Chinese Cherry (Cerasus pseudocerasus Lindl.) ARF7 Participates in Root Development and Responds to Drought and Low Phosphorus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. RNA Extraction and Gene Isolation
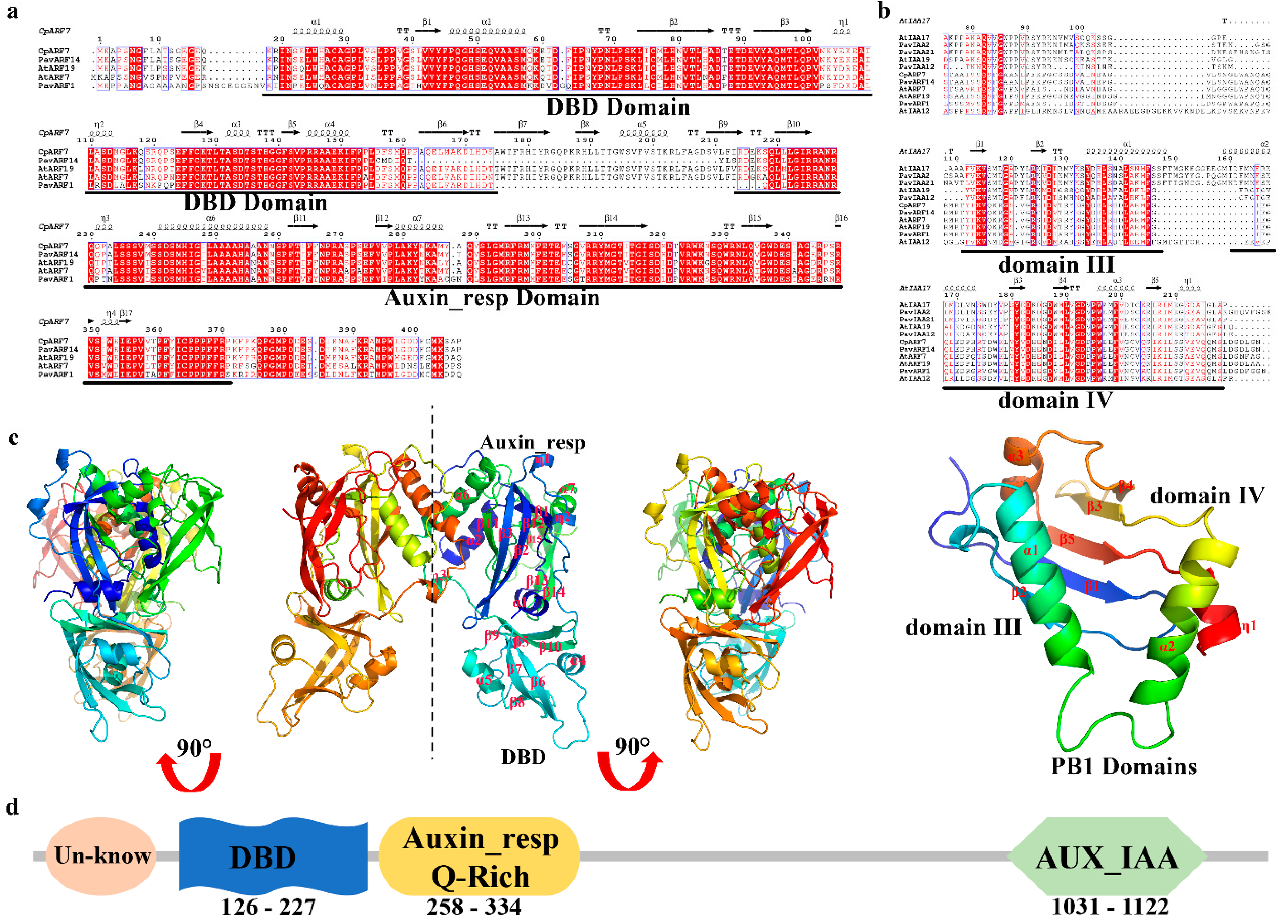
2.3. Sequence Analysis of the CpARF7
2.4. Genetic Transformation of Tomato
2.5. Vegetative Growth and Root System Parameter Analyses
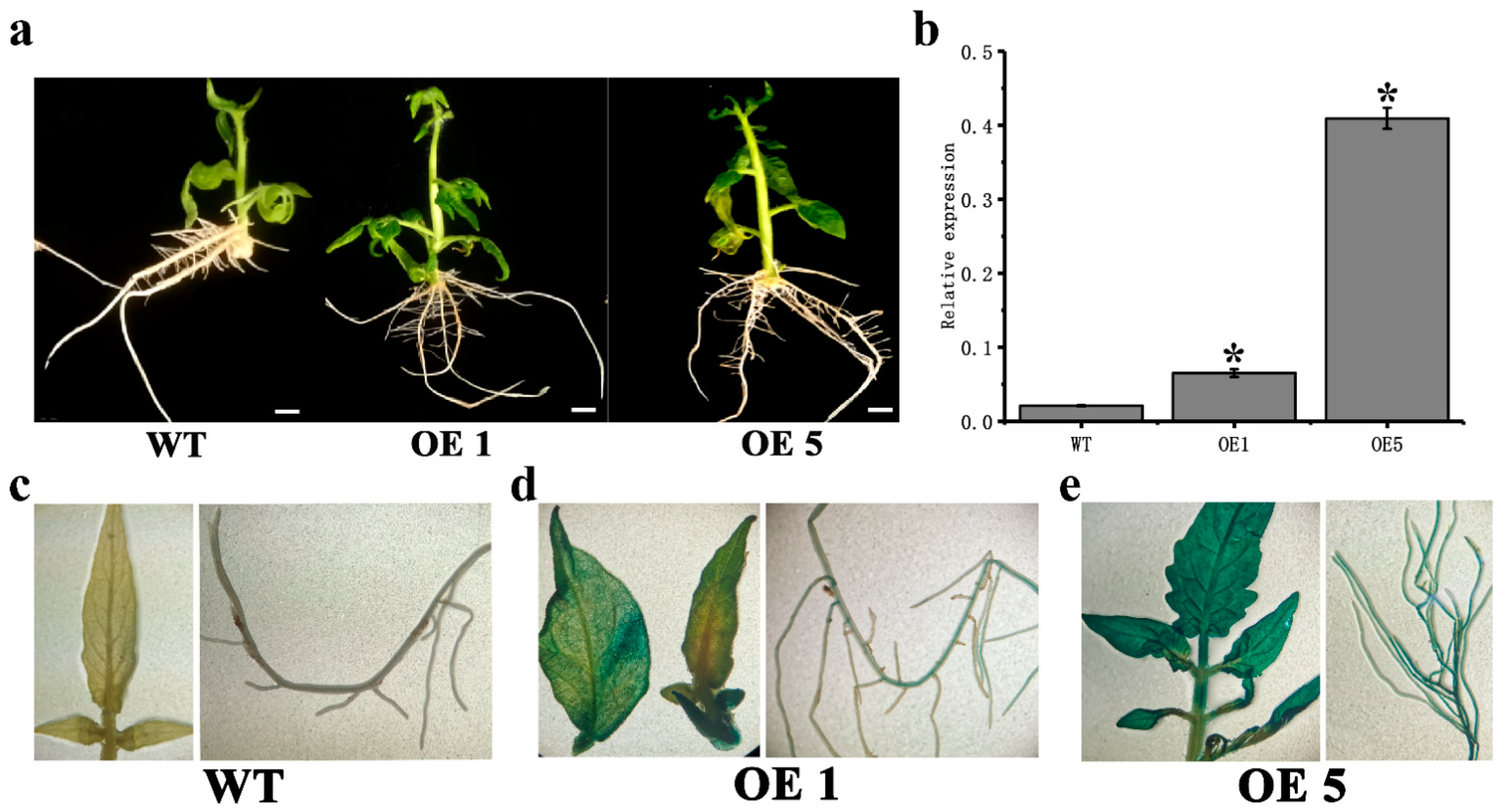
2.6. Real-Time Quantitative PCR and GUS Staining
2.7. Statistical Analysis
3. Results
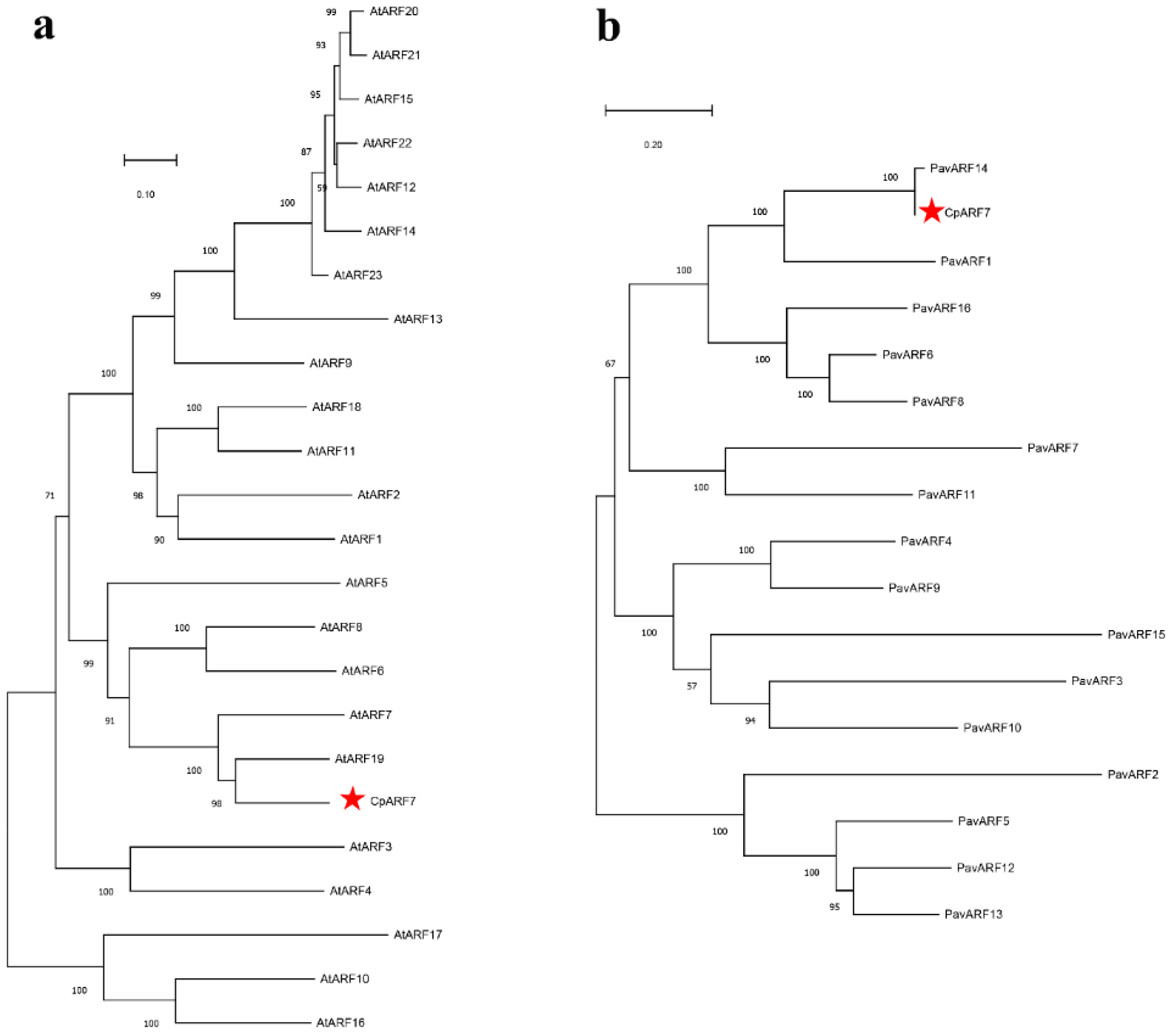
3.1. CpARF7 in the ARF Family
3.2. CpARF7 Promotes Root Growth
3.3. CpARF7 Can Improve the Drought Resistance of Transformed Tomato
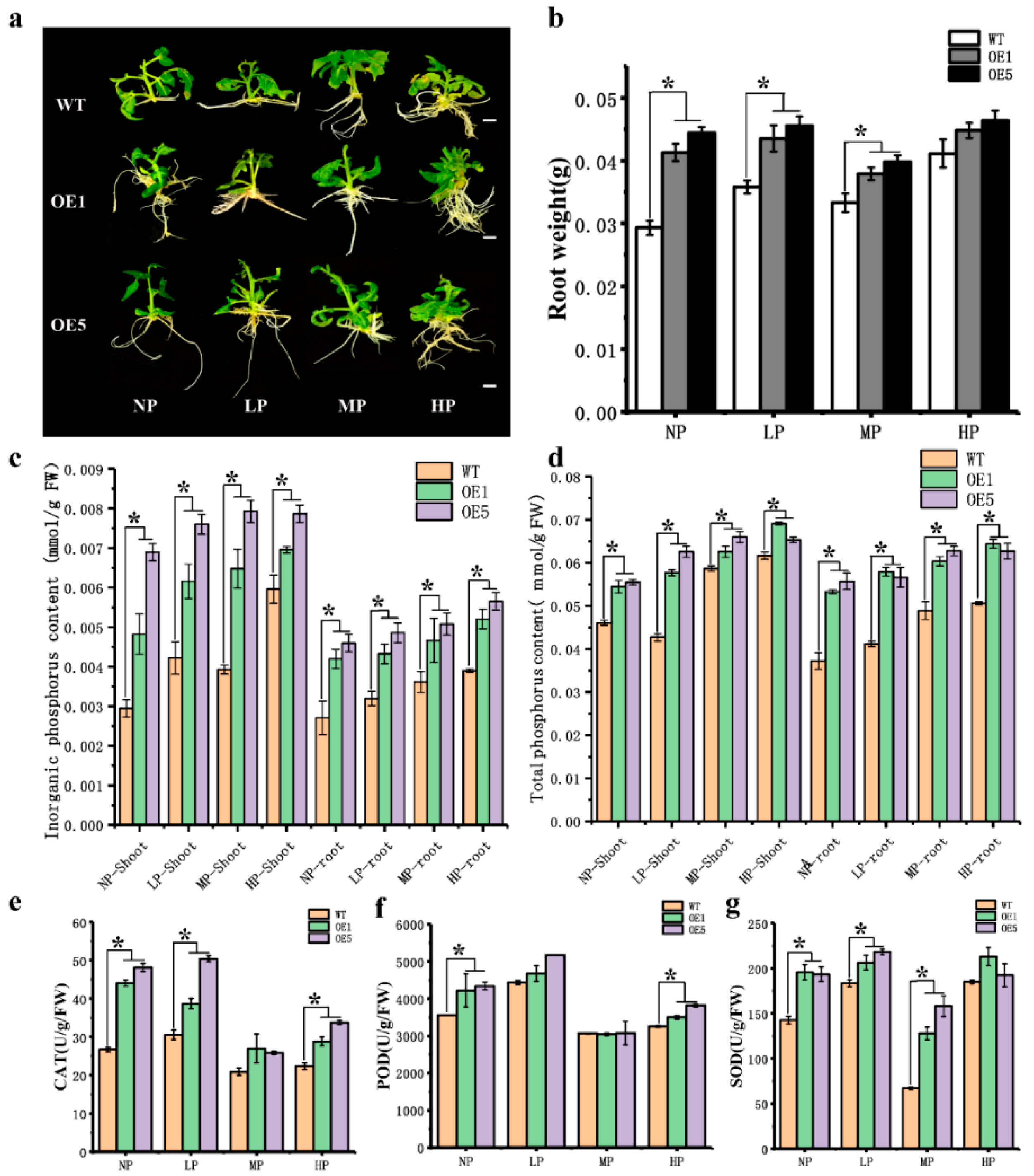
3.4. Overexpression of CpARF7 Improves the Phosphorus Absorption Capacity of Roots
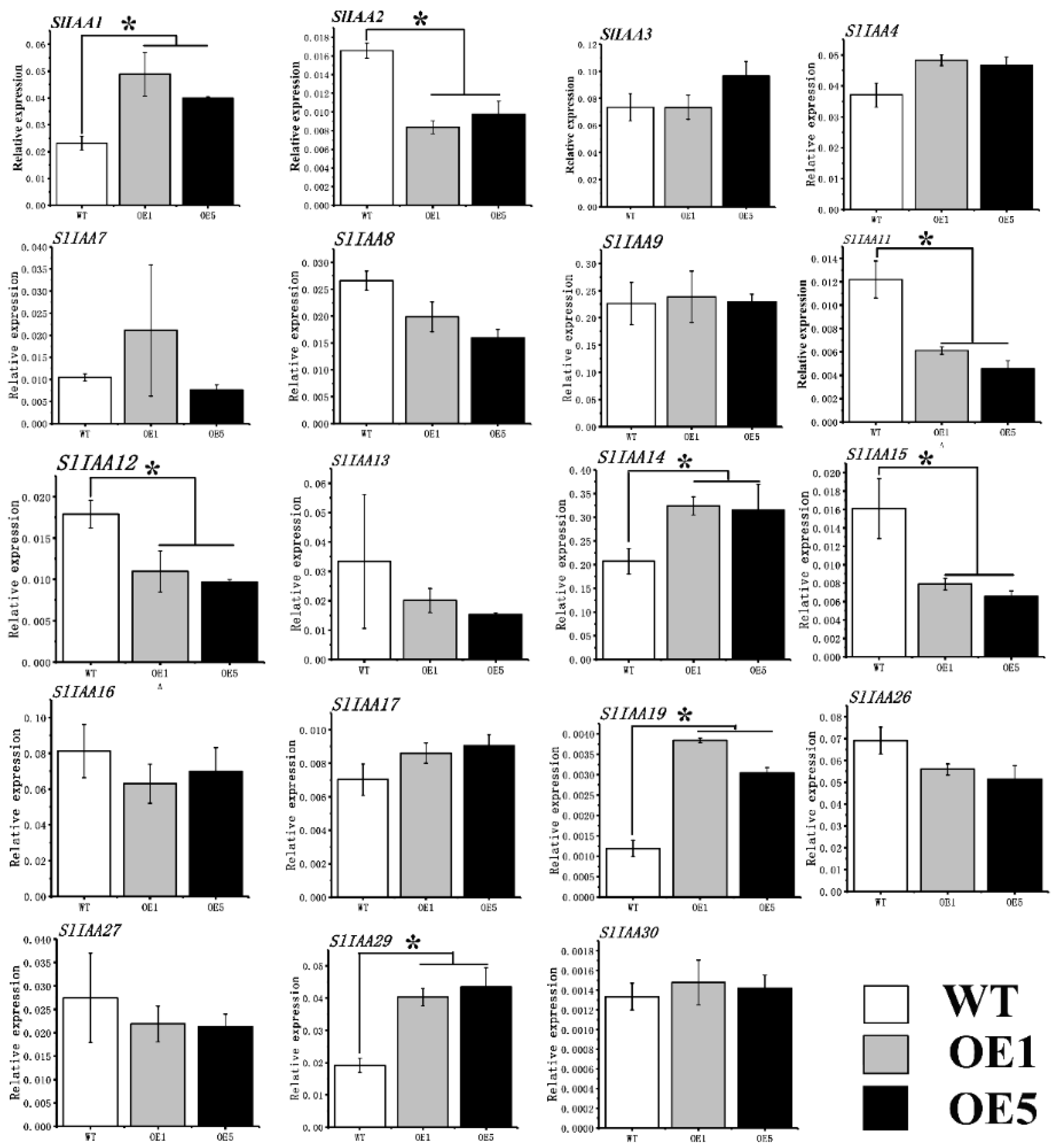
3.5. The Effect of Overexpression of CpARF7 on Downstream Gene Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, J.; Chen, T.; Wang, Y.; Chen, Q.; Sun, B.; Luo, Y.; Zhang, Y.; Tang, H.; Wang, X. Genetic diversity and domestication footprints of Chinese cherry [cerasus pseudocerasus (Lindl.) g.don] as revealed by nuclear microsatellites. Front. Plant Sci. 2018, 9, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Motte, H.; Vanneste, S.; Beeckman, T. Molecular and Environmental Regulation of Root Development. Annu. Rev. Plant Biol. 2019, 70, 465–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saini, S.; Sharma, I.; Kaur, N.; Pati, P.K. Auxin: A master regulator in plant root development. Plant Cell Rep. 2013, 32, 741–757. [Google Scholar] [CrossRef]
