Characteristics of the Complete Chloroplast Genome of Pourthiaea (Rosaceae) and Its Comparative Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Data Acquisition
2.2. Analysis of the Basic Characteristics of Chloroplast Genomes
2.3. Detection of Repetitive Sequences and SSRs
2.4. Analysis of the IR/SC Boundary Region
2.5. Analysis of Genomic Differences
2.6. Analysis of Nucleotide Polymorphisms
2.7. Phylogenetic Analysis
3. Results and Analysis
3.1. Basic Characteristics of Chloroplast Genomes
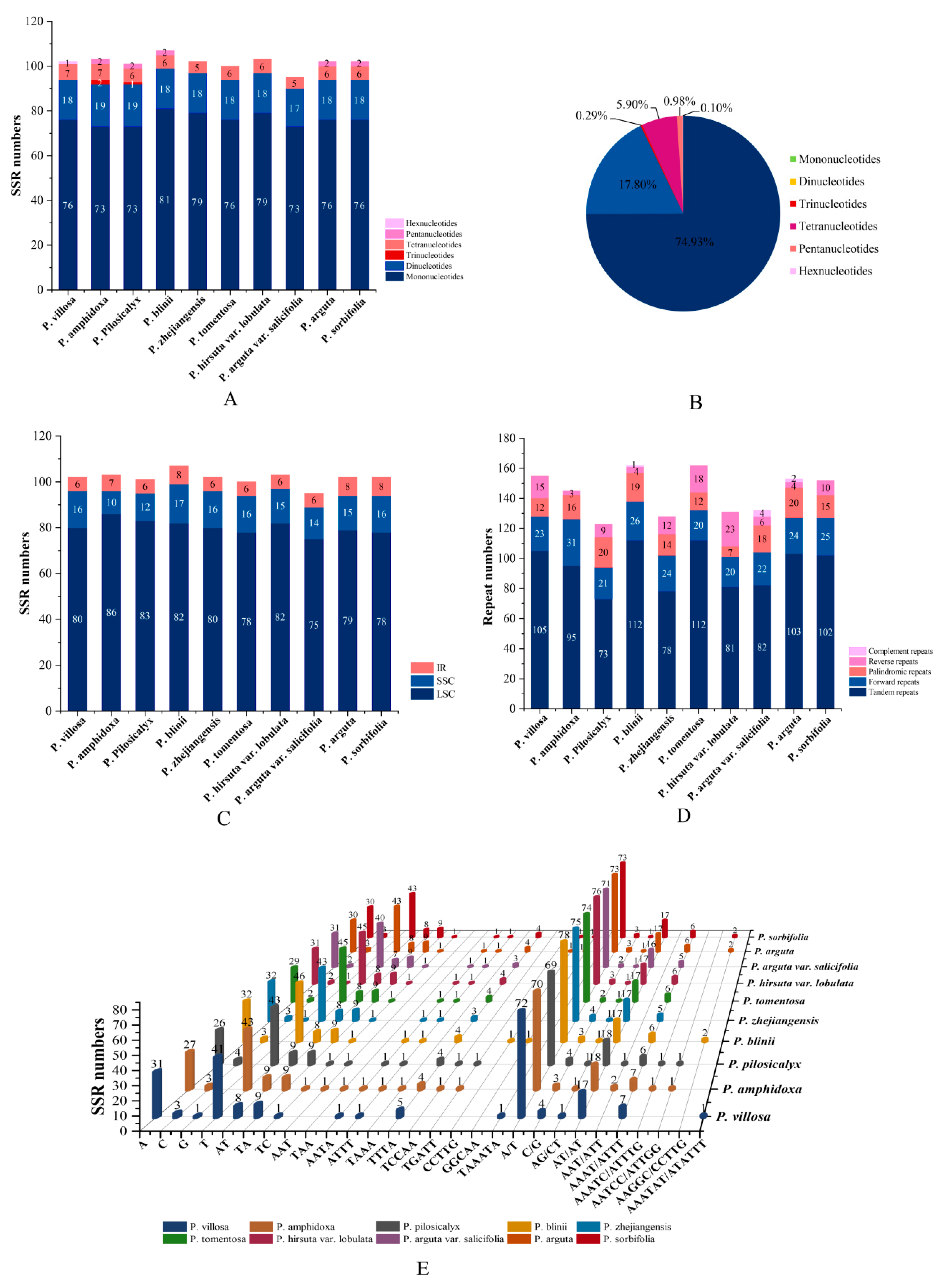
3.2. Repetitive Sequences and SSR Analysis
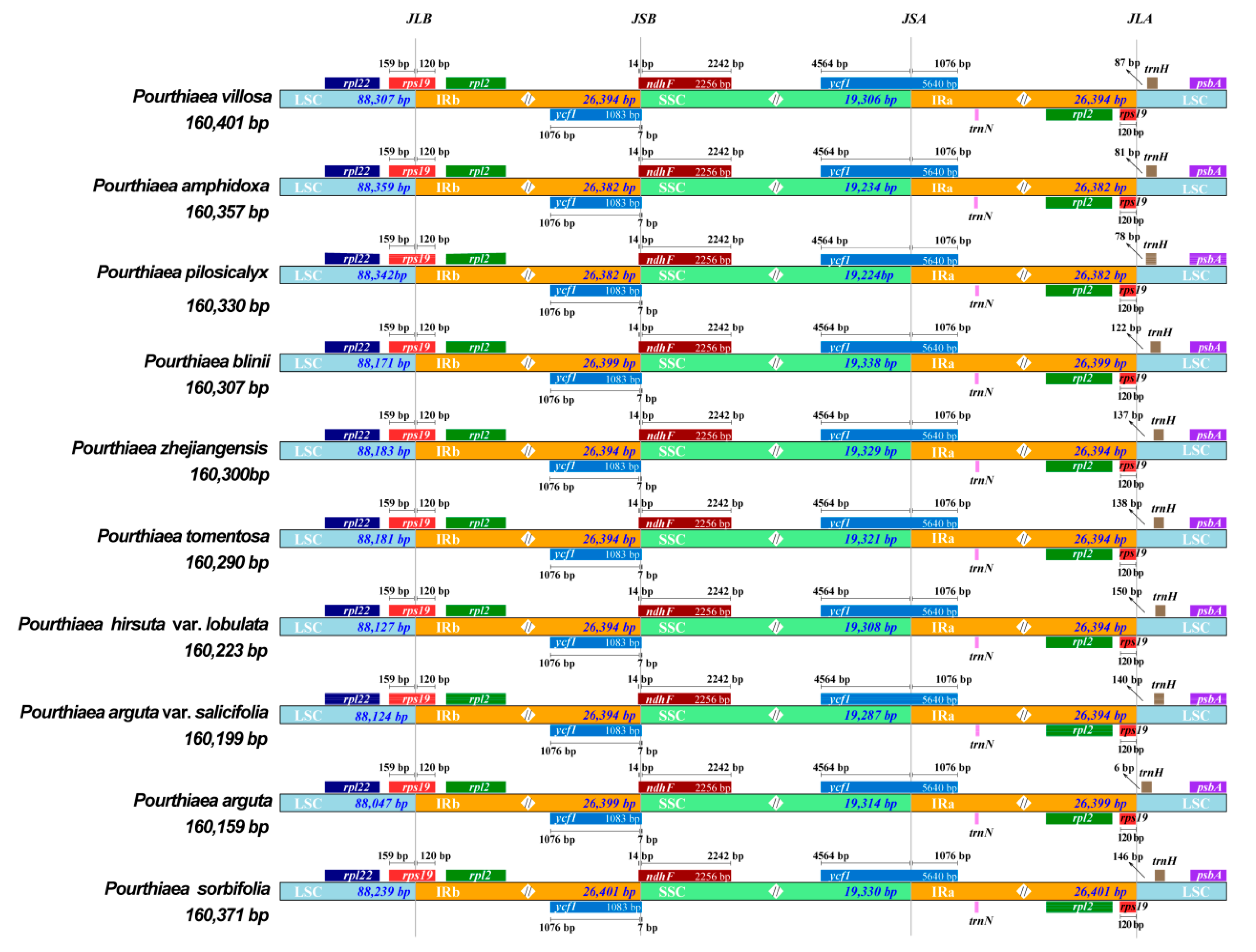
3.3. Analysis of the IR/SC Boundary Region
3.4. Analysis of Genomic Differences
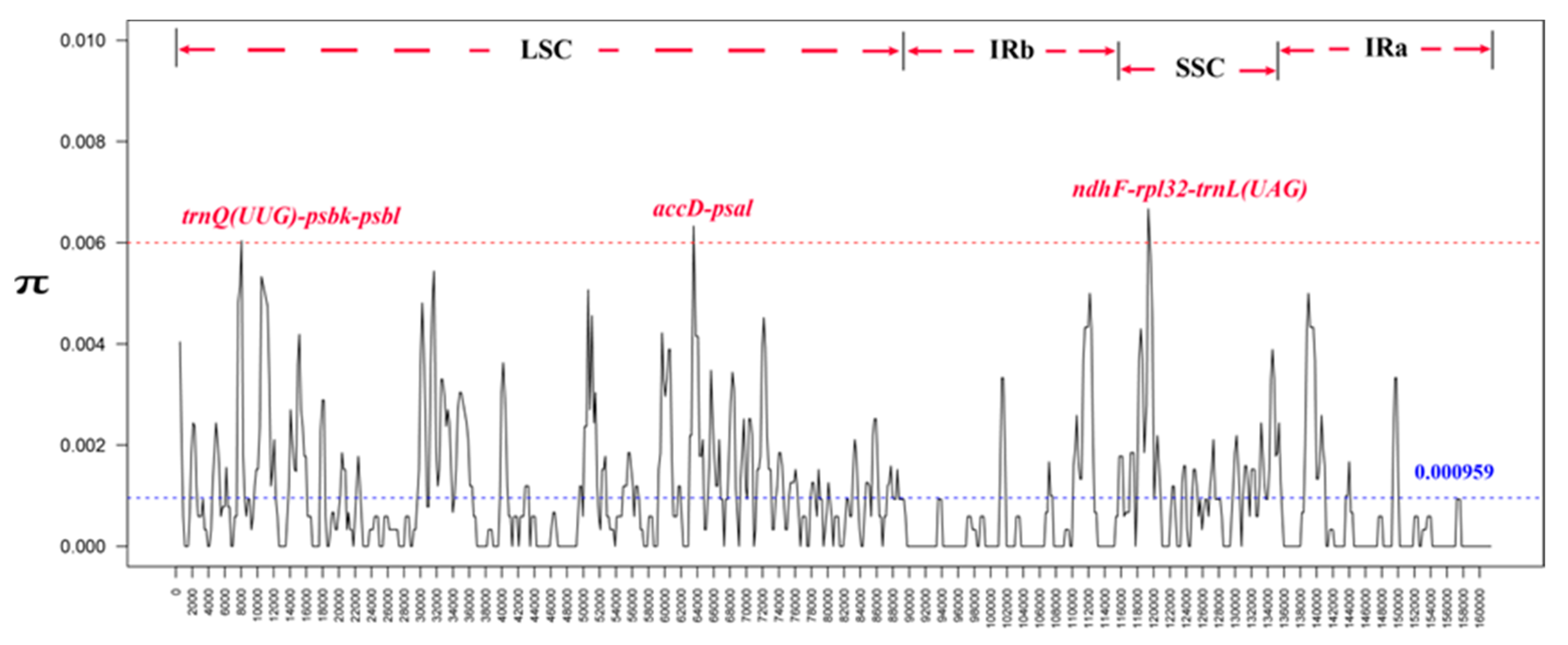
3.5. Analysis of Nucleotide Polymorphisms
3.6. Phylogenetic Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| Genus | Latin Name | Accession Number |
|---|---|---|
| Pourthiaea | P. villosa | MN061989 |
| P. amphidoxa | MN061992 | |
| P. pilosicalyx | MN216024 | |
