Pigment-Related Mutations Greatly Affect Berry Metabolome in San Marzano Tomatoes
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Fruit Sampling
2.3. Metabolite Detection and Quantification
2.4. Statistical and Bioinformatic Analyses
3. Results
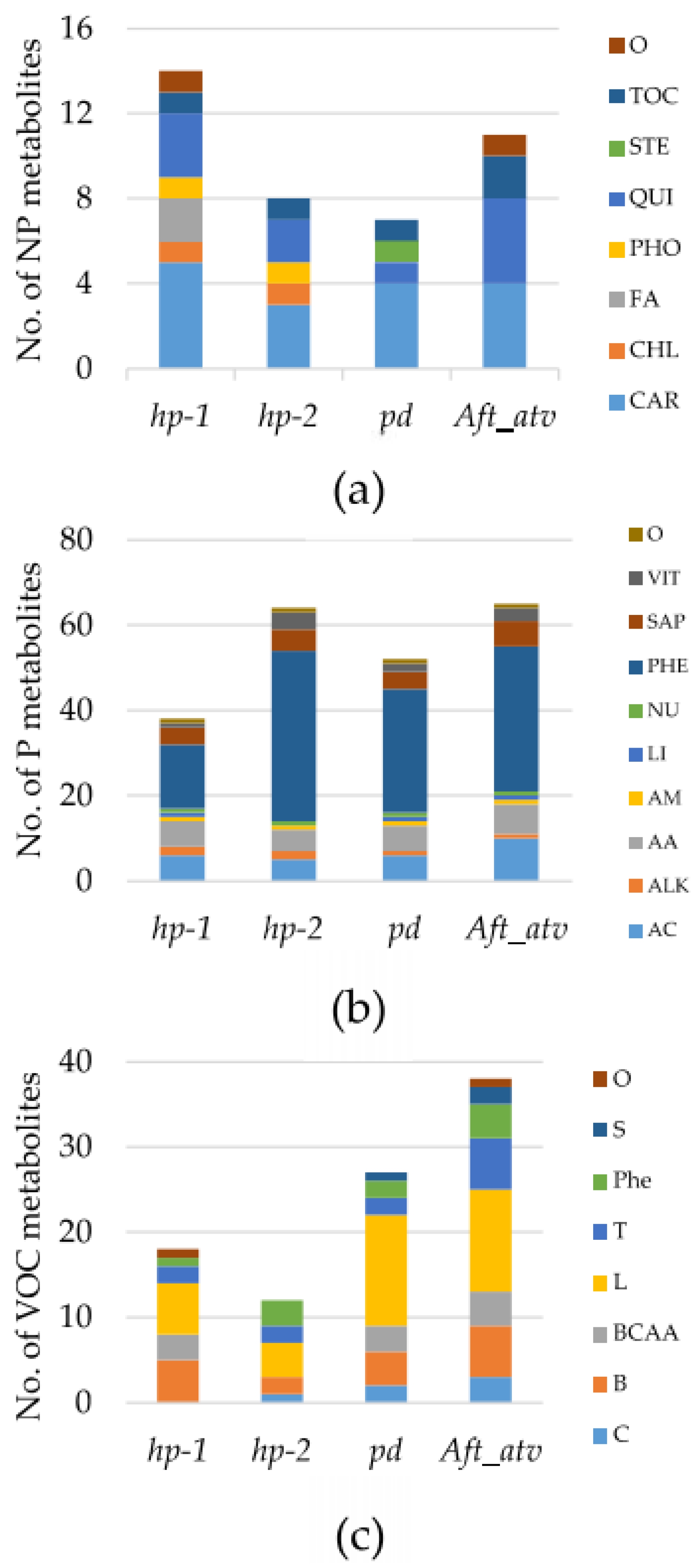
3.1. Metabolite Variation in the Studied Lines
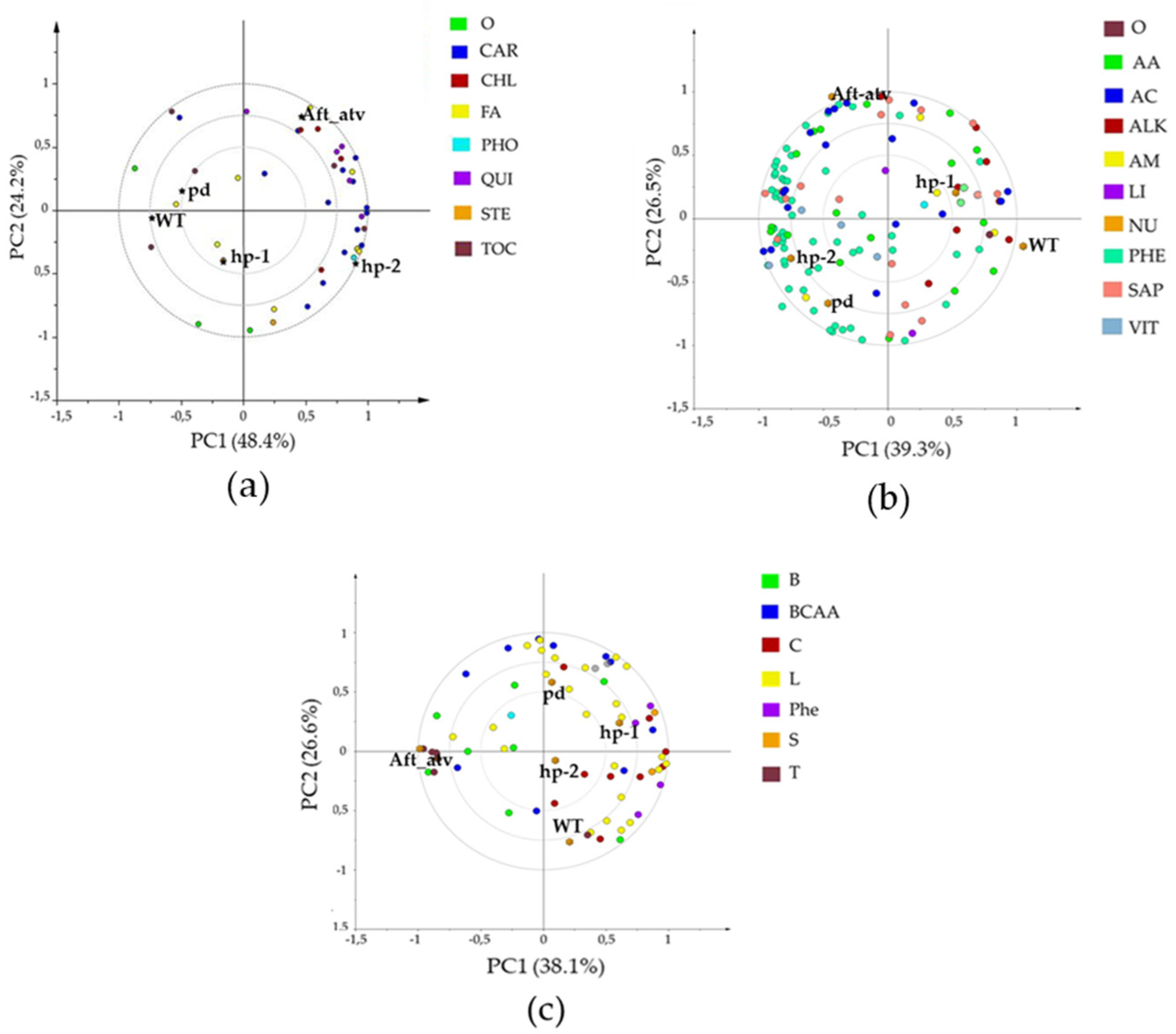
3.2. Multivariate Analysis of NP, P, and VOC Metabolites in the Four Mutated Lines
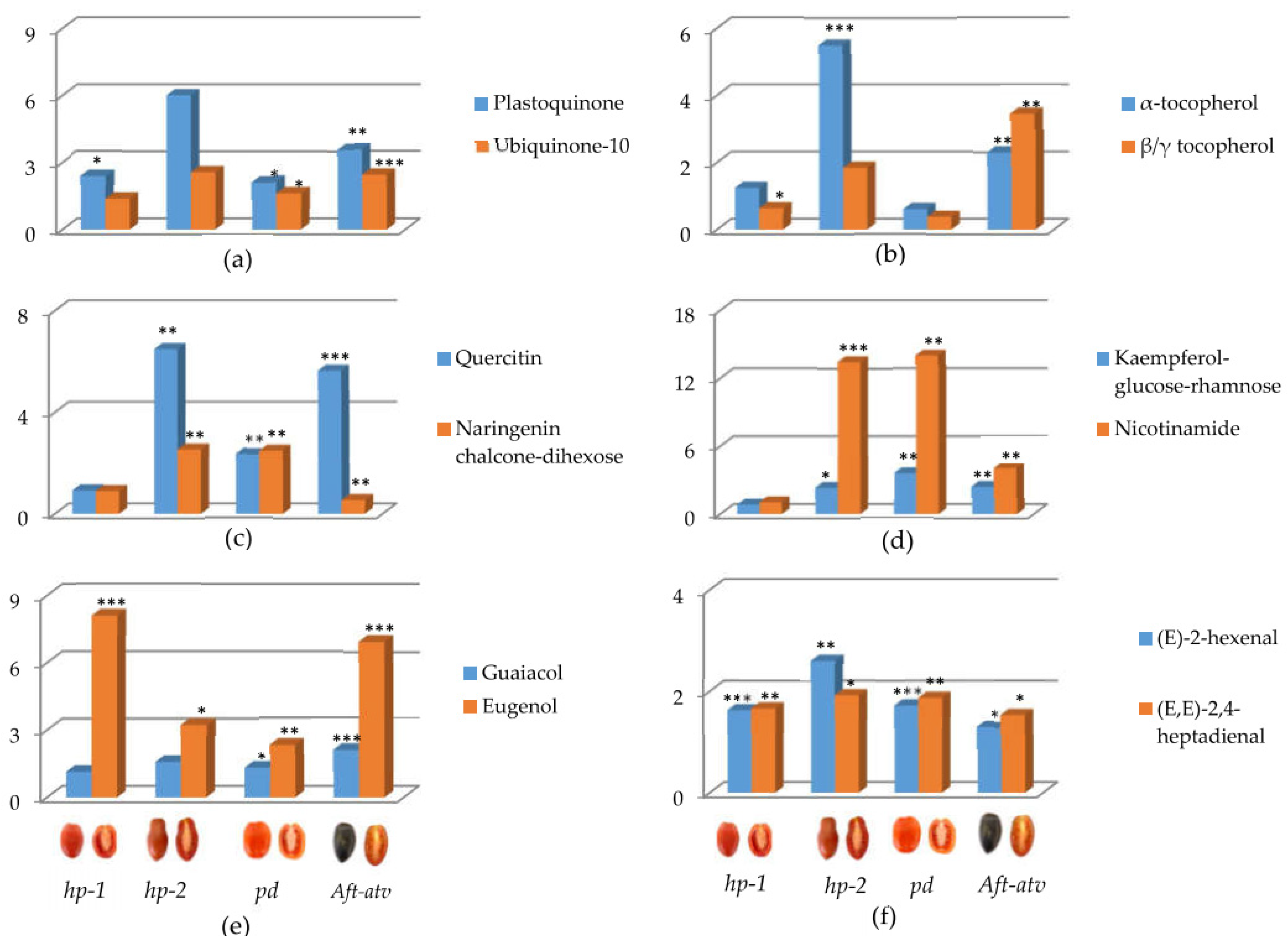
3.3. Univariate Analysis of NP, P, and VOC Metabolites in the Four Mutated Lines
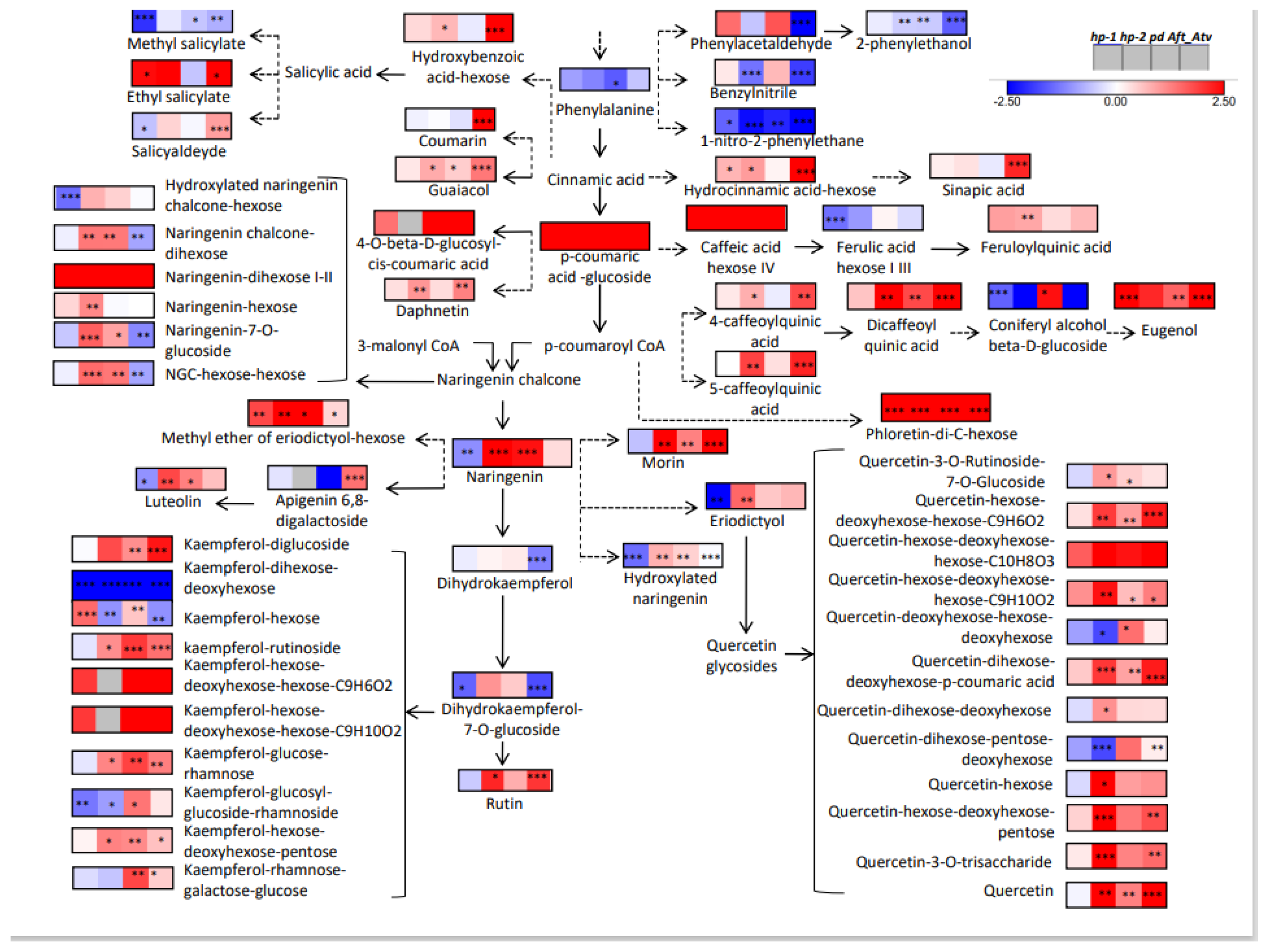
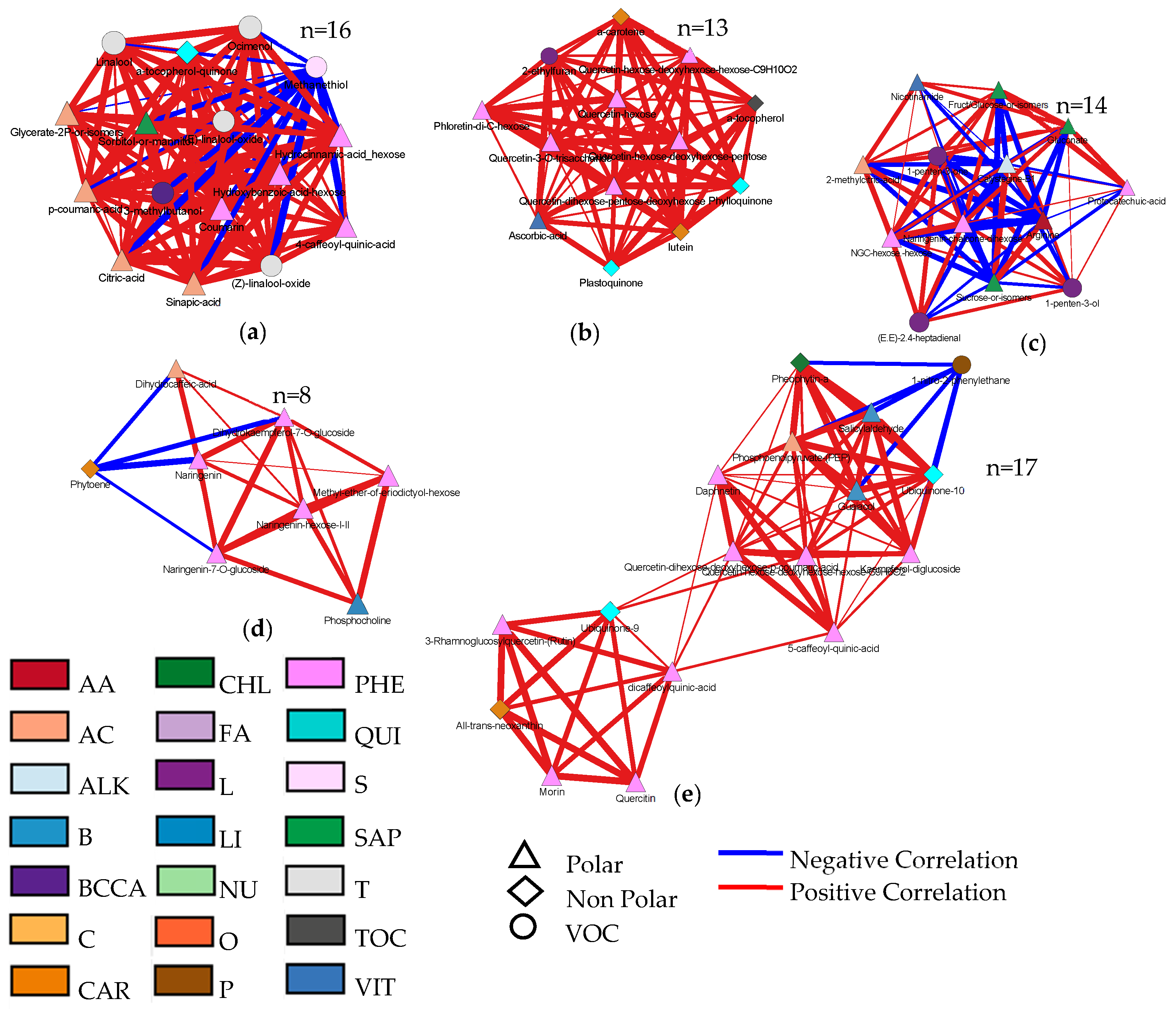
3.4. Metabolic Maps and Correlation Analysis of Fruit Metabolites
4. Discussion
4.1. hp-1 Showed the Highest Variation for NP Metabolites
4.2. Tomato Pigmentation Mutants Show Wide Variation in P Compounds
4.3. Benzenoids, but Not Carotenoid-Derived VOCs, Are Different in the Fruit of the Studied Lines
4.4. Bioinformatics to Evaluate Correlations between Metabolites
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Moco, S.; Capanoglu, E.; Tikunov, Y.; Bino, R.J.; Boyacioglu, D.; Hall, R.D.; Vervoort, J.; De Vos, R.C. Tissue specialization at the metabolite level is perceived during the development of tomato fruit. J. Exp. Bot. 2007, 58, 4131–4146. [Google Scholar] [CrossRef]
- Slimestad, R.; Verheul, M. Review of flavonoids and other phenolics from fruits of different tomato (Lycopersicon esculentum Mill.) cultivars. J. Sci. Food Agric. 2009, 89, 1255–1270. [Google Scholar] [CrossRef]
- Baldina, S.; Picarella, M.E.; Troise, A.D.; Pucci, A.; Ruggieri, V.; Ferracane, R.; Barone, A.; Fogliano, V.; Mazzucato, A. Metabolite profiling of Italian tomato landraces with different fruit types. Front. Plant Sci. 2016, 7, 664. [Google Scholar] [CrossRef]
- Hövelmann, Y.; Steinert, K.; Hübner, F.; Humpf, H.U. Identification of a novel N-caprylhistamine-β-glucoside from tomato fruits and LC-MS/MS-based food screening for imidazole alkaloids. Food Chem. 2020, 312, 126068. [Google Scholar] [CrossRef]
- Buttery, R.G.; Ling, L.C.; Light, D.M. Tomato leaf volatile aroma components. J. Agric. Food Chem. 1987, 35, 1039–1042. [Google Scholar] [CrossRef]
