CRISPR-Cas9 Gene Editing for Fruit and Vegetable Crops: Strategies and Prospects
Abstract
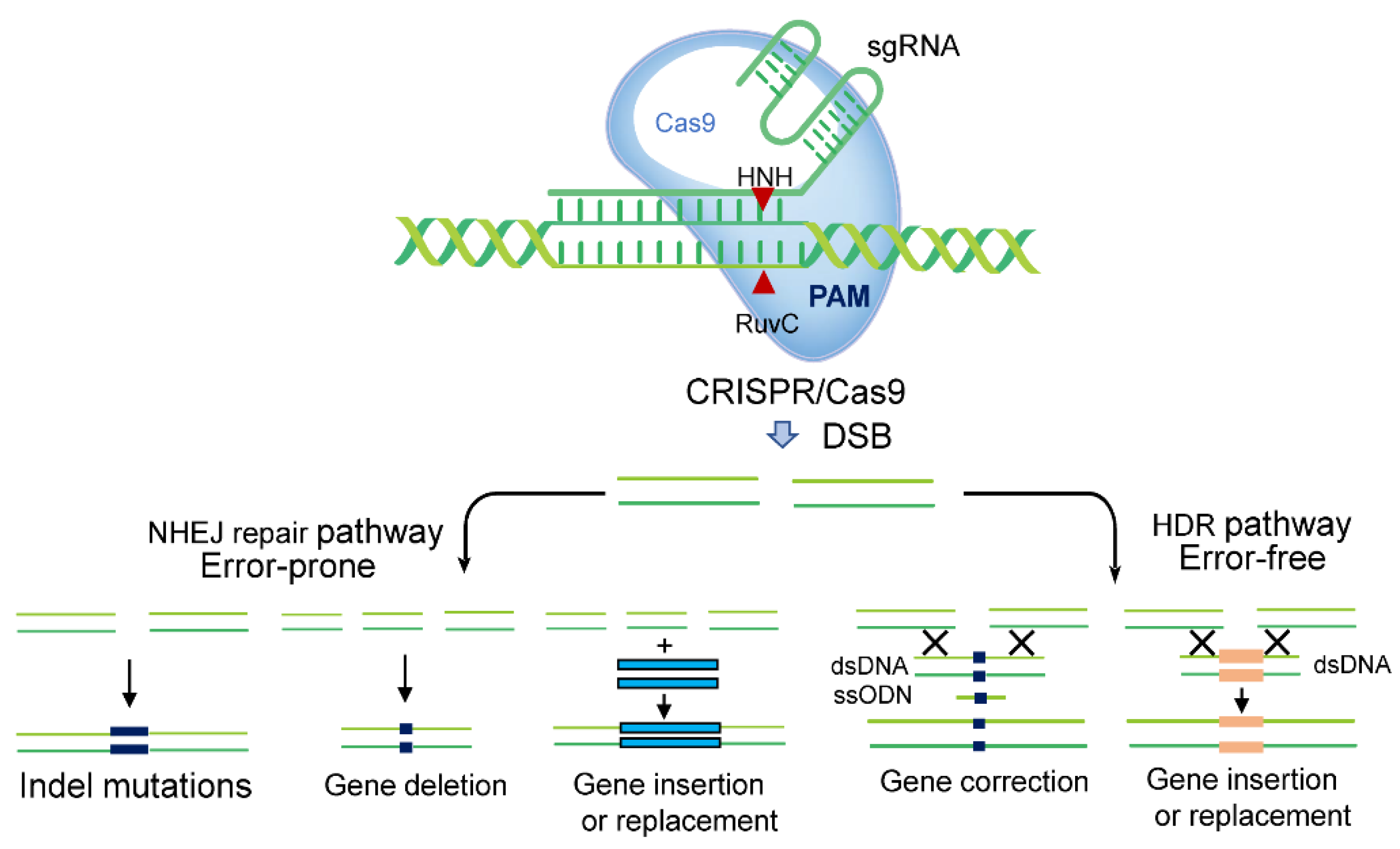
:1. Introduction
1.1. The Discovery and Development of CRISPR Technology
1.2. Development of the CRISPR-Cas System in Plant Studies
1.3. CRISPR-Cas9 in Fruit and Vegetable Crop Improvement
1.3.1. Improvement of Biotic Stress Resistance
1.3.2. Abiotic Stress Resistance Improvement
1.3.3. Herbicide Resistance Improvement
1.3.4. Fruit and Vegetable Quality Improvement
1.3.5. Application of CRISPR-Cas9 to Crop Domestication
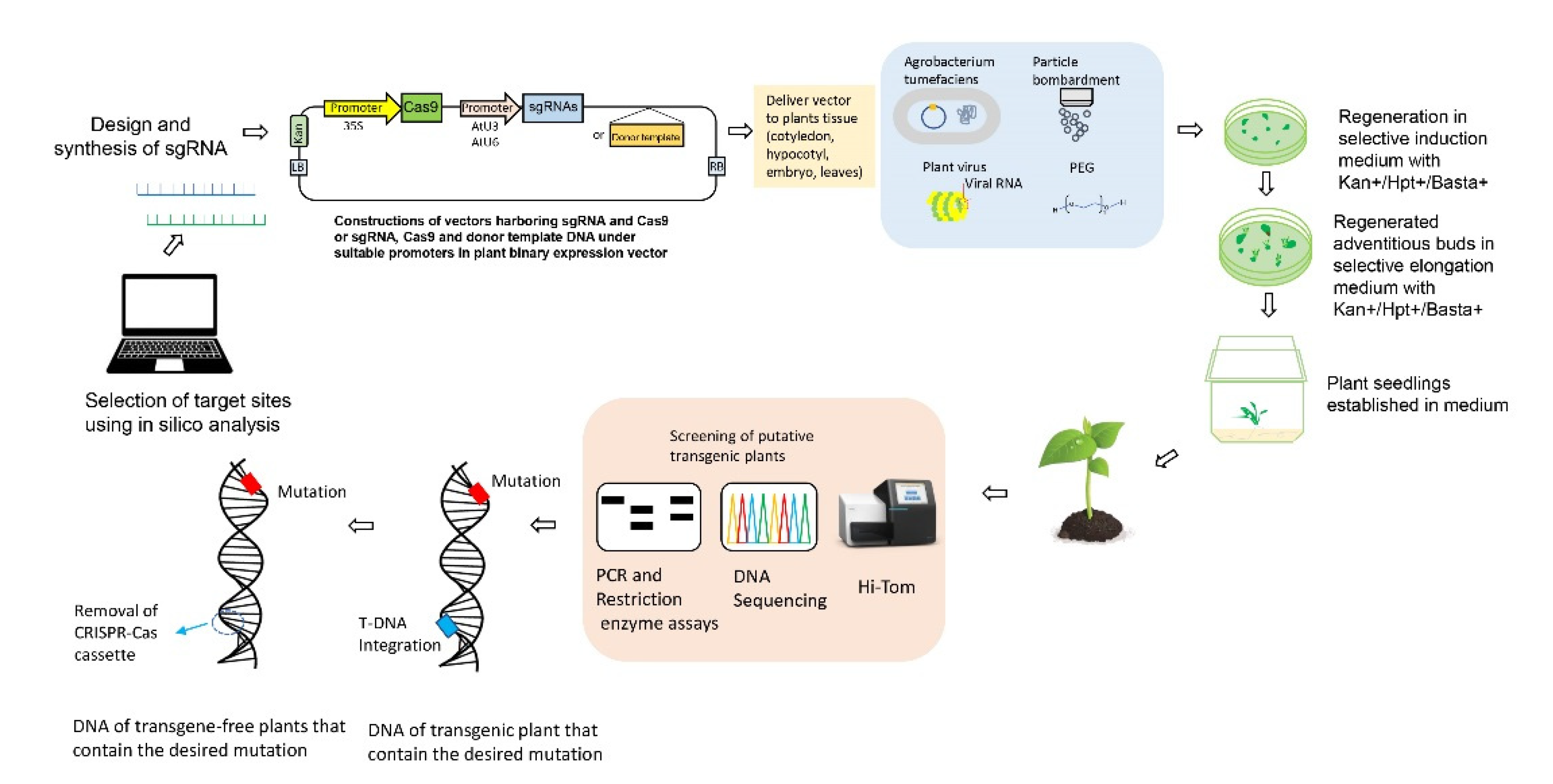
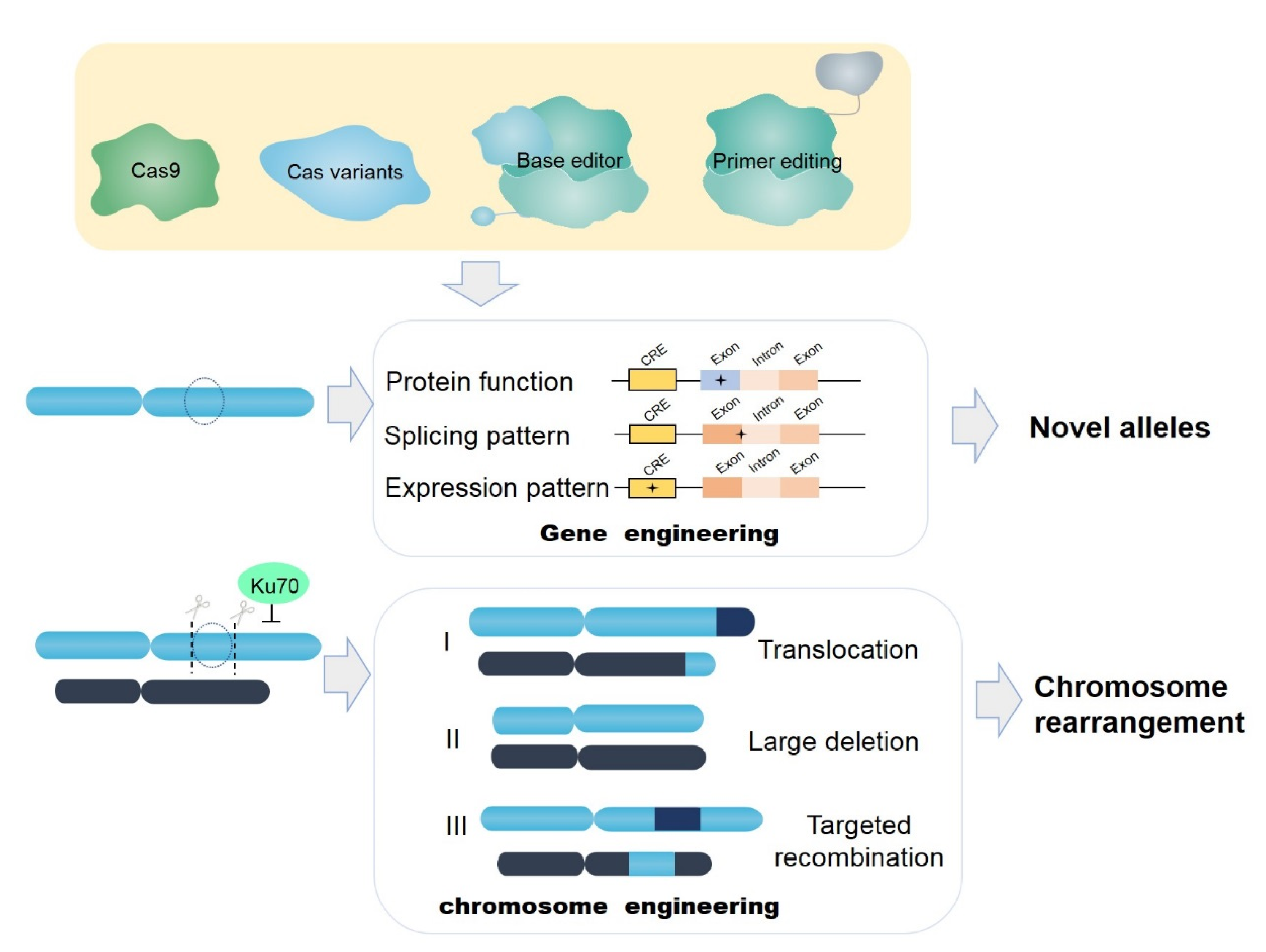
1.4. Improvements to CRISPR-Cas9 Gene-Editing Systems
1.4.1. Production of Non-GM Plants Using CRISPR-Cas9 Gene Editing
1.4.2. Novel Variants of Cas Protein and Applications
1.5. Regulatory Framework of CRISPR-Cas-Edited Crops
2. Future Challenges in the Application of CRISPR-Cas Gene Editing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The Epigenome and Transcriptional Dynamics of Fruit Ripening. Ann. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Karkute, S.G.; Singh, A.K.; Gupta, O.P.; Singh, P.M.; Singh, B. CRISPR/Cas9 Mediated Genome Engineering for Improvement of Horticultural Crops. Front. Plant Sci. 2017, 8, 1635. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.S.; Purugganan, M.D. Evolution of crop species: Genetics of domestication and diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Bigliardi, B.; Galati, F. Innovation trends in the food industry: The case of functional foods. Trends Food Sci. Technol. 2013, 31, 118–129. [Google Scholar] [CrossRef]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech. 2017, 7, 239. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.G.; Redenbaugh, K. Commercialization of a tomato with an antisense polygalacturonase gene: The FLAVR SAVR™ tomato story. Euphytica 1994, 79, 293–297. [Google Scholar] [CrossRef]
