Identification and Characterization of Trihelix Transcription Factors and Expression Changes during Flower Development in Pineapple
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of AcTrihelix Genes in Pineapple
2.2. Phylogenetic, Conserved Motif, and Gene Structure Analyses
2.3. cis-Element Identification, Chromosome Location, Duplication Events, and Collinear Analysis
2.4. Expression Profiling of AcTrihelix Genes by RNA-Seq
2.5. Plant Materials and Treatments
2.6. RNA Extraction and Real-Time Quantitative PCR (RT-qPCR) Analysis
3. Results
3.1. Identification and Characterization of Trihelix Proteins in Pineapple
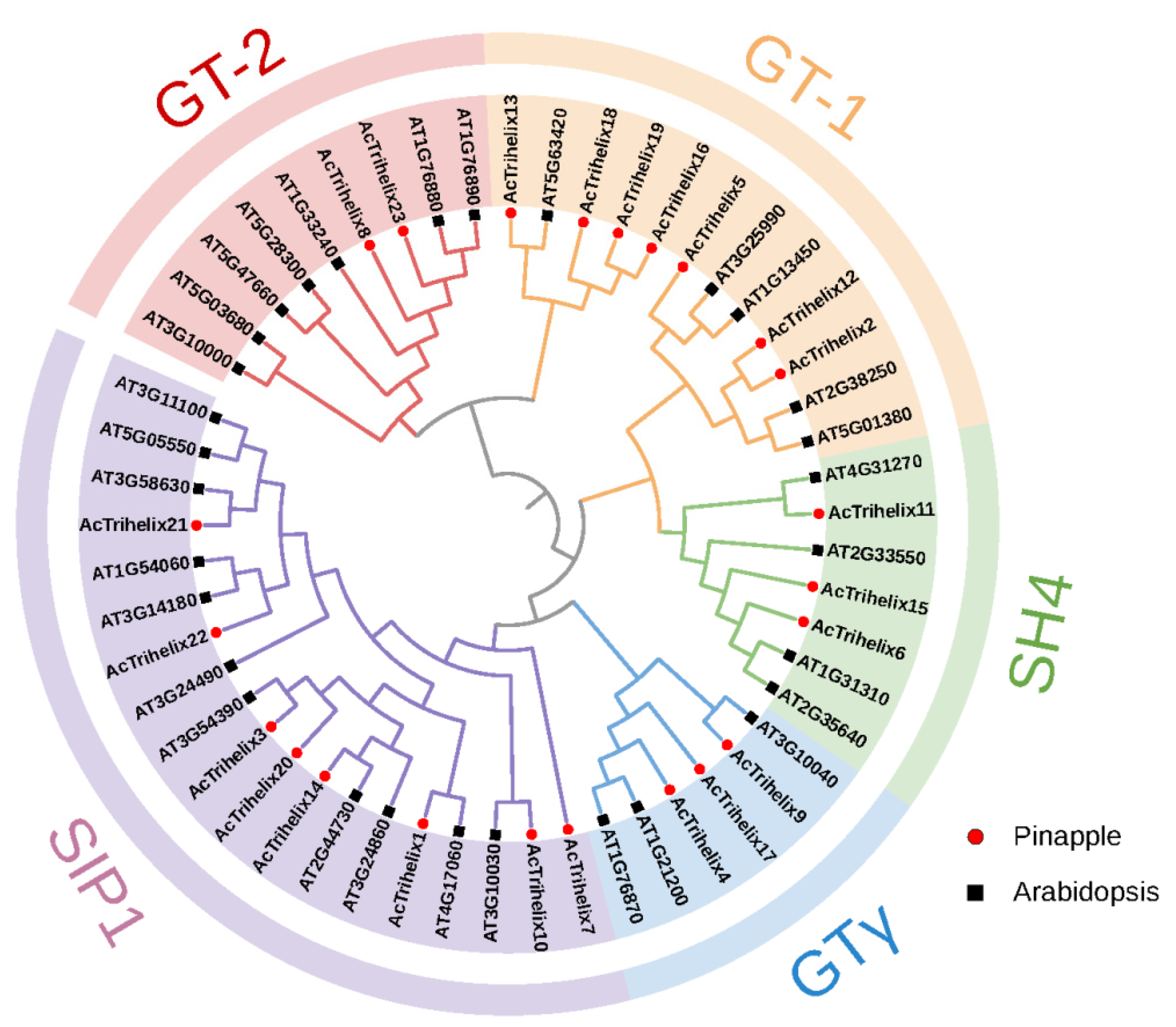
3.2. Phylogenetic Analysis
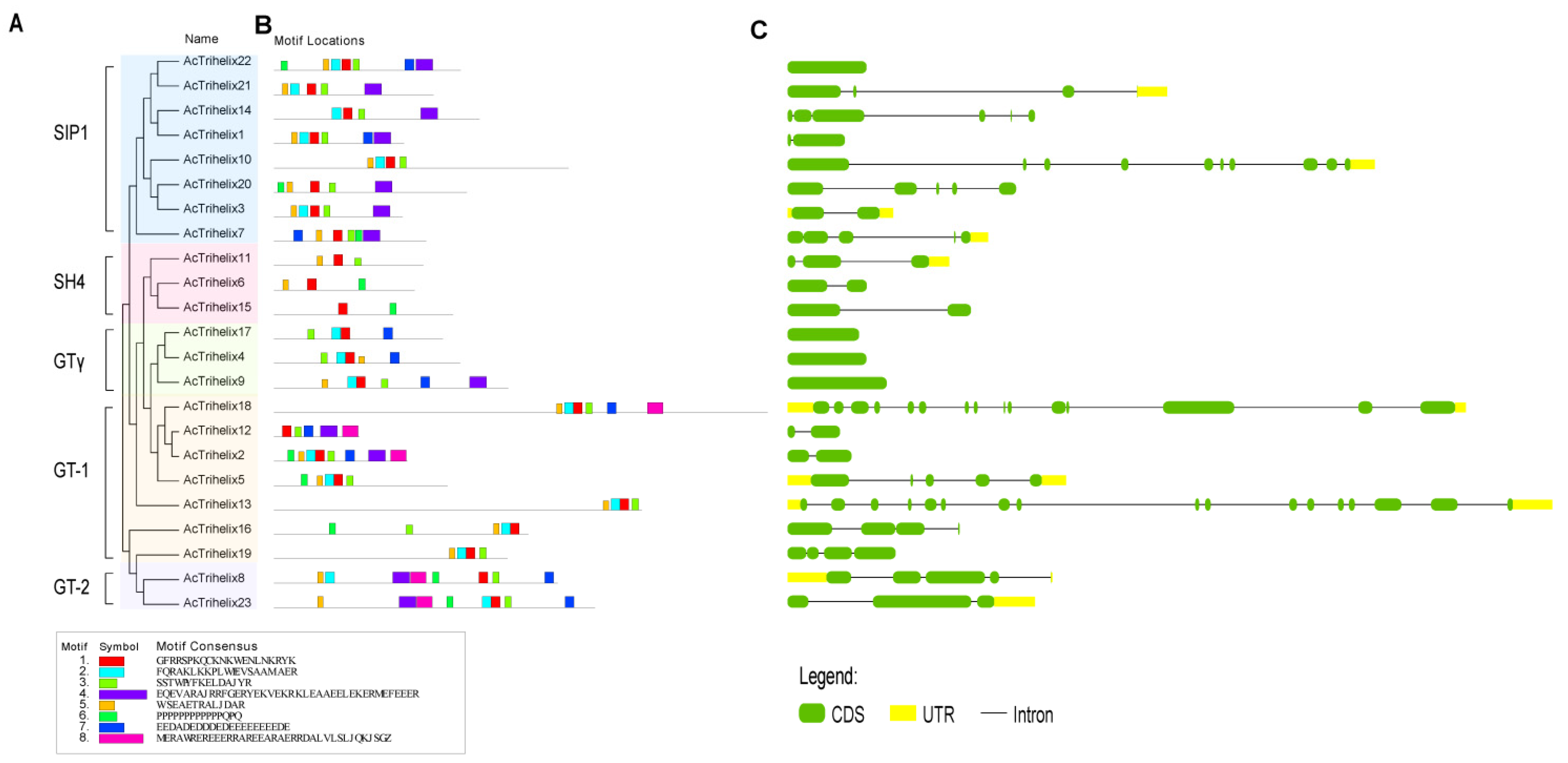
3.3. Gene Structures, Conserved Domains, and Motif Analysis
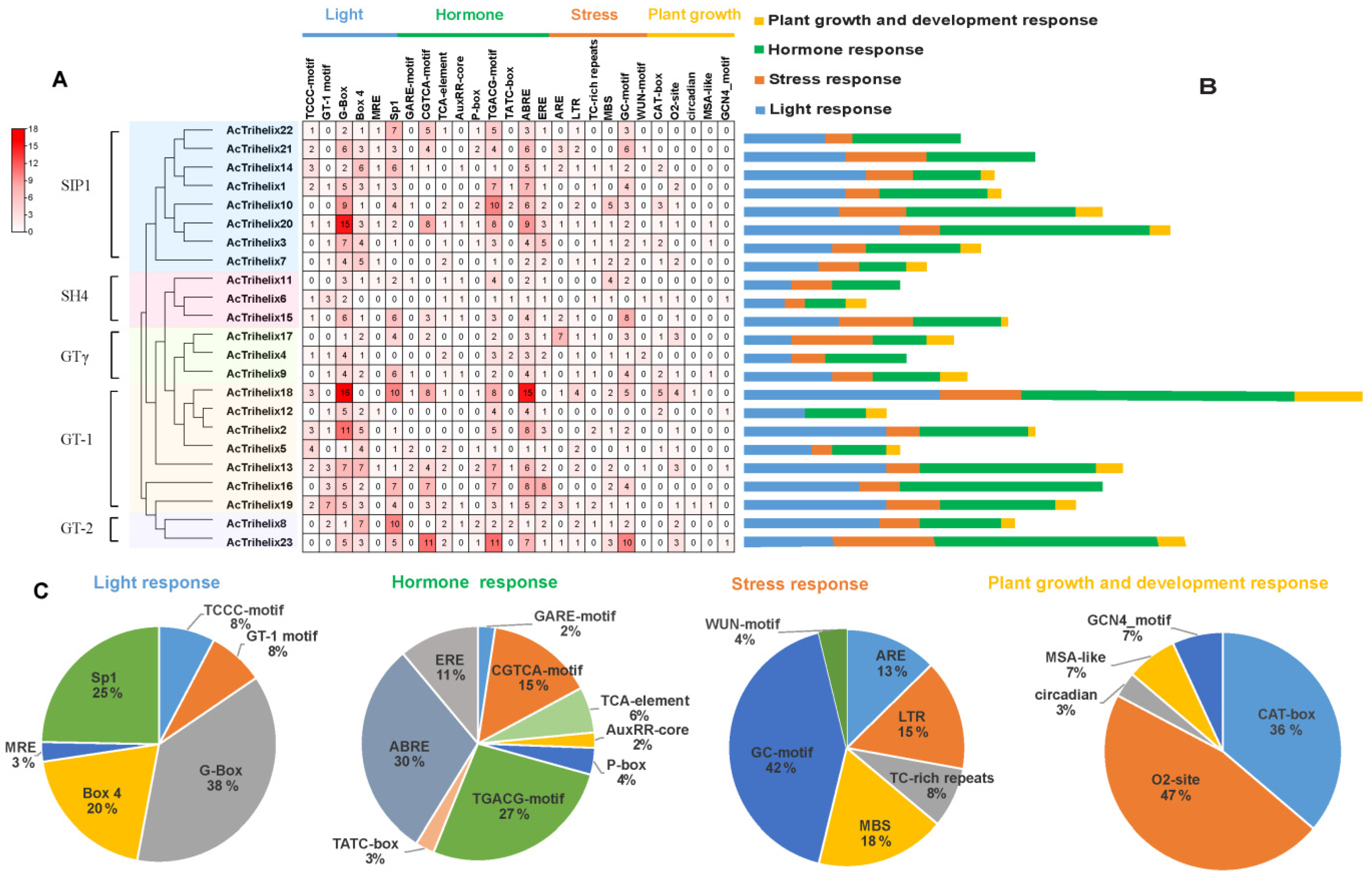
3.4. cis-Element Analysis of the AcTrihelix Promoters
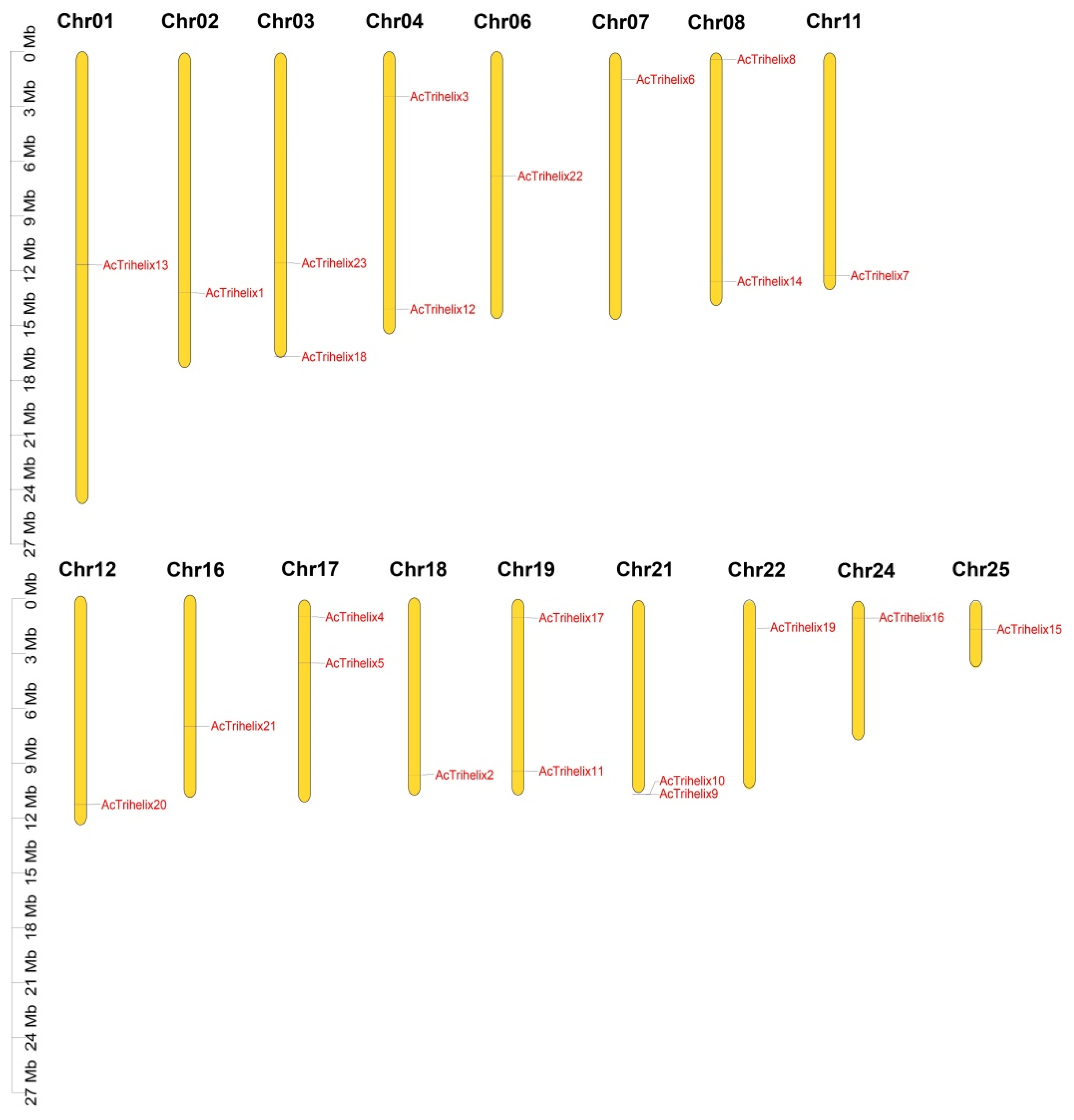
3.5. Chromosomal Location and Gene Duplication of AcTrihelix Gene
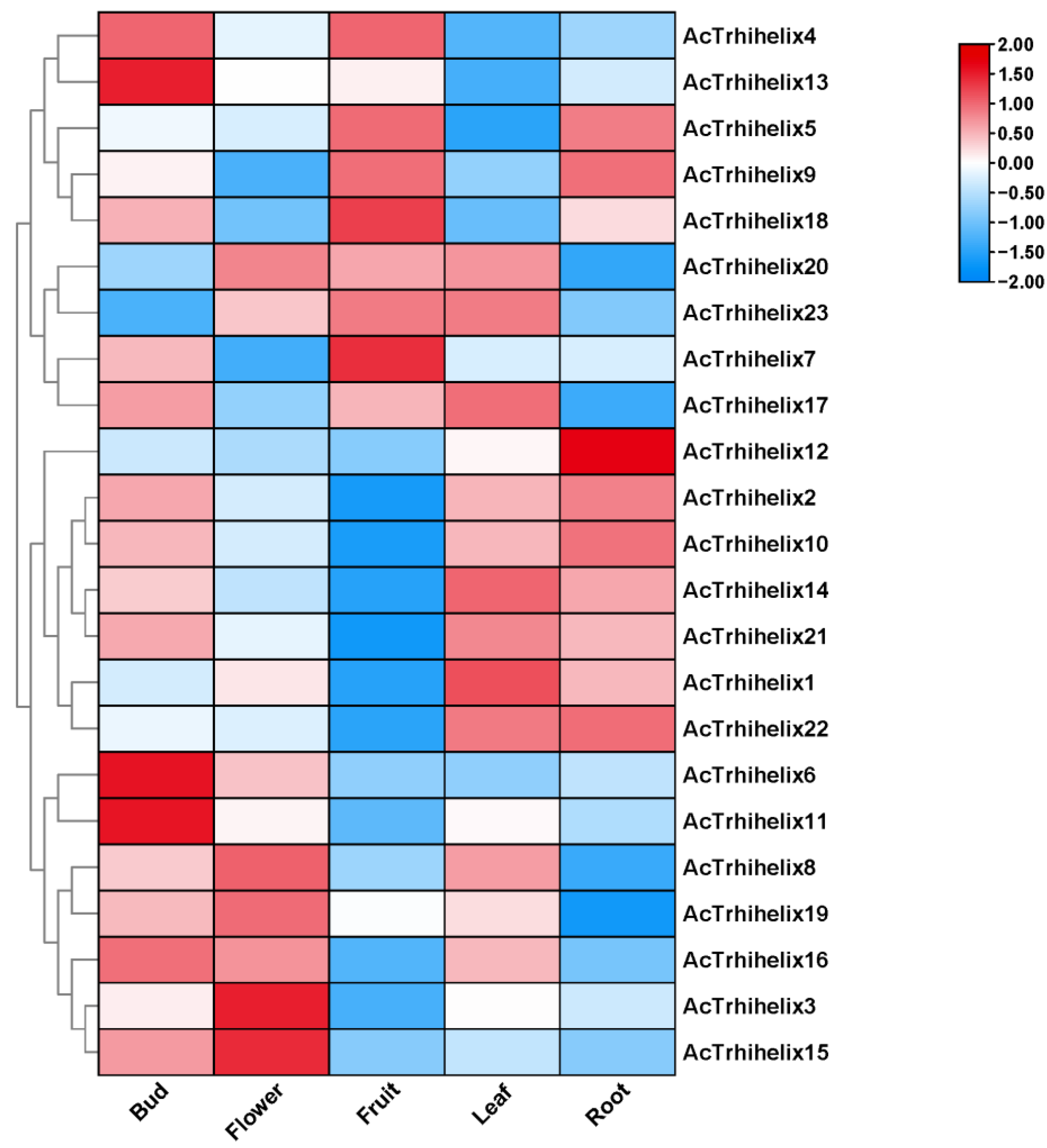
3.6. Expression Profiles of AcTrihelix in Different Tissues in Pineapple
3.7. Expression Profiles of AcTrihelix in Flower Induction and Flowering Process in Pineapple
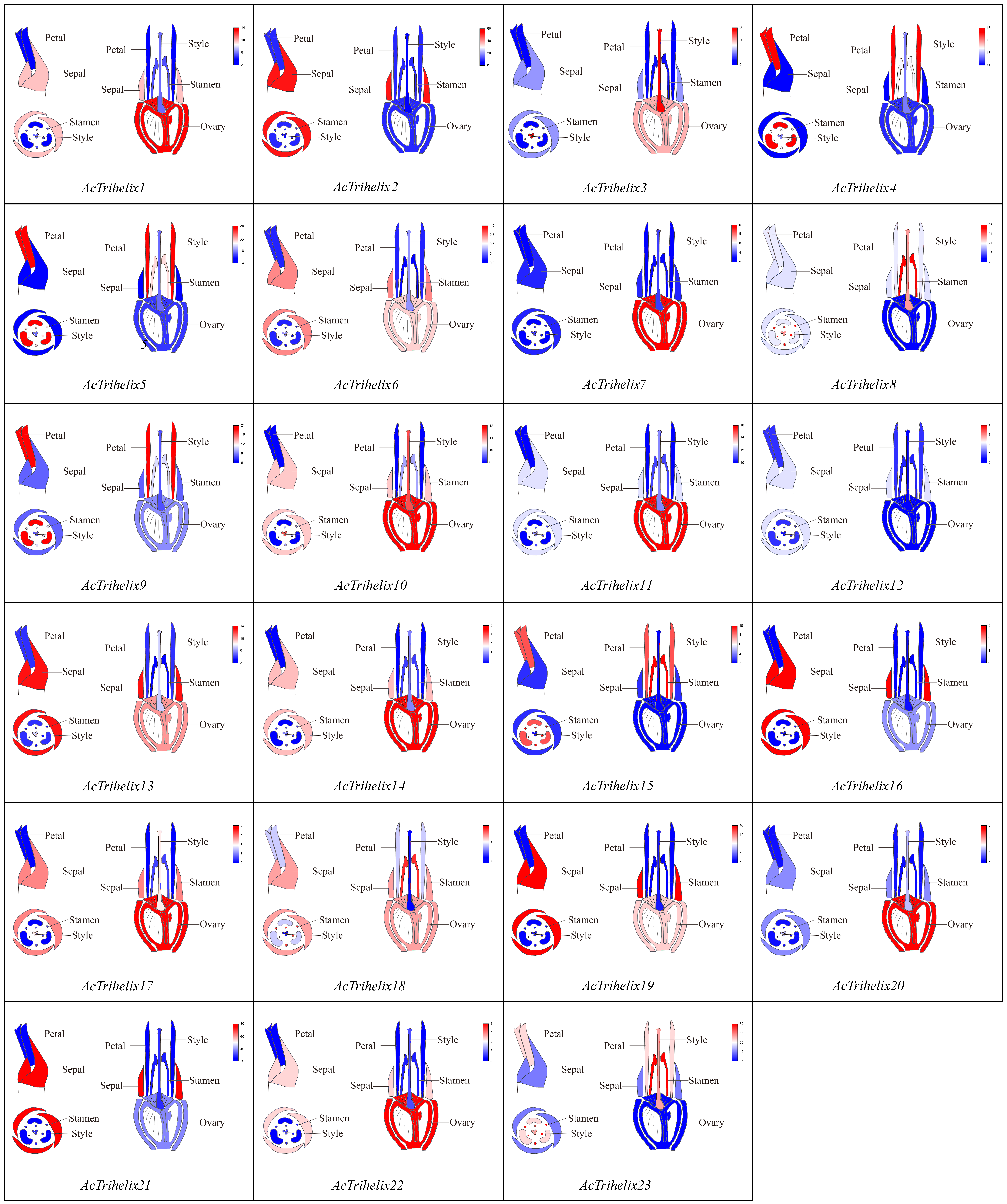
3.8. Expression Profiles of AcTrihelix in Floral Tissue and Organ
3.9. qRT-PCR Assays of the Expression Patterns of AcTrihelix in Flowering Process and Floral Organ
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.M.; Zhang, M.H.; Sun, X.; Mao, X.; Wang, J.; Liu, H.L.; Zheng, H.L.; Zhen, Z.; Zhao, H.W.; Zou, D.T. Genome-Wide Characterization and Identification of Trihelix Transcription Factor and Expression Profiling in Response to Abiotic Stresses in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2019, 20, 251. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors-light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef]