- Rambla, J.L.; Medina, A.; Fernández-del-Carmen, A.; Barrantes, W.; Grandillo, S.; Cammareri, M.; Lòpez-Casado, G.; Rodrigo, G.; Alonso, A.; Garcìa-Mrtìnez, S.; et al. Identification, introgression, and validation of fruit volatile QTLs from a red-fruited wild tomato species. J. Exp. Bot. 2017, 68, 429–442. [Google Scholar] [CrossRef]
- Li, J.; Di, T.; Bai, J. Distribution of volatile compounds in different fruit structures in four tomato cultivars. Molecules 2019, 24, 2594. [Google Scholar] [CrossRef]
- Tilesi, F.; Lombardi, A.; Mazzucato, A. Scientometric and Methodological Analysis of the Recent Literature on the Health-Related Effects of Tomato and Tomato Products. Foods 2021, 10, 1905. [Google Scholar] [CrossRef]
- Friedman, M. Chemistry, biochemistry, and dietary role of potato polyphenols. A review. J. Agric. Food Chem. 1997, 45, 1523–1540. [Google Scholar] [CrossRef]
- Martínez-Valverde, I.; Periago, M.J.; Provan, G.; Chesson, A. Phenolic compounds, lycopene and antioxidant activity in commercial varieties of tomato (Lycopersicum esculentum). J. Sci. Food Agric. 2002, 82, 323–330. [Google Scholar] [CrossRef]
- Frusciante, L.; Carli, P.; Ercolano, M.R.; Pernice, R.; Di Matteo, A.; Fogliano, V.; Pellegrini, N. Antioxidant nutritional quality of tomato. Mol. Nutr. Food Res. 2007, 51, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, E.A.; Nisperos-Carriedo, M.O.; Moshonas, M.G. Quantitative analysis of flavor and other volatiles and for certain constituents of two tomato cultivars during ripening. J. Am. Soc. Hortic. Sci. 1991, 116, 265–269. [Google Scholar] [CrossRef]
- Tikunov, Y.M.; Molthoff, J.; de Vos, R.C.; Beekwilder, J.; van Houwelingen, A.; van der Hooft, J.J.; Nijenhuis-de Vries, M.; Labrie, C.W.; Verkerke, W.; van de Geest, H.; et al. Non-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. Plant Cell 2013, 25, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Tieman, D.; Zhu, G.; Resende, M.F., Jr.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Ortiz Beltran, K.S.; Taylor, M.; Zhang, B.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Butelli, E.; Titta, L.; Giorgio, M.; Mock, H.P.; Matros, A.; Peterek, S.; Schijlen, E.G.W.M.; Hall, R.D.; Bovy, A.G.; Luo, J.; et al. Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 2008, 26, 1301–1308. [Google Scholar] [CrossRef]
