | 23.7 ± 4.4 (17.0–30.3) | 10.7 ± 1.7 (8.8–11.7) | 20.5 ± 2.2 (17.0–26.0) |
| Anal body diam | 8.1 ± 1.0 (5.7–9.8) | 7.6 ± 0.4 (7.2–8.0) | 8.5 ± 1.2 (6.9–10.4) |
| Spicules | - | 16.2 ± 0.5 (15.7–16.7) | - |
| Gubernaculum | - | 3.8 ± 0.1 (3.8–3.9) | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosmaninho, T.; Mota, M.; Inácio, M.L.; Eisenback, J.D.; Gutiérrez-Gutiérrez, C. Six First Reports of Pin Nematodes from Portugal, with an Update of the Systematics, Genetic Diversity, and Phylogeny of the Genus Paratylenchus (Nematoda: Tylenchulidae). Horticulturae 2022, 8, 343. https://doi.org/10.3390/horticulturae8040343
Rosmaninho T, Mota M, Inácio ML, Eisenback JD, Gutiérrez-Gutiérrez C. Six First Reports of Pin Nematodes from Portugal, with an Update of the Systematics, Genetic Diversity, and Phylogeny of the Genus Paratylenchus (Nematoda: Tylenchulidae). Horticulturae. 2022; 8(4):343. https://doi.org/10.3390/horticulturae8040343
Chicago/Turabian StyleRosmaninho, Teresa, Manuel Mota, Maria L. Inácio, Jonathan D. Eisenback, and Carlos Gutiérrez-Gutiérrez. 2022. "Six First Reports of Pin Nematodes from Portugal, with an Update of the Systematics, Genetic Diversity, and Phylogeny of the Genus Paratylenchus (Nematoda: Tylenchulidae)" Horticulturae 8, no. 4: 343. https://doi.org/10.3390/horticulturae8040343
APA StyleRosmaninho, T., Mota, M., Inácio, M. L., Eisenback, J. D., & Gutiérrez-Gutiérrez, C. (2022). Six First Reports of Pin Nematodes from Portugal, with an Update of the Systematics, Genetic Diversity, and Phylogeny of the Genus Paratylenchus (Nematoda: Tylenchulidae). Horticulturae, 8(4), 343. https://doi.org/10.3390/horticulturae8040343