- Green, P.J.; Kay, S.A.; Chua, N.H. Sequence-specific interactions of a pea nuclear factor with light-responsive elements upstream of the rbcS-3A gene. EMBO J. 1987, 6, 2543–2549. [Google Scholar] [CrossRef] [PubMed]
- Nagano, Y. Several Features of the GT-Factor Trihelix Domain Resemble Those of the Myb DNA-Binding Domain. Plant Physiol. 2000, 124, 491–494. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.T.; Liu, M.Y.; Sun, W.J.; Huang, L.; Wu, Q.; Bu, T.L.; Li, C.L.; Chen, H. Genome-wide identification and expression analysis of the trihelix transcription factor family in tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 344. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Ma, X.; Yu, G.H.; Wang, Q.; Wang, L.; Kong, L.R.; Kim, W.; Wang, H.W. Evolutionary history of trihelix family and their functional diversification. Plant Physiol. 2000, 124, 491–493. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.Y.; Shi, X.X.; He, L.; Guo, Y.; Zang, D.D.; Li, H.Y.; Zhang, W.H.; Wang, Y.C. Arabidopsis thaliana Trihelix Transcription Factor AST1 Mediates Salt and Osmotic Stress Tolerance by Binding to a Novel AGAG-Box and Some GT Motifs. Plant Cell Physiol. 2018, 59, 946–965. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, D.C.; Shan, T.F.; Xu, S.B.; Qin, R.Y.; Li, H.; Negm, M.; Wu, D.X.; Li, J. The trihelix transcription factor OsGTγ-2 is involved adaption to salt stress in rice. Plant Mol. Biol. 2020, 103, 545–560. [Google Scholar] [CrossRef]