| P. blinii | MN061990 | |
| P. zhejiangensis | MN061988 | |
| P. tomentosa | MN061995 | |
| P. hirsuta var. lobulata | MN061986 | |
| P. arguta var. salicifolia | MN061987 | |
| P. arguta | MN061991 | |
| P. sorbifolia | MN061994 | |
| Stranvaesia | S. nussia | MK920284 |
| S. bodinieri | MK920276 | |
| S. oblanceolata | MK920280 | |
| Photinia | Ph. serratifolia | MN577865 |
| Ph. lochengensis | MN577888 | |
| Ph. lanuginosa | MN577890 | |
| Ph. × fraseri | MZ128520 | |
| Ph. crassifolia | MZ984217 | |
| Ph. prunifolia | MK920279 | |
| Ph. glabra | MZ984218 | |
| Ph. integrifolia | MN577879 | |
| Ph. taishunensis | MK920278 | |
| Ph. beckii | MN577889 | |
| Ph. glomerata | OM772659 | |
| Ph. prionophylla | MN577891 | |
| Ph. davidsoniae | MT230547 | |
| Eriobotrya | E. henryi | NC_045345 |
| Rhaphiolepis | R. lanceolata | MN577867 |
References
- Plants of the World Online Kew Science. Available online: https://powo.science.kew.org/taxon/urn:lsid:ipni.org:names:33978-1 (accessed on 24 November 2022).