- Azari, R.; Tadmor, Y.; Meir, A.; Reuveni, M.; Evenor, D.; Nahon, S.; Shlomo, H.; Chen, L.; Levin, I. Light signaling genes and their manipulation towards modulation of phytonutrient content in tomato fruits. Biotechnol. Adv. 2010, 28, 108–118. [Google Scholar] [CrossRef]
- Davuluri, G.R.; Van Tuinen, A.; Fraser, P.D.; Manfredonia, A.; Newman, R.; Burgess, D.; Brummell, D.A.; King, S.R.; Palys, J.; Uhlig, J.; et al. Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat. Biotechnol. 2005, 23, 890–895. [Google Scholar] [CrossRef]
- Reynard, G.B. Origin of Webb Special (Black Queen) in tomato. Rep. Tomato Genet. Coop. 1956, 40, 44–64. [Google Scholar]
- Lieberman, M.; Segev, O.; Gilboa, N.; Lalazar, A.; Levin, I. The tomato homolog of the gene encoding UV-damaged DNA binding protein 1 (DDB1) underlined as the gene that causes the high pigment-1 mutant phenotype. Theor. Appl. Genet. 2004, 108, 1574–1581. [Google Scholar] [CrossRef]
- Soressi, G.P. New spontaneous or chemically-induced fruit ripening tomato mutants. Rep. Tomato Genet. Coop. 1975, 25, 21–22. [Google Scholar]
- Mustilli, A.C.; Fenzi, F.; Ciliento, R.; Alfano, F.; Bowler, C. Phenotype of the tomato high pigment-2 mutant is caused by a mutation in the tomato homolog of DEETIOLATED1. Plant Cell 1999, 11, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, D.F.; Gahrtz, M.; Maxwell, B.B.; Cook, R.K.; Kan, J.M.; Alonso, J.M.; Ecker, R.J.; Chory, J. De-etiolated 1 and damaged DNA binding protein 1 interact to regulate Arabidopsis photomorphogenesis. Curr. Biol. 2002, 12, 1462–1472. [Google Scholar] [CrossRef]
- Levin, I.; De Vos, C.R.; Tadmor, Y.; Bovy, A.; Lieberman, M.; Oren Shamir, M.; Segev, O.; Kolotilin, I.; Menachem, K.; Ovadia, R.; et al. High pigment tomato mutants—More than just lycopene (a review). Isr. J. Plant Sci. 2006, 54, 179–190. [Google Scholar] [CrossRef]
- Minoggio, M.; Bramati, L.; Simonetti, P.; Gardana, C.; Iemoli, L.; Santangelo, E.; Mauri, P.L.; Spigno, P.; Soressi, G.P.; Pietta, P.G. Polyphenol pattern and antioxidant activity of different tomato lines and cultivars. Ann. Nutr. Metab. 2003, 47, 64–69. [Google Scholar] [CrossRef]