- Fang, Y.J.; Xie, K.B.; Hou, X.; Hu, H.H.; Xiong, L.Z. Systematic analysis of GT factor family of rice reveals a novel subfamily involved in stress responses. Mol. Genet. Genom. 2010, 283, 157–169. [Google Scholar] [CrossRef]
- Xie, Z.M.; Zou, H.F.; Lei, G.; Wei, W.; Zhou, Q.Y.; Niu, C.F.; Liao, Y.; Tian, A.G.; Ma, B.; Zhang, W.K.; et al. Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS ONE 2009, 4, e6898. [Google Scholar] [CrossRef]
- Yu, C.Y.; Song, L.L.; Song, J.W.; Ouyang, B.; Guo, L.J.; Shang, L.L.; Wang, T.T.; Li, H.X.; Zhang, J.H.; Ye, Z.B. ShCIGT, a Trihelix family gene, mediates cold and drought tolerance by interacting with SnRK1 in tomato. Plant Sci. 2018, 270, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Park, H.C.; Kim, M.L.; Kang, Y.H.; Jeon, J.M.; Yoo, J.H.; Kim, M.C.; Park, C.Y.; Jeong, J.C.; Moon, B.C.; Lee, J.H.; et al. Pathogen- and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 2004, 135, 2150–2161. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Hong, G.F.; Han, B. Transcript abundance of rml1, encoding a putative GT1-like factor in rice, is up-regulated by Magnaporthe grisea and down-regulated by light. Gene 2004, 324, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Barr, M.; Willmann, M.R.; Jenik, P.D. Is there a role for trihelix transcription factors in embryo maturation? Plant Signal. Behav. 2012, 7, 205–209. [Google Scholar] [CrossRef]
- Gao, M.J.; Li, X.; Lui, X.; Lui, H.; Gropp, G.M.; Lydiate, D.D.; Wei, S.; Hegedus, D.D. ASIL1 is required for proper timing of seed filling in Arabidopsis. Plant Signal Behav. 2011, 6, 1886–1888. [Google Scholar] [CrossRef]