- Liu, B.; Hong, D. A taxonomic revision of the Pourthiaea villosa complex (Rosaceae). Phytotaxa 2016, 244, 201. [Google Scholar] [CrossRef]
- Editing Committee of Flora of China, Chinese Academy of Sciences. Flora of China; Science Press: Beijing, China, 1974; Volume 36, pp. 216–260. [Google Scholar]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Xiang, Y.; Huang, C.-H.; Hu, Y.; Wen, J.; Li, S.; Yi, T.; Chen, H.; Xiang, J.; Ma, H. Evolution of Rosaceae Fruit Types Based on Nuclear Phylogeny in the Context of Geological Times and Genome Duplication. Mol. Biol. Evol. 2017, 34, 262–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-D.; Jin, J.-J.; Chen, S.-Y.; Chase, M.W.; Soltis, D.E.; Li, H.-T.; Yang, J.-B.; Li, D.-Z.; Yi, T.-S. Diversification of Rosaceae since the Late Cretaceous based on plastid phylogenomics. New Phytol. 2017, 214, 1355–1367. [Google Scholar] [CrossRef]
- Verbylaitė, R.; Ford-Lloyd, B.; Newbury, J. The phylogeny of woody Maloideae (Rosaceae) using chloroplast trnL-trnF sequence data. Biologija 2006, 1, 60–63. [Google Scholar]
- Li, Q.Y.; Guo, W.; Liao, W.B.; Macklin, J.A.; Li, J.H. Generic limits of Pyrinae: Insights from nuclear ribosomal DNA sequences. Bot. Stud. 2012, 53, 151–164. [Google Scholar]
- Sun, J.; Shi, S.; Li, J.; Yu, J.; Wang, L.; Yang, X.; Guo, L.; Zhou, S. Phylogeny of Maleae (Rosaceae) Based on Multiple Chloroplast Regions: Implications to Genera Circumscription. BioMed. Res. Int. 2018, 2018, 7627191. [Google Scholar] [CrossRef]
- Lindley, J. Photinia arbutifolia: Californian Hawthorn or Photinia. Bot. Regist. 1820, 6, 491. [Google Scholar]
- Lindley, J. Observations on the natural Group of Plants called Pomaceae. Trans. Linn. Soc. Lond. 1821, 13, 88–106. [Google Scholar] [CrossRef]
- DeCandolle, A.P. Prodromus Systematis Naturalis Regni Vegetabilis; Treuttel et Würts: Paris, France, 1825. [Google Scholar]
- Guo, W.; Fan, Q.; Zhang, X.-Z.; Liao, W.-B.; Wang, L.-Y.; Wu, W.; Potter, D. Molecular reappraisal of relationships between Photinia, Stranvaesia and Heteromeles (Rosaceae, Maleae). Phytotaxa 2020, 447, 103–115. [Google Scholar] [CrossRef]
- Rehder, A. Manual of Cultivated Trees and Shrubs Hardy in North America; Macmillan: New York, NY, USA, 1940. [Google Scholar] [CrossRef]
- Potter, D.; Eriksson, T.; Evans, R.C.; Oh, S.; Smedmark, J.; Morgan, D.R.; Kerr, M.; Robertson, K.R.; Arsenault, M.; Dickinson, T.A.; et al. Phylogeny and classification of Rosaceae. Plant Syst. Evol. 2007, 266, 5–43. [Google Scholar] [CrossRef]
- Robertson, K.R.; Phipps, J.B.; Rohrer, J.R.; Smith, P.G. A Synopsis of Genera in Maloideae (Rosaceae). Syst. Bot. 1991, 16, 376. [Google Scholar] [CrossRef]
- Campbell, C.S.; Evans, R.C.; Morgan, D.R.; Dickinson, T.A.; Arsenault, M.P. Phylogeny of subtribe Pyrinae (formerly the Maloideae, Rosaceae): Limited resolution of a complex evolutionary history. Plant Syst. Evol. 2007, 266, 119–145. [Google Scholar] [CrossRef]