- Meier, M.; Liu, Y.; Lay-Pruitt, K.S.; Takahashi, H.; von Wirén, N. Auxin-mediated root branching is determined by the form of available nitrogen. Nat. Plants 2020, 6, 1136–1145. [Google Scholar] [CrossRef] [PubMed]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guilfoyle, T.J.; Hagen, G. Auxin response factors. Curr. Opin. Plant Biol. 2007, 10, 453–460. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 2006, 88, 360–371. [Google Scholar] [CrossRef] [Green Version]
- Xing, H.; Pudake, R.N.; Guo, G.; Xing, G.; Hu, Z.; Zhang, Y.; Sun, Q.; Ni, Z. Genome-wide identification and expression profiling of auxin response factor (ARF) gene family in maize. BMC Genomics 2011, 12, 178. [Google Scholar] [CrossRef] [Green Version]
- Li, S.B.; Ouyang, W.Z.; Hou, X.J.; Xie, L.L.; Hu, C.G.; Zhang, J.Z. Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 2015, 6, 119. [Google Scholar] [CrossRef] [Green Version]
- Diao, D.; Hu, X.; Guan, D.; Wang, W.; Yang, H.; Liu, Y. Genome-wide identification of the ARF (auxin response factor) gene family in peach and their expression analysis. Mol. Biol. Rep. 2020, 47, 4331–4344. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Qiu, Z.; Wen, Z.; Zhang, H.; Li, Z.; Hong, Y.; Qiao, G. Genome-Wide Identification of ARF Gene Family Suggests a Functional Expression Pattern during Fruitlet Abscission in Prunus avium L. Int. J. Mol. Sci. 2021, 22, 11968. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Dinh, T.T.; Li, D.; Shi, B.; Li, Y.; Cao, X.; Guo, L.; Pan, Y.; Jiao, Y.; Chen, X. AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 2014, 80, 629–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.; Liu, R.; Gu, W.; Dong, X. The Solanum lycopersicum auxin response factor SlARF2 participates in regulating lateral root formation and flower organ senescence. Plant Sci. 2017, 256, 103–111. [Google Scholar] [CrossRef]
- Yamauchi, T.; Tanaka, A.; Inahashi, H.; Nishizawa, N.K.; Tsutsumi, N.; Inukai, Y.; Nakazono, M. Fine control of aerenchyma and lateral root development through AUX/IAA- And ARF-dependent auxin signaling. Proc. Natl. Acad. Sci. USA. 2019, 116, 20770–20775. [Google Scholar] [CrossRef] [Green Version]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M.; Chan, A.; Chang, C.; Ecker, J.R.; Hughes, B.; Lui, A.; Nguyen, D.; et al. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [Green Version]
- Okushima, Y.; Fukaki, H.; Onoda, M.; Theologis, A.; Tasaka, M. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 2007, 19, 118–130. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Jones, B.; Li, Z.; Frasse, P.; Delalande, C.; Regad, F.; Chaabouni, S.; Latché, A.; Pech, J.C.; Bouzayen, M. The tomato Aux/IAA transcription factor IAA9 is involved in fruit development and leaf morphogenesis. Plant Cell 2005, 17, 2676–2692. [Google Scholar] [CrossRef] [Green Version]
- Ayyaz, A.; Miao, Y.; Hannan, F.; Islam, F.; Zhang, K.; Xu, J.; Farooq, M.A.; Zhou, W. Drought tolerance in Brassica napus is accompanied with enhanced antioxidative protection, photosynthetic and hormonal regulation at seedling stage. Physiol. Plant. 2021, 172, 1133–1148. [Google Scholar] [CrossRef]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.K. Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef]