- Tigchelaar, E.C.; Tomes, M.L.; Erickson, H.T.; Graham, T.O.; Barman, R.J. Pigment diluter (pd), a new plant and fruit color mutant. Rep. Tomato Genet. Coop. 1970, 20, 64. [Google Scholar]
- Giorgiev, C. Anthocyanin fruit tomato. Rep. Tomato Genet. Coop. 1972, 22, 10. [Google Scholar]
- Rick, C.M.; Uhlig, J.W.; Jones, A.D. High alpha-tomatine content in ripe fruit of Andean Lycopersicon esculentum var. cerasiforme: Developmental and genetic aspects. Proc. Natl. Acad. Sci. USA 1994, 91, 12877–12881. [Google Scholar] [CrossRef]
- O’Donnell, P.J.; Schmelz, E.; Block, A.; Miersch, O.; Wasternack, C.; Jones, J.B.; Klee, H.J. Multiple hormones act sequentially to mediate a susceptible tomato pathogen defense response. Plant Physiol. 2003, 133, 1181–1189. [Google Scholar] [CrossRef]
- Mes, P.J.; Boches, P.; Myers, J.R.; Durst, R. Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hortic. Sci. 2008, 133, 262–269. [Google Scholar] [CrossRef]
- Boches, P.; Myers, J. The anthocyanin fruit tomato gene (Aft) is associated with a DNA polymorphism in a MYB transcription factor. HortScience 2007, 42, 856. [Google Scholar]
- Yan, S.; Chen, N.; Huang, Z.; Li, D.; Zhi, J.; Yu, B.; Liu, X.; Cao, B.; Qiu, Z. Anthocyanin Fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020, 225, 2048–2063. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant 2020, 13, 42–58. [Google Scholar] [CrossRef] [PubMed]
- Colanero, S.; Tagliani, A.; Perata, P.; Gonzali, S. Alternative splicing in the anthocyanin fruit gene encoding an R2R3 MYB transcription factor affects anthocyanin biosynthesis in tomato fruits. Plant Commun. 2020, 1, 100006. [Google Scholar] [CrossRef]
- Kerckhoffs, L.H.J.; Schreuder, M.E.L.; Tuinen, A.V.; Koornneef, M.; Kendrick, R.E. Phytochrome control of anthocyanin biosynthesis in tomato seedlings: Analysis using photomorphogenic mutants. Photochem. Photobiol. 1997, 65, 374–381. [Google Scholar] [CrossRef]
- Rick, C.M.; Reeves, A.F.; Zobel, R.W. Inheritance and linkage relations of four new mutants. Rep. Tomato Genet. Coop. 1968, 18, 34–35. [Google Scholar]
- Cao, X.; Qiu, Z.; Wang, X.; Van Giang, T.; Liu, X.; Wang, J.; Wang, X.; Gao, J.; Guo, Y.; Du, Y.; et al. A putative R3 MYB repressor is the candidate gene underlying atroviolacium, a locus for anthocyanin pigmentation in tomato fruit. J. Exp. Bot. 2017, 68, 5745–5758. [Google Scholar] [CrossRef]