- Griffith, M.E.; Conceio, A.D.S.; Smyth, D.R. PETAL LOSS gene regulates initiation and orientation of second whorl organs in the Arabidopsis flower. Development 1999, 126, 5635–5644. [Google Scholar] [CrossRef]
- Brewer, P.B.; Howles, P.A.; Dorian, K.; Kristen, G.M.E.; Ishida, T.; Kaplan-Levy, R.N.; Kilinc, A.; Smyth, D.R. PETAL LOSS, a trihelix transcription factor gene, regulates perianth architecture in the Arabidopsis flower. Development 2004, 131, 4035–4045. [Google Scholar] [CrossRef]
- Li, X.; Qin, G.J.; Chen, Z.L.; Gu, H.Y.; Qu, L.J. A gain-of-function mutation of transcriptional factor PTL results in curly leaves, dwarfism and male sterility by affecting auxin homeostasis. Plant Mol. Biol. 2008, 66, 315–327. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.L.; Qi, S.L.; Touqeer, A.; Li, H.Y.; Zhang, X.L.; Liu, X.F.; Wu, S. SlGT11 controls floral organ patterning and floral determinacy in tomato. BMC Plant Biol. 2020, 20, 562. [Google Scholar] [CrossRef]
- Xu, H.; Yu, Q.; Shi, Y.; Hua, X.; Tang, H.; Yang, L.; Ming, R.; Zhang, J. PGD: Pineapple Genomics Database. Hortic. Res. 2018, 5, 66. [Google Scholar] [CrossRef] [PubMed]
- Poole, R.L. The TAIR database. Methods Mol. Biol. 2007, 406, 179–212. [Google Scholar] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.; Chen, Y.; Lu, C.; Hwang, J.K. Prediction of protein subcellular localization. Proteins 2006, 64, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Kohli, D.K.; Bachhawat, A.K. CLOURE: Clustal Output Reformatter, a program for reformatting ClustalX/ClustalW outputs for SNP analysis and molecular systematics. Nucleic Acids Res. 2003, 31, 3501–3502. [Google Scholar] [CrossRef]