- Wieczorek, P.; Wrzesińska, B.; Obrepalska-Steplowska, A. Assessment of reference gene stability influenced by extremely divergent disease symptoms in Solanum lycopersicum L. J. Virol. Methods 2013, 194, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Chen, Y.; Chen, Y.; Shin, J.H.; Mila, I.; Audran, C.; Zouine, M.; Pirrello, J.; Bouzayen, M. The tomato Ethylene Response Factor Sl-ERF.B3 integrates ethylene and auxin signaling via direct regulation of Sl-Aux/IAA27. New Phytol. 2018, 219, 631–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.A.; Bouzayen, M.; Zouine, M. Auxin response factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef] [Green Version]
- Santhi, C.K.V.; Rajesh, M.K.; Ramesh, S.V.; Muralikrishna, K.S.; Gangaraj, K.P.; Gupta, P.; Dash, P.K. Genome-wide exploration of auxin response factors (ARFs) and their expression dynamics in response to abiotic stresses and growth regulators in coconut (Cocos nucifera L.). Plant Gene 2021, 28, 100344. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A sweetpotato auxin response factor gene (IbARF5) is involved in carotenoid biosynthesis and salt and drought tolerance in transgenic Arabidopsis. Front. Plant Sci. 2018, 9, 01307. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Sun, C.; Xu, Y.; Chen, Y.; Yu, C.; Qian, Q.; Jiang, D.A.; Qi, Y. Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 2014, 201, 91–103. [Google Scholar] [CrossRef]
