Digitalization of Clubroot Disease Index, a Long Overdue Task
Abstract
:1. Clubroot, an Introduction
2. Clubroot Disease Index and Plasmodiophora brassicae Pathotyping
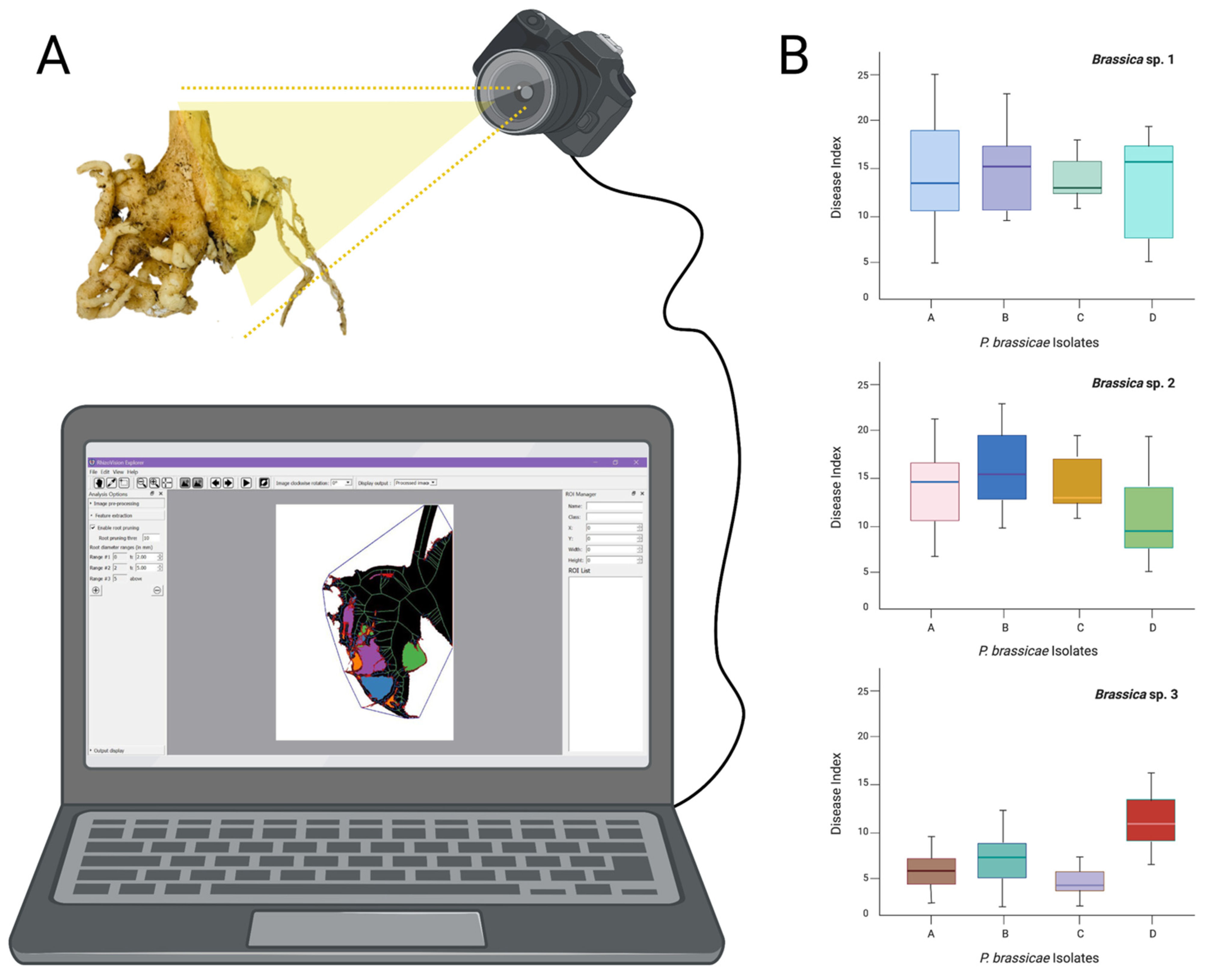
3. Moving to the Digital World—Which Are the Best Alternatives?
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Dixon, G.R. The biology of Plasmodiophora brassicae Wor.–A review of recent advances. Acta Hortic. 2006, 706, 271–282. [Google Scholar] [CrossRef]
- Dixon, G.R. The occurrence and economic impact of Plasmodiophora brassicae and clubroot disease. J. Plant Growth Regul. 2009, 28, 194–202. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.; Turnbull, G.D.; Fredua-Agyeman, R.; Hollman, K.; Kaus, S. Characterization of clubroot (Plasmodiophora brassicae) from canola (Brassica napus) in the Peace Country of Alberta, Canada. Can. J. Plant Pathol. 2020, 43, 155–161. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Howard, R.J.; Hartman, M.; Turkington, T.K. Progress towards the sustainable management of clubroot [Plasmodiophora brassicae] of canola in the Canadian prairies. Prairie Soils Crop. 2011, 4, 114–121. [Google Scholar] [CrossRef]
- Somé, A.; Manzanares, M.J.; Laurens, F.; Baron, F.; Thomas, G.; Rouxel, F. Variation for virulence on Brassica napus L. amongst Plasmodiophora brassicae collections from France and derived single-spore isolates. Plant Pathol. 1996, 45, 432–439. [Google Scholar] [CrossRef]
- Williams, P.H. A system for the determination of races of Plasmodiophora brassicae that infect cabbage and rutabaga. Phytopathology 1966, 56, 624–626. [Google Scholar]
- Buczacki, S.T.; Toxopeus, H.; Mattusch, P.; Johnston, T.D.; Dixon, G.R.; Hobolth, L.A. Study of physiologic specialization in Plasmodiophora brassicae: Proposals for attempted rationalization through an international approach. Trans. Br. Mycol. Soc. 1975, 65, 295–303. [Google Scholar] [CrossRef]
- Strelkov, S.E.; Hwang, S.F.; Manolii, V.P.; Cao, T.; Fredua-Agyeman, R.; Harding, M.W.; Peng, G.; Gossen, B.D.; Mcdonald, M.R.; Feindel, D. Virulence and pathotype classification of Plasmodiophora brassicae populations collected from clubroot resistant canola (Brassica napus) in Canada. Can. J. Plant Pathol. 2018, 40, 284–298. [Google Scholar] [CrossRef]
- Pang, W.; Liang, Y.; Zhan, Z.; Li, X.; Piao, Z. Development of a Sinitic Clubroot Differential Set for the Pathotype Classification of Plasmodiophora brassicae. Front. Plant Sci. 2020, 11, 1360. [Google Scholar] [CrossRef]
- Griffiths, M. A 3D Print Repository for Plant Phenomics. Plant Phenomics 2020. [Google Scholar] [CrossRef]
- Lobet, G.; Draye, X. Novel scanning procedure enabling the vectorization of entire rhizotron-grown root systems. Plant Methods 2013, 9, 1. [Google Scholar] [CrossRef] [Green Version]
- Himmelbauer, M.L. Estimating length, average diameter and surface area of roots using two different image analyses systems. Plant Soil 2004, 260, 111–120. [Google Scholar] [CrossRef]
- Mattupalli, C.; Seethepalli, A.; York, L.M.; Young, C.A. Digital imaging to evaluate root system architectural changes associated with soil biotic factors. Phytobiomes J. 2019, 3, 102–111. [Google Scholar] [CrossRef] [Green Version]
- Kimura, K.; Kikuchi, S.; Yamasaki, S. Accurate root length measurement by image analysis. Plant Soil 1999, 216, 117–127. [Google Scholar] [CrossRef]