- Colanero, S.; Perata, P.; Gonzali, S. The atroviolacea gene encodes an R3-MYB protein repressing anthocyanin synthesis in tomato plants. Front. Plant Sci. 2018, 9, 830. [Google Scholar] [CrossRef]
- Gonzali, S.; Mazzucato, A.; Perata, P. Purple as a tomato: Towards high anthocyanin tomatoes. Trends Plant Sci. 2009, 14, 237–241. [Google Scholar] [CrossRef]
- Dono, G.; Picarella, M.E.; Pons, C.; Santangelo, E.; Monforte, A.; Granell, A.; Mazzucato, A. Characterization of a repertoire of tomato fruit genetic variants in the San marzano genetic background. Sci. Hortic. 2020, 261, 108927. [Google Scholar] [CrossRef]
- Long, M.; Millar, D.J.; Kimura, Y.; Donovan, G.; Rees, J.; Fraser, P.D.; Bramley, P.M.; Bolwell, G.P. Metabolite profiling of carotenoid and phenolic pathways in mutant and transgenic lines of tomato: Identification of a high antioxidant fruit line. Phytochemistry 2006, 67, 1750–1757. [Google Scholar] [CrossRef]
- Bovy, A.; Schijlen, E.; Hall, R.D. Metabolic engineering of flavonoids in tomato (Solanum lycopersicum): The potential for metabolomics. Metabolomics 2006, 3, 399–412. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, G.; Reuveni, M.; Evenor, D.; Oren-Shamir, M.; Ovadia, R.; Sapir-Mir, M.; Bootbool Man, A.; Nahon, S.; Shlomo, H.; Chen, L.; et al. ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. Theor. Appl. Genet. 2012, 124, 295–307. [Google Scholar] [CrossRef] [PubMed]
- Perez-Fons, L.; Wells, T.; Corol, D.I.; Ward, J.L.; Gerrish, C.; Beale, M.H.; Seymour, G.B.; Bramley, P.M.; Fraser, P.D. A genome-wide metabolomic resource for tomato fruit from Solanum pennellii. Sci. Rep. 2014, 4, 3859. [Google Scholar] [CrossRef]
- Su, X.; Griffin, J.; Xu, J.; Ouyang, P.; Zhao, Z.; Wang, W. Identification and quantification of anthocyanins in purple-fleshed sweet potato leaves. Heliyon 2019, 5, e01964. [Google Scholar] [CrossRef] [PubMed]
- Blando, F.; Berland, H.; Maiorano, G.; Durante, M.; Mazzucato, A.; Picarella, M.E.; Andersen, Ø.M. Nutraceutical characterization of anthocyanin-rich fruits produced by “Sun Black” tomato line. Front. Nutr. 2019, 6, 133. [Google Scholar] [CrossRef]