- Jones, V.A.S.; Dolan, L. The evolution of root hairs and rhizoids. Ann. Bot. 2012, 110, 205–212. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.B.; Sperry, J.S.; Dawson, T.E. Root water uptake and transport: Using physiological processes in global predictions. Trends Plant Sci. 2000, 5, 482–488. [Google Scholar] [CrossRef]
- Rellán-Álvarez, R.; Lobet, G.; Dinneny, J.R. Environmental Control of Root System Biology. Annu. Rev. Plant Biol. 2016, 67, 619–642. [Google Scholar] [CrossRef] [Green Version]
- Moubayidin, L.; Di Mambro, R.; Sabatini, S. Cytokinin-auxin crosstalk. Trends Plant Sci. 2009, 14, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Luijten, M.; Heidstra, R. Arabidopsis Root Development. Annu. Plant Rev. 2009, 37, 1–38. [Google Scholar] [CrossRef]
- Lau, S.; Shao, N.; Bock, R.; Jürgens, G.; De Smet, I. Auxin signaling in algal lineages: Fact or myth? Trends Plant Sci. 2009, 14, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Finet, C.; Fourquin, C.; Vinauger, M.; Berne-Dedieu, A.; Chambrier, P.; Paindavoine, S.; Scutt, C.P. Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 2010, 63, 952–959. [Google Scholar] [CrossRef]
- Kato, H.; Nishihama, R.; Weijers, D.; Kohchi, T. Evolution of nuclear auxin signaling: Lessons from genetic studies with basal land plants. J. Exp. Bot. 2018, 69, 291–301. [Google Scholar] [CrossRef]
- Wu, W.; Liu, Y.; Wang, Y.; Li, H.; Liu, J.; Tan, J.; He, J.; Bai, J.; Ma, H. Evolution analysis of the Aux/IAA gene family in plants shows dual origins and variable nuclear localization signals. Int. J. Mol. Sci. 2017, 18, 2107. [Google Scholar] [CrossRef] [Green Version]