- Nakai, T. Praecursores ad Floram Sylvaticam Koreanam. VI. (Pomaceae). Bot. Mag. 1916, 30, 15–33. [Google Scholar] [CrossRef]
- IKetani, H.; Ohashi, H. Pourthiaea (Rosaceae) distinct from Photinia. J. Jpn. Bot. 1991, 66, 352–355. [Google Scholar]
- Iketani, H.; Ohashi, H. Pourthiaea Decne. In Flora of Japan (Agiospermae Dicotyledoneae Archichlamydeae); Iwatsuki, K., Boufford, D.E., Ohba, H., Eds.; Kodansha: Tokyo, Japan, 2001; Volume IIb, p. 116. [Google Scholar]
- Guo, W.; Yu, Y.; Shen, R.-J.; Liao, W.-B.; Chin, S.-W.; Potter, D. A phylogeny of Photinia sensu lato (Rosaceae) and related genera based on nrITS and cpDNA analysis. Plant Syst. Evol. 2011, 291, 91–102. [Google Scholar] [CrossRef]
- Wang, L. Taxonomic Revision of Stranvaesia and Photinia (Rosaceae); Sun Yat-sen University: Guangzhou, China, 2018. [Google Scholar] [CrossRef]
- Liu, B.; Hong, D.; Zhou, S.; Xu, C.; Dong, W.; Johnson, G.; Wen, J. Phylogenomic analyses of the Photinia complex support the recognition of a new genus Phippsiomeles and the resurrection of a redefined Stranvaesia in Maleae (Rosaceae). J. Syst. Evol. 2019, 57, 678–694. [Google Scholar] [CrossRef]
- Liu, B.-B.; Hong, D.-Y. A taxonomic revision of four complexes in the genus Pourthiaea (Rosaceae). Phytotaxa 2017, 325, 1. [Google Scholar] [CrossRef]
- Lou, Y.-L.; Jin, Z.-T.; Ma, D.-K.; Liu, B.-B. A comprehensive checklist of the deciduous photinia genus Pourthiaea (Maleae, Rosaceae), with emphasis on their validity and typification. PhytoKeys 2022, 202, 1–33. [Google Scholar] [CrossRef]
- Xing, S.C.; Liu, C.J. Progress in Chloroplast Genome Analysis. Prog. Biochem. Biophys. 2008, 35, 21–28. [Google Scholar] [CrossRef]
- Jansen, R.K.; Saski, C.; Lee, S.-B.; Hansen, A.K.; Daniell, H. Complete Plastid Genome Sequences of Three Rosids (Castanea, Prunus, Theobroma): Evidence for At Least Two Independent Transfers of rpl 22 to the Nucleus. Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Raubeson, L.A.; Boore, J.L.; Depamphilis, C.W.; Chumley, T.W.; Haberle, R.C.; Wyman, S.K.; Alverson, A.J.; Peery, R.; Herman, S.J.; et al. Methods for Obtaining and Analyzing Whole Chloroplast Genome Sequences. Methods Enzymol. 2005, 395, 348–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dong, W.; Zhou, S. Structural Mutations and Reorganizations in Chloroplast Genomes of Flowering Plants. Acta Bot. Boreali-Occident. Sin. 2012, 32, 1282–1288. [Google Scholar] [CrossRef]
- Goremykin, V.V.; Hirsch-Ernst, K.; Wölfl, S.; Hellwig, F.H. Analysis of the Amborella trichopoda Chloroplast Genome Sequence Suggests That Amborella Is Not a Basal Angiosperm. Mol. Biol. Evol. 2003, 20, 1499–1505. [Google Scholar] [CrossRef] [PubMed]