- Hall, B.G. Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Rao, X.Y.; Huang, X.L.; Zhou, Z.C.; Lin, X. An improvement of the 2−ΔΔCT method for quantitative real-time polymerase chain reaction data analysis. Biostat. Bioinform. Biomath. 2013, 3, 71–85. [Google Scholar]
- Azam, S.M.; Liu, Y.H.; Rahman, Z.U.; Ali, H.; Yan, C.; Wang, L.L.; Priyadarshani, V.G.N.; Hu, B.Y.; Xiong, J.Y.; Qin, Y. Identification, Characterization and Expression Profiles of Dof Transcription Factors in Pineapple (Ananas comosus L). Trop. Plant Biol. 2018, 11, 49–64. [Google Scholar] [CrossRef]
- Lalitha, S. Primer premier 5. Biotech. Softw. Internet Rep. 2000, 1, 270–272. [Google Scholar] [CrossRef]
- Gao, H.Y.; Huang, R.; Liu, J.; Gao, Z.M.; Zhao, H.S.; Li, X.P. Genome-Wide Identification of Trihelix Genes in Moso Bamboo (Phyllostachys edulis) and Their Expression in Response to Abiotic Stress. J. Plant Growth Regul. 2019, 38, 1127–1140. [Google Scholar] [CrossRef]
- Tong, Y.; Huang, H.; Wang, Y.H. Genome-Wide Analysis of the Trihelix Gene Family and Their Response to Cold Stress in Dendrobium officinale. Sustainability 2021, 13, 2826. [Google Scholar] [CrossRef]
- Yu, C.Y.; Cai, X.F.; Ye, Z.B.; Li, H.X. Genome-wide identification and expression profiling analysis of trihelix gene family in tomato. Biochem. Biophys. Res. Commun. 2015, 468, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Shen, W.Y.; Shaheen, S.; Li, Y.Y.; Liu, Z.R.; Wang, Z. Genome-wide identification and analysis of the trihelix transcription factors in sunflower. Biol. Plant. 2021, 65, 80–87. [Google Scholar] [CrossRef]