- Guilfoyle, T.J.; Hagen, G. Getting a grasp on domain III/IV responsible for Auxin Response Factor-IAA protein interactions. Plant Sci. 2012, 190, 82–88. [Google Scholar] [CrossRef]
- Finet, C.; Berne-Dedieu, A.; Scutt, C.P.; Marlétaz, F. Evolution of the ARF gene family in land plants: Old domains, new tricks. Mol. Biol. Evol. 2013, 30, 45–56. [Google Scholar] [CrossRef] [Green Version]
- Ulmasov, T.; Hagen, G.; Guilfoyle, T.J. Activation and repression of transcription by auxin response factors. Proc. Natl. Acad. Sci. USA 1999, 96, 5844–5849. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, N.; Xu, H.; Jiang, S.; Fang, H.; Su, M.; Zhang, Z.; Zhang, T.; Chen, X. Auxin regulates anthocyanin biosynthesis through the Aux/IAA–ARF signaling pathway in apple. Hortic. Res. 2018, 5, 59. [Google Scholar] [CrossRef] [Green Version]
- Orosa-Puente, B.; Leftley, N.; von Wangenheim, D.; Banda, J.; Srivastava, A.K.; Hill, K.; Truskina, J.; Bhosale, R.; Morris, E.; Srivastava, M.; et al. Root branching toward water involves posttranslational modification of transcription factor ARF7. Science 2018, 362, 1407–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.E.; Muto, H.; Higuchi, K.; Matamura, T.; Tatematsu, K.; Koshiba, T.; Yamamoto, K.T. Disruption and overexpression of auxin response factor 8 gene of Arabidopsis affect hypocotyl elongation and root growth habit, indicating its possible involvement in auxin homeostasis in light condition. Plant