- Clegg, M.T.; Gaut, B.S.; Learn, G.H., Jr.; Morton, B.R. Rates and patterns of chloroplast DNA evolution. Proc. Natl. Acad. Sci. USA 1994, 91, 6795–6801. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. OrganellarGenomeDRAW—A suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Wu, L.; Cui, Y.; Wang, Q.; Xu, Z.; Wang, Y.; Lin, Y.; Song, J.; Yao, H. Identification and phylogenetic analysis of five Crataegus species (Rosaceae) based on complete chloroplast genomes. Planta 2021, 254, 14. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Dong, Z.; Zhang, R.; Shi, M.; Song, Y.; Xin, Y.; Li, F.; Ma, J.; Xin, P. The complete plastid genome of the endangered shrub Brassaiopsis angustifolia (Araliaceae): Comparative genetic and phylogenetic analysis. PLoS ONE 2022, 17, e0269819. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Amiryousefi, A.; Hyvönen, J.; Poczai, P. IRscope: An online program to visualize the junction sites of chloroplast genomes. Bioinformatics 2018, 34, 3030–3031. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Leseberg, C.H.; Duvall, M.R. The Complete Chloroplast Genome of Coix lacryma-jobi and a Comparative Molecular Evolutionary Analysis of Plastomes in Cereals. J. Mol. Evol. 2009, 69, 311–318. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.-L.; Wang, R.-N.; Zhang, N.-Y.; Fan, W.-B.; Fang, M.-F.; Li, Z.-H. Molecular Evolution of Chloroplast Genomes of Orchid Species: Insights into Phylogenetic Relationship and Adaptive Evolution. Int. J. Mol. Sci. 2018, 19, 716. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef]
- Zhu, T.; Zhang, L.; Chen, W.; Yin, J.; Li, Q. Analysis of Chloroplast Genomes in 1342 Plants. Genom. Appl. Biol. 2017, 36, 4323–4333. [Google Scholar] [CrossRef]
- Raman, G.; Park, K.T.; Kim, J.-H.; Park, S. Characteristics of the completed chloroplast genome sequence of Xanthium spinosum: Comparative analyses, identification of mutational hotspots and phylogenetic implications. BMC Genom. 2020, 21, 855. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Zhang, L.; Qi, J.; Zhang, L. Complete Chloroplast Genome Sequence of Hibiscus cannabinus and Comparative Analysis of the Malvaceae Family. Front. Genet. 2020, 11, 227. [Google Scholar] [CrossRef]
- Bi, Y. Comparative Chloroplast Genomics of the Genus LILIUM; Jilin Agricultural University: Changchun, China, 2017. [Google Scholar]
- Wwi, X.; Zhang, Q.; Liu, W.; Liu, N.; Zang, Y.; Xu, M.; Liu, S.; Zang, Y.; Ma, X. Phylogenetic Relationship Analysis of Common Apricot (Prunus armeniaca L.) Revealed by Chloroplast SSR Haplotypes. J. Plant Genet. Resour. 2018, 19, 705–712. [Google Scholar] [CrossRef]
- Wang, H.; Lou, X.; Zhang, Z. Application in Germplasm Resource Research Using Chloroplast Simple Sequence Repeat. Mol. Plant Breed. 2006, 3, 92–98. [Google Scholar]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H.; Sun, C.; Li, X.; Li, C.; et al. The Complete Chloroplast Genome Sequence of the Medicinal Plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef]