- Wang, W.; Peng, W.; Liu, T.K.; Ren, H.; Hou, X. Genome-wide analysis and expression divergence of the trihelix family in Brassica rapa: Insight into the evolutionary patterns in plants. Sci. Rep. 2017, 7, 6463. [Google Scholar] [CrossRef]
- Osorio, M.B.; Bücker-Neto, L.; Castilhos, G.; Turchetto-Zolet, A.C.; Wiebke-Strohm, B.; Bodanese-Zanettini, M.H.; Margis-Pinheiro, M. Identification and in silico characterization of soybean trihelix-GT and bHLH transcription factors involved in stress responses. Genet. Mol. Biol. 2012, 35, 233–246. [Google Scholar] [CrossRef]
- Li, K.L.; Zhang, Y.B.; Shi, M.X.; Chen, S.S.; Yang, M.F.; Ding, Y.Q.; Peng, Y.S.; Dong, Y.B.; Yang, H.; Li, Z.H.; et al. Genome-wide identification and expression profile analysis of trihelix transcription factor family genes in response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genom. 2021, 22, 738. [Google Scholar] [CrossRef]
- Gao, M.J.; Lydiate, D.J.; Li, X.; Lui, H.; Gjetvaj, B.; Hegedus, D.D.; Rozwadowski, K. Repression of seed maturation genes by a trihelix transcriptional repressor in arabidopsis seedlings. Plant Cell 2009, 21, 54–71. [Google Scholar] [CrossRef]
- Paterson, A.H.; Freeling, M.; Tang, H.; Wang, X. Insights from the comparison of plant genome sequences. Annu. Rev. Plant Biol. 2010, 61, 349–372. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Y.; Li, W.; Lin, Y.; Wang, C.; Xu, R.; Zhang, L.F. Genome-wide characterization and expression analysis of soybean trihelix gene family. PeerJ 2020, 8, e8753. [Google Scholar] [CrossRef] [PubMed]
- Quon, T.; Lampugnani, E.R.; Smyth, D.R. PETAL LOSS and ROXY1 Interact to Limit Growth within and between Sepals but to Promote Petal Initiation in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 152. [Google Scholar] [CrossRef]