- Millstone, E.; Stirling, A.; Glover, D. Regulating Genetic Engineering: The Limits and Politics of Knowledge. Issues Sci. Technol. 2015, 31, 23–26. [Google Scholar]
- Bawa, A.S.; Anilakumar, K.R. Genetically modified foods: Safety, risks and public concerns-a review. J. Food Sci. Technol. 2013, 50, 1035–1046. [Google Scholar] [CrossRef] [Green Version]
- Waltz, E. Gene-edited CRISPR mushroom escapes US regulation. Nature 2016, 7599, 293. [Google Scholar] [CrossRef] [Green Version]
- Waltz, E. With a free pass, CRISPR-edited plants reach market in record time. Nat. Biotechnol. 2018, 36, 6–7. [Google Scholar] [CrossRef]
- Jansen, R.; Embden, J.D.V.; Gaastra, W.; Schouls, L.M. Identification of genes that are associated with DNA repeats in prokaryotes. Mol. Microbiol. 2002, 43, 1565–1575. [Google Scholar] [CrossRef] [PubMed]
- Bolotin, A. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology 2005, 151, 2551–2561. [Google Scholar] [CrossRef] [Green Version]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Pourcel, C. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology 2005, 151, 653–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaboli, S.; Babazada, H. CRISPR Mediated Genome Engineering and its Application in Industry. Curr. Issues Mol. Biol. 2018, 26, 81–92. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef]
- Rouillon, C.; Zhou, M.; Zhang, J.; Politis, A.; Beilsten-Edmands, V.; Cannone, G.; Graham, S.; Robinson, C.V.; Spagnolo, L.; White, M.F. Structure of the CRISPR interference complex CSM reveals key similarities with cascade. Mol. Cell 2013, 52, 124–134. [Google Scholar] [CrossRef] [Green Version]
- Gasiunas, G.; Barrangou, R.; Horvath, P.; Siksnys, V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc. Natl. Acad. Sci. USA 2012, 109, 15539–15540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Yin, K.; Gao, C.; Qiu, J.L. Progress and prospects in plant genome editing. Nat. Plants 2017, 3, 17107. [Google Scholar] [CrossRef] [PubMed]