- Duan, Y.; Zhang, K. Comparative analysis and phylogenetic evolution of the complete chloroplast genome of Ammopiptanthus. Acta Bot. Boreali-Occident. Sin. 2020, 40, 1323–1332. [Google Scholar] [CrossRef]
- Wang, R.-J.; Cheng, C.-L.; Chang, C.-C.; Wu, C.-L.; Su, T.-M.; Chaw, S.-M. Dynamics and evolution of the inverted repeat-large single copy junctions in the chloroplast genomes of monocots. BMC Evol. Biol. 2008, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Mehmood, F.; Abdullah, S.I.; Ahmed, I.; Waheed, M.T.; Mirza, B. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics 2020, 112, 1522–1530. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zuo, L.; Lu, D.; Lu, B.; Yang, M.; Wang, J. Comparative analysis of chloroplast genomes of five Robinia species: Genome comparative and evolution analysis. Gene 2019, 689, 141–151. [Google Scholar] [CrossRef]
- Li, J.; Jiang, M.; Wang, L.; Yu, J.; Chen, H.; Liu, J.; Liu, C. The complete chloroplast genome of Photinia davidsoniae: Molecular structures and comparative analysis. Mitochondrial DNA Part B Resour. 2021, 6, 1431–1439. [Google Scholar] [CrossRef]
- Abdullah; Mehmood, F.; Shahzadi, I.; Waseem, S.; Mirza, B.; Ahmed, I.; Waheed, M.T. Chloroplast genome of Hibiscus rosa-sinensis (Malvaceae): Comparative analyses and identification of mutational hotspots. Genomics 2020, 112, 581–591. [Google Scholar] [CrossRef]
- Menezes, A.P.A.; Resende-Moreira, L.C.; Buzatti, R.S.O.; Nazareno, A.G.; Carlsen, M.; Lobo, F.P.; Kalapothakis, E.; Lovato, M.B. Chloroplast genomes of Byrsonima species (Malpighiaceae): Comparative analysis and screening of high divergence sequences. Sci. Rep. 2018, 8, 2210. [Google Scholar] [CrossRef]
- McPhee, D.J.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete Chloroplast Genome of Medicinal Plant Lonicera japonica: Genome Rearrangement, Intron Gain and Loss, and Implications for Phylogenetic Studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef]
- Iram, S.; Hayat, M.Q.; Tahir, M.; Gul, A.; Abdullah; Ahmed, I. Chloroplast Genome Sequence of Artemisia scoparia: Comparative Analyses and Screening of Mutational Hotspots. Plants 2019, 8, 476. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; He, P.; Li, P.; Lee, J.; Soltis, D.E.; Fu, C. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genom. 2018, 19, 235. [Google Scholar] [CrossRef]
- Du, Z.; Yang, L.; Zhang, C.; Xu, H.; Chen, Z. Characteristics of the complete chloroplast genome of Dendrobium ochreatum and its comparative analysis. Chin. J. Trop. Crops 2021, 42, 3111–3119. [Google Scholar] [CrossRef]