- Zhang, H.; Pan, X.; Liu, S.; Lin, W.Q.; Li, Y.H.; Zhang, X.M. Genome-wide analysis of AP2/ERF transcription factors in pineapple reveals functional divergence during flowering induction mediated by ethylene and floral organ development. Genomics 2020, 113, 474–489. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Gene ID | Protein/AA | MW (kDa) | pI | Instability Index | GRAVY | Subcellular Localization |
|---|---|---|---|---|---|---|---|
| AcTrihelix1 | Aco001232.1 | 310 | 35.37 | 5.22 | 67.04 | −1.11 | N |
| AcTrihelix2 | Aco001562.1 | 318 | 37.37 | 7.17 | 76.45 | −1.23 | N |
| AcTrihelix3 | Aco002322.1 | 306 | 33.89 | 8.78 | 46.02 | −0.86 | N |
| AcTrihelix4 | Aco003181.1 | 444 | 50.89 | 6.03 | 52.11 | −0.93 | N |
| AcTrihelix5 | Aco003514.1 | 414 | 47.30 | 6.10 | 56.72 | −0.83 | N |
| AcTrihelix6 | Aco005017.1 | 335 | 38.13 | 8.82 | 62.67 | −1.03 | N |
| AcTrihelix7 | Aco005686.1 | 363 | 42.40 | 4.82 | 71.97 | −1.37 | N |
| AcTrihelix8 | Aco007563.1 | 677 | 73.09 | 8.84 | 72.72 | −0.91 | N |
| AcTrihelix9 | Aco007761.1 | 559 | 60.94 | 5.42 | 66.22 | −0.95 | N |
| AcTrihelix10 | Aco007763.1 | 703 | 76.76 | 6.54 | 41.54 | −0.58 | N |
| AcTrihelix11 | Aco008315.1 | 356 | 39.37 | 8.41 | 52.18 | −0.84 | N |
| AcTrihelix12 | Aco011052.1 | 203 | 24.60 | 9.21 | 90.86 | −1.36 | N |
| AcTrihelix13 | Aco011421.1 | 878 | 97.27 | 8.92 | 42.96 | −0.31 | N |
| AcTrihelix14 | Aco011649.1 | 490 | 53.03 | 11.33 | 74.69 | −0.43 | N |
| AcTrihelix15 | Aco012997.1 | 427 | 46.46 | 10.74 | 97.54 | −0.85 | N |
| AcTrihelix16 | Aco013248.1 | 607 | 64.26 | 6.76 | 66.99 | −0.61 | N |