- Baltes, N.J.; Gil-Humanes, J.; Cermak, T.; Atkins, P.A.; Voytas, D.F. DNA replicons for plant genome engineering. Plant cell 2014, 26, 151–163. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Ishikawa, Y.; Osakabe, K.; Nakayama, S.; Kaya, H.; Araki, T.; Shibahara, K.-I.; Abe, K.; Ichikawa, H.; Valentine, L.; et al. Increased frequency of homologous recombination and T-DNA integration in Arabidopsis CAF-1 mutants. EMBO J. 2006, 25, 5579–5590. [Google Scholar] [CrossRef] [PubMed]
- Gil-Humanes, J.; Wang, Y.; Liang, Z.; Shan, Q.; Ozuna, C.V.; Sánchez-León, S.; Baltes, N.J.; Starker, C.; Barro, F.; Gao, C.; et al. High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J. 2017, 89, 1251–1262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayathilaka, K.; Sheridan, S.D.; Bold, T.D.; Bochenska, K.; Logan, H.L.; Weichselbaum, R.R.; Bishop, D.K.; Connell, P.P. A chemical compound that stimulates the human homologous recombination protein RAD51. Proc. Natl. Acad. Sci. USA 2008, 105, 15848–15853. [Google Scholar] [CrossRef] [Green Version]
- Donohoue, P.D.; Barrangou, R.; May, A.P. Advances in Industrial Biotechnology Using CRISPR-Cas Systems. Trends Biotechnol. 2018, 36, 134–146. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2019, 97, 795. [Google Scholar] [CrossRef] [Green Version]
- Liang, W.; van Wersch, S.; Tong, M.; Li, X. TIR-NB-LRR immune receptor SOC3 pairs with truncated TIR-NB protein CHS1 or TN2 to monitor the homeostasis of E3 ligase SAUL1. New Phytol. 2019, 221, 2054–2066. [Google Scholar] [CrossRef]
- Mao, Y.; Yang, X.; Zhou, Y.; Zhang, Z.; Botella, J.R.; Zhu, J.-K. Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol. 2018, 19, 149. [Google Scholar] [CrossRef]