- Lu, R.-S.; Li, P.; Qiu, Y.-X. The Complete Chloroplast Genomes of Three Cardiocrinum (Liliaceae) Species: Comparative Genomic and Phylogenetic Analyses. Front. Plant Sci. 2016, 7, 2054. [Google Scholar] [CrossRef] [PubMed]
- Celiński, K.; Kijak, H.; Wiland-Szymańska, J. Complete Chloroplast Genome Sequence and Phylogenetic Inference of the Canary Islands Dragon Tree (Dracaena draco L.). Forests 2020, 11, 309. [Google Scholar] [CrossRef]
- Tang, P.; Peng, C. The rate and mode of chloroplast genome evolution. Bull. Biol. 2010, 45, 8–10. [Google Scholar] [CrossRef]
- Liang, F.; Wen, X.; Gao, H.; Zhang, Y. Analysis of Chloroplast Genomes Features of Asteraceae Species. Genom. Appl. Biol. 2018, 37, 5437–5447. [Google Scholar] [CrossRef]
- Liu, L.-X.; Du, Y.-X.; Folk, R.A.; Wang, S.-Y.; Soltis, D.E.; Shang, F.-D.; Li, P. Plastome Evolution in Saxifragaceae and Multiple Plastid Capture Events Involving Heuchera and Tiarella. Front. Plant Sci. 2020, 11, 361. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Kuzoff, R.K. Discordance between Nuclear and Chloroplast Phylogenies in the Heuchera Group (Saxifragaceae). Evolution 1995, 49, 727–742. [Google Scholar] [CrossRef]
- Liu, B.; Ren, C.; Kwak, M.; Hodel, R.G.; Xu, C.; He, J.; Zhou, W.; Huang, C.; Ma, H.; Qian, G.; et al. Phylogenomic conflict analyses in the apple genus Malus s.l. reveal widespread hybridization and allopolyploidy driving diversification, with insights into the complex biogeographic history in the Northern Hemisphere. J. Integr. Plant Biol. 2022, 64, 1020–1043. [Google Scholar] [CrossRef]
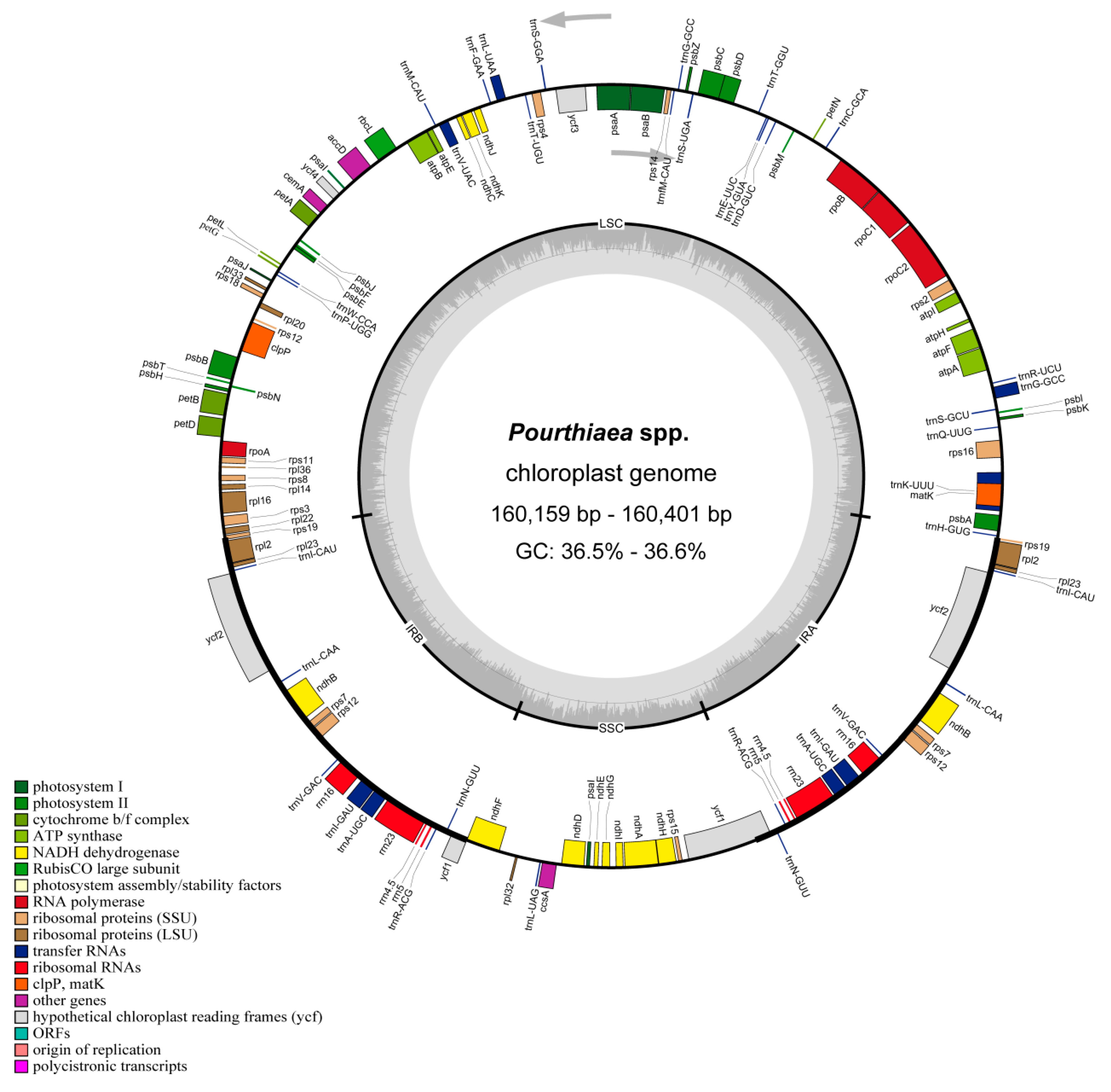

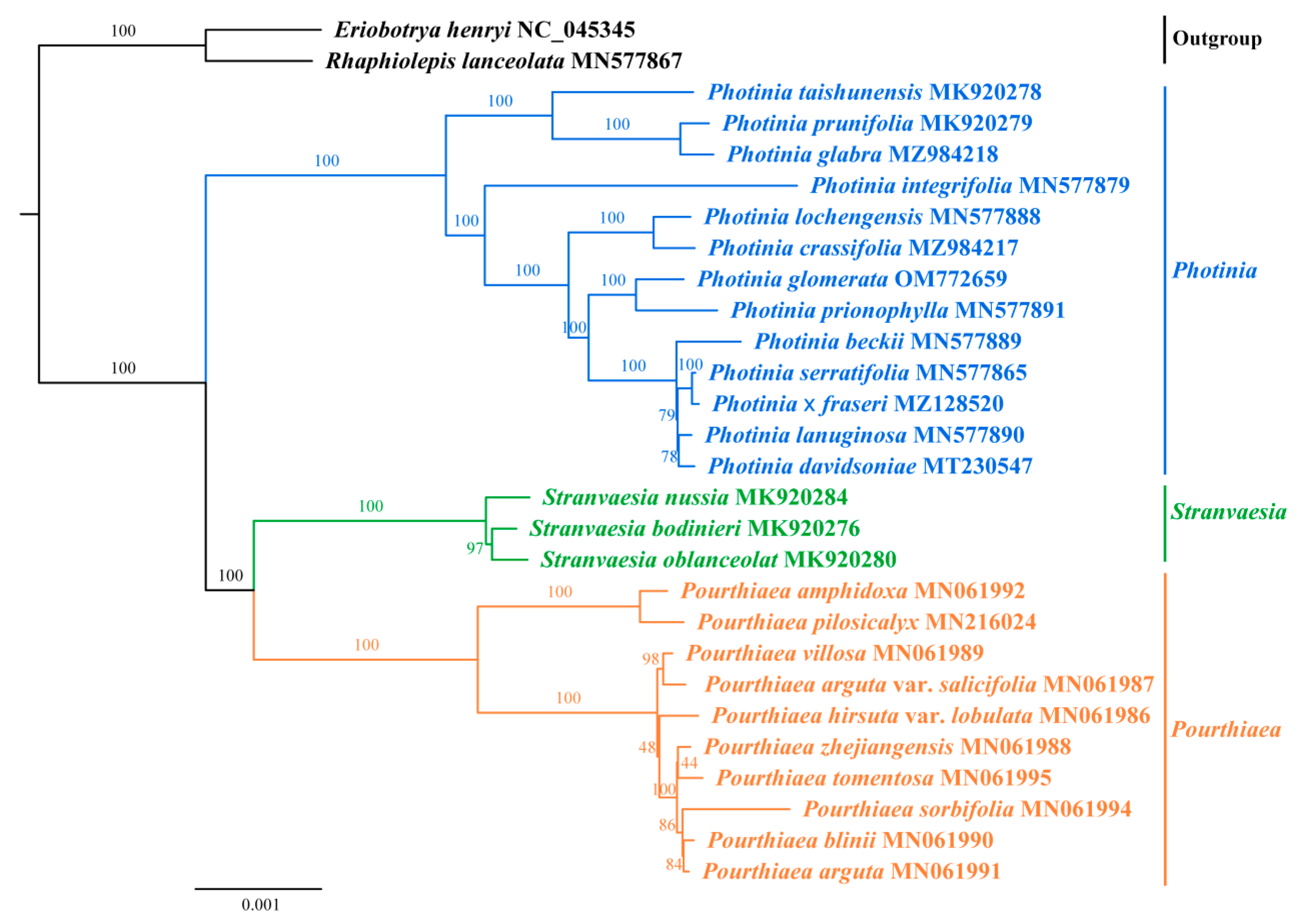
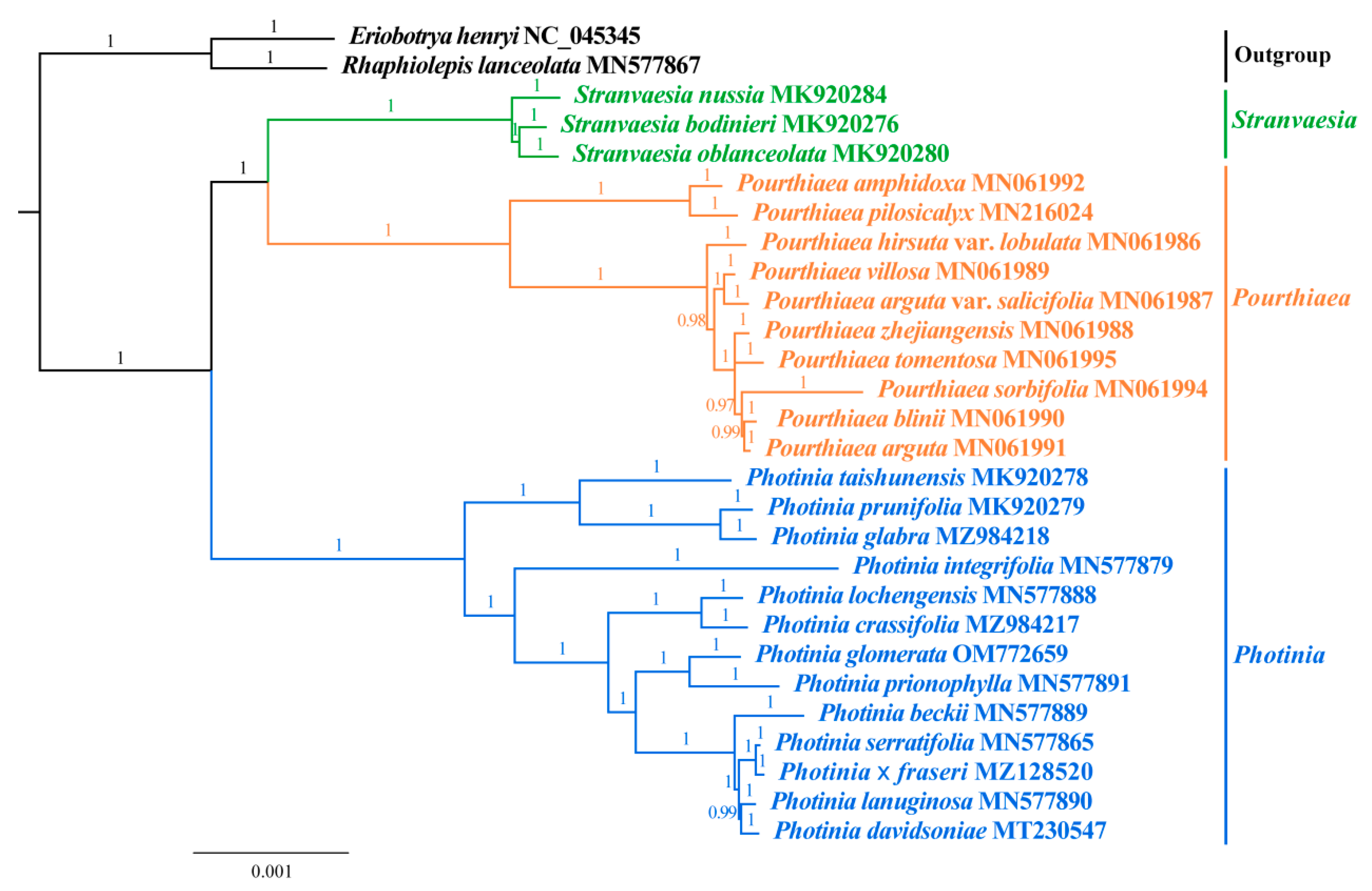
| Species | Size (bp) | Gene Number | GC Content (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LSC | SSC | IR | Total | CDS | tRNA | rRNA | Total | LSC | SSC | IR | Mean | |
| P. villosa | 88,307 | 19,306 | 26,394 | 160,401 | 83 | 37 | 8 | 128 | 36.5 | 30.3 | 42.7 | 36.5 |
| P. amphidoxa | 88,359 | 19,234 | 26,382 | 160,357 | 83 | 37 | 8 | 128 | 34.1 | 30.4 | 42.6 | 36.5 |
| P. pilosicalyx | 88,342 | 19,306 | 26,341 | 160,330 | 84 | 37 | 8 | 129 | 34.4 | 30.3 | 42.7 | 36.6 |
| P. blinii | 88,171 | 19,338 | 26,399 | 160,307 | 83 | 38 | 8 | 129 | 34.2 | 30.2 | 42.7 | 36.5 |
| P. zhejiangensis | 88,183 | 19,329 | 26,394 | 160,300 | 83 | 37 | 8 | 128 | 36.6 | 30.2 | 42.6 | 36.6 |
| P. tomentosa | 88,181 | 19,321 | 26,394 | 160,290 | 83 | 37 | 8 | 128 | 34.1 | 30.3 | 42.6 | 36.5 |
| P. hirsuta var. lobulata | 88,127 | 19,308 | 26,394 | 160,223 | 83 | 38 | 8 | 129 | 34.2 | 30.3 | 42.7 | 36.5 |
| P. arguta var. salicifolia | 88,124 | 19,287 | 26,394 | 160,199 | 83 | 37 | 8 | 128 | 34.3 | 30.4 | 42.7 | 36.6 |
| P. arguta | 88,047 | 19,314 | 26,399 | 160,159 | 83 | 38 | 8 | 129 | 34.2 | 30.3 | 42.7 | 36.5 |
| P. sorbifolia | 88,239 | 19,330 | 26,401 | 160,371 | 83 | 37 | 8 | 128 | 34.2 | 30.3 | 42.6 | 36.5 |