| AcTrihelix17 | Aco015633.1 | 403 | 44.50 | 5.55 | 48.58 | −0.96 | N |
| AcTrihelix18 | Aco017242.1 | 1179 | 129.66 | 6.25 | 53.97 | −0.23 | Pla |
| AcTrihelix19 | Aco017510.1 | 557 | 59.58 | 8.72 | 68.35 | −0.62 | N |
| AcTrihelix20 | Aco018180.1 | 460 | 50.38 | 9.28 | 57.04 | −0.86 | N |
| AcTrihelix21 | Aco021658.1 | 380 | 42.09 | 9.76 | 52.18 | −0.74 | N |
| AcTrihelix22 | Aco023568.1 | 445 | 48.57 | 7.14 | 72.58 | −1.12 | N |
| AcTrihelix23 | Aco024437.1 | 766 | 81.19 | 5.57 | 73.19 | −0.87 | N |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Ouyang, Y.; Wei, Y.; Kou, J.; Zhang, X.; Zhang, H. Identification and Characterization of Trihelix Transcription Factors and Expression Changes during Flower Development in Pineapple. Horticulturae 2022, 8, 894. https://doi.org/10.3390/horticulturae8100894
Wang J, Ouyang Y, Wei Y, Kou J, Zhang X, Zhang H. Identification and Characterization of Trihelix Transcription Factors and Expression Changes during Flower Development in Pineapple. Horticulturae. 2022; 8(10):894. https://doi.org/10.3390/horticulturae8100894
Chicago/Turabian StyleWang, Jing, Yanwei Ouyang, Yongzan Wei, Jingjing Kou, Xiaohan Zhang, and Hongna Zhang. 2022. "Identification and Characterization of Trihelix Transcription Factors and Expression Changes during Flower Development in Pineapple" Horticulturae 8, no. 10: 894. https://doi.org/10.3390/horticulturae8100894
APA StyleWang, J., Ouyang, Y., Wei, Y., Kou, J., Zhang, X., & Zhang, H. (2022). Identification and Characterization of Trihelix Transcription Factors and Expression Changes during Flower Development in Pineapple. Horticulturae, 8(10), 894. https://doi.org/10.3390/horticulturae8100894