- Dono, G.; Rambla, J.L.; Frusciante, S.; Granell, A.; Diretto, G.; Mazzucato, A. Color Mutations Alter the Biochemical Composition in the San Marzano Tomato Fruit. Metabolites 2020, 10, 110. [Google Scholar] [CrossRef]
- Sulli, M.; Mandolino, G.; Sturaro, M.; Onofri, C.; Diretto, G.; Parisi, B.; Giuliano, G. Molecular and biochemical characterization of a potato collection with contrasting tuber carotenoid content. PLoS ONE 2017, 12, e0184143. [Google Scholar] [CrossRef]
- Diretto, G.; Rubio-Moraga, A.; Argandoña, J.; Castillo, P.; Gómez-Gómez, L.; Ahrazem, O. Tissue-specific accumulation of sulfur compounds and saponins in different parts of garlic cloves from purple and white ecotypes. Molecules 2017, 22, 1359. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Sanders, M.A.; Bijl, M.A.; Delwel, R.; Horsman, S.; Moorhouse, M.J.; van der Spek, P.J.; Lowenberg, B.; Valk, P.J. HeatMapper: Powerful combined visualization of gene expression profile correlations, genotypes, phenotypes and sample characteristics. BMC Bioinform. 2007, 7, 337. [Google Scholar] [CrossRef][Green Version]
- Thompson, A. A comparison of fruit quality constituents of normal and high pigment tomatoes. Proc. Natl. Acad. Sci. USA 1961, 78, 464–473. [Google Scholar]
- Bino, R.J.; De Vos, C.R.; Lieberman, M.; Hall, R.D.; Bovy, A.; Jonker, H.H.; Tikunov, Y.; Lommen, A.; Moco, S.; Levin, I. The light-hyperresponsive high pigment-2 dg mutation of tomato: Alterations in the fruit metabolome. New Phytol. 2005, 166, 427–438. [Google Scholar] [CrossRef] [PubMed]
- Nisar, N.; Li, L.; Lu, S.; Khin, N.C.; Pogson, B.J. Carotenoid metabolism in plants. Mol. Plant 2015, 8, 68–82. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C. Chemistry and biology of vitamin E. Mol. Nutr. Food Res. 2005, 49, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Zanfini, A.; Corbini, G.; la Rosa, C.; Dreass, E. Antioxidant activity of tomato lipophilic extracts and interactions between carotenoids and α-tocopherol in synthetic mixtures. LWT Food Sci. Technol. 2010, 43, 67–72. [Google Scholar] [CrossRef]
- Verhoeyen, M.E.; Bovy, A.; Collins, G.; Muir, S.; Robinson, S.; De Vos, C.H.R.; Colliver, S. Increasing antioxidant levels in tomatoes through modification of the flavonoid biosynthetic pathway. J. Exp. Bot. 2002, 53, 2099–2106. [Google Scholar] [CrossRef] [PubMed]