J. 2004, 40, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Zhang, N.; Zhang, J.; Jin, X.; Zhu, X.; Ma, R.; Li, S.; Lui, S.; Yue, Y.; Si, H. Knockdown of MicroRNA160a/b by STTM leads to root architecture changes via auxin signaling in Solanum tuberosum. Plant Physiol. Biochem. 2021, 166, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Qi, Y.; Wang, S.; Shen, C.; Zhang, S.; Chen, Y.; Xu, Y.; Liu, Y.; Wu, Y.; Jiang, D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012, 193, 109–120. [Google Scholar] [CrossRef]
- Shen, X.; He, J.; Ping, Y.; Guo, J.; Hou, N.; Cao, F.; Li, X.; Geng, D.; Wang, S.; Chen, P.; et al. The positive feedback regulatory loop of miR160-Auxin Response Factor 17-HYPONASTIC LEAVES 1 mediates drought tolerance in apple trees. Plant Physiol. 2021. [Google Scholar] [CrossRef]
- Li, H.; Zhang, X.; Tong, B.; Wang, Y.; Yang, C. Expression analysis of the BpARF genes in Betula platyphylla under drought stress. Plant Physiol. Biochem. 2020, 148, 273–281. [Google Scholar] [CrossRef]
- Killi, D.; Raschi, A.; Bussotti, F. Lipid peroxidation and chlorophyll fluorescence of photosystem ii performance during drought and heat stress is associated with the antioxidant capacities of C3 sunflower and C4 maize varieties. Int. J. Mol. Sci. 2020, 21, 4846. [Google Scholar] [CrossRef]
- Su, L.; Dai, Z.; Li, S.; Xin, H. A novel system for evaluating drought-cold tolerance of grapevines using chlorophyll fluorescence. BMC Plant Biol. 2015, 15, 82. [Google Scholar] [CrossRef] [Green Version]