| Category | Group of Genes | Genes Names | Amount |
|---|---|---|---|
| Photosynthesis gene | Photosystems I | psaA, psaB, psaJ, psaI (×2) | 5 |
| Photosystems II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbJ, psbK, psbI, psbM, psbN, psbT, psbZ | 14 | |
| Cytochrome b/f complex | petA, petB *, petD *, petG, petL, petN | 6 | |
| ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI | 6 | |
| NADH dehydrogenase | ndhA *, ndhB * (×2), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | 12 | |
| Rubisco Large subunit | rbcL | 1 | |
| Self-replication gene | RNA polymerase | rpoA, rpoB, rpoC1 *, rpoC2 | 4 |
| Ribosomal proteins (SSU) | rps2, rps3, rps4, rps7 (×2), rps8, rps11, rps12 ** (×2), rps14, rps15, rps16 *, rps18, rps19 (×2) | 16 | |
| Ribosomal proteins (LSU) | rpl2 * (×2), rpl14, rpl16, rpl20, rpl22, rpl23(×2), rpl32, rpl33, rpl36 | 11 | |
| Transfer RNAs | trnA-UGC * (×2), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-GCC * (×2), trnH-GUG, trnI-CAU (×2), trnI-GAU * (×2), trnK-UUU *, trnL-CAA (×2), trnL-UAA *, trnL-UAG, trnM-CAU, trnN-GUU (×2), trnP-UGG, trnQ-UUG, trnR-ACG (×2), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC (×2), trnV-UAC*, trnW-CCA, trnY-GUA | 37 | |
| Ribosomal RNAs | rrn4.5 (×2), rrn5 (×2), rrn16 (× 2), rrn23 (×2) | 8 | |
| Other genes | Maturase | matK | 1 |
| Envelop membrane protein | cemA | 1 | |
| Subunit of acetyl-CoA-carboxylase | accD | 1 | |
| c-type cytochrome synthesis gene | ccsA | 1 | |
| Proteolysis | clpP ** | 1 | |
| Hypothetical chloroplast reading frames (ycf) | Ycf1 (×2), ycf2 (×2), ycf3 **, ycf4 | 6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, Z.; Zhao, W.; Xin, Y.; Shen, W.; Wang, F.; Li, Q.; Tu, Y.; Zhang, H.; Dong, Z.; Xin, P. Characteristics of the Complete Chloroplast Genome of Pourthiaea (Rosaceae) and Its Comparative Analysis. Horticulturae 2022, 8, 1144. https://doi.org/10.3390/horticulturae8121144
Cao Z, Zhao W, Xin Y, Shen W, Wang F, Li Q, Tu Y, Zhang H, Dong Z, Xin P. Characteristics of the Complete Chloroplast Genome of Pourthiaea (Rosaceae) and Its Comparative Analysis. Horticulturae. 2022; 8(12):1144. https://doi.org/10.3390/horticulturae8121144
Chicago/Turabian StyleCao, Zhengying, Wenzhi Zhao, Yaxuan Xin, Weixiang Shen, Fei Wang, Qishao Li, Yuxiang Tu, Haorong Zhang, Zhanghong Dong, and Peiyao Xin. 2022. "Characteristics of the Complete Chloroplast Genome of Pourthiaea (Rosaceae) and Its Comparative Analysis" Horticulturae 8, no. 12: 1144. https://doi.org/10.3390/horticulturae8121144
APA StyleCao, Z., Zhao, W., Xin, Y., Shen, W., Wang, F., Li, Q., Tu, Y., Zhang, H., Dong, Z., & Xin, P. (2022). Characteristics of the Complete Chloroplast Genome of Pourthiaea (Rosaceae) and Its Comparative Analysis. Horticulturae, 8(12), 1144. https://doi.org/10.3390/horticulturae8121144