- Mazzucato, A.; Willems, D.; Bernini, R.; Picarella, M.E.; Santangelo, E.; Ruiu, F.; Tilesi, F.; Soressi, G.P. Novel phenotypes related to the breeding of purple-fruited tomatoes and effect of peel extracts on human cancer cell proliferation. Plant Physiol. Biochem. 2013, 72, 125–133. [Google Scholar] [CrossRef]
- Ververidis, F.; Trantas, E.; Douglas, C.; Vollmer, G.; Kretzschmar, G.; Panopoulos, N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part I: Chemical diversity, impacts on plant biology and human health. Biotechnol. J. Healthc. Nutr. Technol. 2007, 2, 1214–1234. [Google Scholar] [CrossRef]
- Hancock, R.D.; Viola, R. Improving the nutritional value of crops through enhancement of L-ascorbic acid (vitamin C) content: Rationale and biotechnological opportunities. J. Agric. Food Chem. 2005, 53, 5248–5257. [Google Scholar] [CrossRef]
- Klein, B.P.; Perry, A.K. Ascorbic acid and vitamin A activity in selected vegetables from different geographical areas of the United States. J. Food Sci. 1982, 47, 941–945. [Google Scholar] [CrossRef]
- Mageroy, M.H.; Tieman, D.M.; Floystad, A.; Taylor, M.G.; Klee, H.J. A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant J. 2012, 69, 1043–1051. [Google Scholar] [CrossRef]
- Zanor, M.I.; Rambla, J.L.; Chaïb, J.; Steppa, A.; Medina, A.; Granell, A.; Fernie, A.R.; Causse, M. Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J. Exp. Bot. 2009, 60, 2139–2154. [Google Scholar] [CrossRef] [PubMed]
- Koeduka, T.; Fridman, E.; Gang, D.R.; Vassão, D.G.; Jackson, B.L.; Kish, C.M.; Orlova, I.; Spassova, S.M.; Lewis, N.G.; Noel, J.P.; et al. Eugenol and isoeugenol, characteristic aromatic constituents of spices, are biosynthesized via reduction of a coniferyl alcohol ester. Proc. Natl. Acad. Sci. USA 2006, 103, 10128–10133. [Google Scholar] [CrossRef] [PubMed]
- Klee, H.J. Improving the flavor of fresh fruits: Genomics, biochemistry, and biotechnology. New Phytol. 2010, 187, 44–56. [Google Scholar] [CrossRef] [PubMed]