- Shen, C.; Wang, S.; Zhang, S.; Xu, Y.; Qian, Q.; Qi, Y.; Jiang, D.A. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 2013, 36, 607–620. [Google Scholar] [CrossRef]
- Liu, X.; Chu, S.; Sun, C.; Xu, H.; Zhang, J.; Jiao, Y.; Zhang, D. Genome-wide identification of low phosphorus responsive microRNAs in two soybean genotypes by high-throughput sequencing. Funct. Integr. Genom. 2020, 20, 825–838. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hou, Q.; Li, X.; Qiu, Z.; Hong, Y.; Tian, T.; Li, S.; Ran, J.; Qiao, G. Chinese Cherry (Cerasus pseudocerasus Lindl.) ARF7 Participates in Root Development and Responds to Drought and Low Phosphorus. Horticulturae 2022, 8, 158. https://doi.org/10.3390/horticulturae8020158
Hou Q, Li X, Qiu Z, Hong Y, Tian T, Li S, Ran J, Qiao G. Chinese Cherry (Cerasus pseudocerasus Lindl.) ARF7 Participates in Root Development and Responds to Drought and Low Phosphorus. Horticulturae. 2022; 8(2):158. https://doi.org/10.3390/horticulturae8020158
Chicago/Turabian StyleHou, Qiandong, Xiaorong Li, Zhilang Qiu, Yi Hong, Tian Tian, Shuang Li, Jiaxin Ran, and Guang Qiao. 2022. "Chinese Cherry (Cerasus pseudocerasus Lindl.) ARF7 Participates in Root Development and Responds to Drought and Low Phosphorus" Horticulturae 8, no. 2: 158. https://doi.org/10.3390/horticulturae8020158
APA StyleHou, Q., Li, X., Qiu, Z., Hong, Y., Tian, T., Li, S., Ran, J., & Qiao, G. (2022). Chinese Cherry (Cerasus pseudocerasus Lindl.) ARF7 Participates in Root Development and Responds to Drought and Low Phosphorus. Horticulturae, 8(2), 158. https://doi.org/10.3390/horticulturae8020158