- Hess, G.T.; Tycko, J.; Yao, D.; Bassik, M.C. Methods and Applications of CRISPR-Mediated Base Editing in Eukaryotic Genomes. Mol. Cell 2017, 68, 26–43. [Google Scholar] [CrossRef]
- Niu, Q.; Wu, S.; Li, Y.; Yang, X.; Liu, P.; Xu, Y.; Lang, Z. Expanding the scope of CRISPR/Cas9-mediated genome editing in plants using an xCas9 and Cas9-NG hybrid. J. Integr. Plant Biol. 2020, 62, 398–402. [Google Scholar] [CrossRef] [Green Version]
- Brooks, C.; Nekrasov, V.; Lippman, Z.B.; Van Eck, J. Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 2014, 166, 1292–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulus, D. Genetic resources and selected conservation methods of tomato. J. Appl. Bot. Food Qual. 2018, 91, 135–144. [Google Scholar]
- Ma, C.; Liu, M.; Li, Q.; Si, J.; Ren, X.; Song, H. Efficient BoPDS Gene Editing in Cabbage by the CRISPR/Cas9 System. Hortic. Plant J. 2019, 5, 164–169. [Google Scholar] [CrossRef]
- Sun, B.; Zheng, A.; Jiang, M.; Xue, S.; Yuan, Q.; Jiang, L.; Chen, Q.; Li, M.; Wang, Y.; Zhang, Y.; et al. CRISPR/Cas9-mediated mutagenesis of homologous genes in Chinese kale. Sci. Rep. 2018, 8, 16786. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Gao, Q.; Zhang, J.; Zong, M.; Zhang, H.; Ren, Y.; Guo, S.; Gong, G.; Liu, F.; et al. Efficient CRISPR/Cas9-based gene knockout in watermelon. Plant Cell Rep. 2017, 36, 399–406. [Google Scholar] [CrossRef]
- Bastet, A.; Zafirov, D.; Giovinazzo, N.; Guyon-Debast, A.; Nogué, F.; Robaglia, C.; Gallois, J.L. Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 2019, 17, 1736–1750. [Google Scholar] [CrossRef] [Green Version]
- Chandrasekaran, J.; Brumin, M.; Wolf, D.; Leibman, D.; Klap, C.; Pearlsman, M.; Sherman, A.; Arazi, T.; Gal-On, A. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 2016, 17, 1140–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, X.; Zhang, H.; Zhang, Y.; Wang, Y.; Gao, C. Establishing a CRISPR–Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 2015, 1, 15144. [Google Scholar] [CrossRef] [PubMed]