- Lebreton, A.; Labbé, C.; De Ronne, M.; Xue, A.G.; Marchand, G.; Bélanger, R.R. Development of a simple hydroponic assay to study vertical and horizontal resistance of soybean and pathotypes of Phytophthora sojae. Plant Dis. 2018, 102, 114–123. [Google Scholar] [CrossRef] [Green Version]
- Wagner, R.E.; Wilkinson, H.T. An aeroponics system for investigating disease development on soybean taproots infected with Phytophthora sojae. Plant Dis. 1992, 76, 610–614. [Google Scholar] [CrossRef]
- Trachsel, S.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 2011, 341, 75–87. [Google Scholar] [CrossRef]
- Cai, G.; Vanderborght, J.; Klotzsche, A.; van der Kruk, J.; Neumann, J.; Hermes, J.N.; Vereecken, H. Construction of minirhizotron facilities for investigating root zone processes. Vadose Zone J. 2016, 15, vzj2016.05.0043. [Google Scholar] [CrossRef] [Green Version]
- Auer, S. A costum-made hydroponic culture system to study plant roots during root infection with Plasmodiophora brassicae. Protocols.io 2021. [Google Scholar] [CrossRef]
- Hund, A.; Trachsel, S.; Stamp, P. Growth of axile and lateral roots of maize: I development of a phenotying platform. Plant Soil 2009, 325, 335–349. [Google Scholar] [CrossRef]
- Botero, A.; García, C.; Gossen, B.D.; Strelkov, S.E.; Todd, C.D.; Bonham-Smith, P.C.; Pérez-López, E. Clubroot disease in Latin America: Distribution and management strategies. Plant Pathol. 2019, 68, 827–833. [Google Scholar] [CrossRef] [Green Version]
| Method | Pros | Cons | Reference |
|---|---|---|---|
| Root excavations and trenching | Natural conditions Restrictions to growth Complete lifespan of plants 3D growth environment Soil conditions | Destructive Time consuming | [16] |
| Shovelomics | Natural conditions Restrictions to growth Complete lifespan of plants 3D growth environment Soil conditions | Destructive Only a part of the root system is analysed | [17] |
| Field minirhizotrons | Soil conditions Natural atmospheric conditions Complete lifespan of plants | Only part of the root system is observable | [18] |
| Hydroponic system | Easy and direct access for the roots Uniform conditions Complete lifespan of plants 3D growth environment | No physical constraints to growth | [19] |
| Aeroponic growth | Easy to move the samples in the system Easy and direct access for the roots Uniform conditions | No physical constraints to growth Artificial soil environment | [16] |
| Growth on filter paper pouches | Easy to handle Clear difference between the filter paper and the roots | Contamination by fungi Artificial root environment No physical constraints to growth 2D growth Shorter cultivation time | [20] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salih, R.; Pérez-López, E. Digitalization of Clubroot Disease Index, a Long Overdue Task. Horticulturae 2021, 7, 241. https://doi.org/10.3390/horticulturae7080241
Salih R, Pérez-López E. Digitalization of Clubroot Disease Index, a Long Overdue Task. Horticulturae. 2021; 7(8):241. https://doi.org/10.3390/horticulturae7080241
Chicago/Turabian StyleSalih, Rasha, and Edel Pérez-López. 2021. "Digitalization of Clubroot Disease Index, a Long Overdue Task" Horticulturae 7, no. 8: 241. https://doi.org/10.3390/horticulturae7080241
APA StyleSalih, R., & Pérez-López, E. (2021). Digitalization of Clubroot Disease Index, a Long Overdue Task. Horticulturae, 7(8), 241. https://doi.org/10.3390/horticulturae7080241