- Sandmann, G.; Römer, S.; Fraser, P.D. Understanding carotenoid metabolism as a necessity for genetic engineering of crop plants. Metab. Eng. 2006, 8, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Enfissi, E.M.; Barneche, F.; Ahmed, I.; Lichtlé, C.; Gerrish, C.; McQuinn, R.P.; Giovannoni, J.J.; Lopez-Juez, E.; Bowler, C.; Bramley, P.M.; et al. Integrative transcript and metabolite analysis of nutritionally enhanced DE-ETIOLATED1 downregulated tomato fruit. Plant Cell 2010, 22, 1190–1215. [Google Scholar] [CrossRef]
- Kendrick, R.E.; Kerckhoffs, L.H.J.; Van Tuinen, A.; Koornneef, M. Photomorphogenic mutants of tomato. Plant Cell Environ. 1997, 20, 746–751. [Google Scholar] [CrossRef]
- Liavonchanka, A.; Feussner, I. Lipoxygenases: Occurrence, functions and catalysis. J. Plant Physiol. 2006, 163, 348–357. [Google Scholar] [CrossRef]
- Zhang, Y.; Butelli, E.; Alseekh, S.; Tohge, T.; Rallapalli, G.; Luo, J.; Kawar, P.G.; Hill, L.; Santino, A.; Fernie, A.R.; et al. Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat. Commun. 2015, 6, 8635. [Google Scholar] [CrossRef]
- Landy, P.; Boucon, C.; Kooyman, G.M.; Musters, P.A.; Rosing, E.A.; De Joode, T.; Laan, J.; Haring, P.G. Sensory and chemical changes in tomato sauces during storage. J. Agric. Food Chem. 2002, 50, 3262–3271. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dono, G.; Rambla, J.L.; Frusciante, S.; Fabene, E.; Gómez-Cadenas, A.; Granell, A.; Diretto, G.; Mazzucato, A. Pigment-Related Mutations Greatly Affect Berry Metabolome in San Marzano Tomatoes. Horticulturae 2022, 8, 120. https://doi.org/10.3390/horticulturae8020120
Dono G, Rambla JL, Frusciante S, Fabene E, Gómez-Cadenas A, Granell A, Diretto G, Mazzucato A. Pigment-Related Mutations Greatly Affect Berry Metabolome in San Marzano Tomatoes. Horticulturae. 2022; 8(2):120. https://doi.org/10.3390/horticulturae8020120
Chicago/Turabian StyleDono, Gabriella, José Luis Rambla, Sarah Frusciante, Eleonora Fabene, Aurelio Gómez-Cadenas, Antonio Granell, Gianfranco Diretto, and Andrea Mazzucato. 2022. "Pigment-Related Mutations Greatly Affect Berry Metabolome in San Marzano Tomatoes" Horticulturae 8, no. 2: 120. https://doi.org/10.3390/horticulturae8020120
APA StyleDono, G., Rambla, J. L., Frusciante, S., Fabene, E., Gómez-Cadenas, A., Granell, A., Diretto, G., & Mazzucato, A. (2022). Pigment-Related Mutations Greatly Affect Berry Metabolome in San Marzano Tomatoes. Horticulturae, 8(2), 120. https://doi.org/10.3390/horticulturae8020120