- Ali, Z.; Ali, S.; Tashkandi, M.; Zaidi, S.S.E.A.; Mahfouz, M.M. CRISPR/Cas9-Mediated Immunity to Geminiviruses: Differential Interference and Evasion. Sci. Rep. 2016, 6, 26912. [Google Scholar] [CrossRef] [Green Version]
- Borrelli, V.M.G.; Brambilla, V.; Rogowsky, P.; Marocco, A.; Lanubile, A. The Enhancement of Plant Disease Resistance Using CRISPR/Cas9 Technology. Frontiers in Plant Science. 2018, 9, 1245. [Google Scholar] [CrossRef] [PubMed]
- Zeilmaker, T.; Ludwig, N.R.; Elberse, J.; Seidl, M.F.; Berke, L.; Van Doorn, A.; Schuurink, R.C.; Snel, B.; Van den Ackerveken, G. DOWNY MILDEW RESISTANT 6 and DMR6-LIKE OXYGENASE 1 are partially redundant but distinct suppressors of immunity in Arabidopsis. Plant J. 2015, 81, 210–222. [Google Scholar] [CrossRef] [PubMed]
- Paula de Toledo Thomazella, D.; Brail, Q.; Dahlbeck, D.; Staskawicz, B. CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. BioRxiv 2016, 064824. [Google Scholar] [CrossRef] [Green Version]
- Nekrasov, V.; Wang, C.; Win, J.; Lanz, C.; Weigel, D.; Kamoun, S. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 2017, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, R.; Hs, A. Resistance-Gene-Mediated Defense Responses against Biotic Stresses in the Crop Model Plant Tomato. J. Plant Pathol. Microbiol. 2017, 8, 2. [Google Scholar]
- Cahya, P.; Barbetti, M.J.; Barker, S.J. A Novel Tomato Fusarium Wilt Tolerance Gene. Front. Microbiol. 2018, 9, 1226. [Google Scholar]
- Zhang, M.; Liu, Q.; Yang, X.; Xu, J.; Liu, G.; Yao, X.; Ren, R.; Xu, J.; Lou, L. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 2020, 39, 589–595. [Google Scholar] [CrossRef] [PubMed]
- Shujuan, Z.; Liu, W.; Ruirui, Z.; Wenqing, Y.; Rui, L.; Yujing, L.; Jiping, S.; Lin, S. Knockout of SlMAPK3 Reduced Disease Resistance to Botrytis cinerea in Tomato Plants. J. Agric. Food Chem. 2018, 66, 8949–8956. [Google Scholar]
- Andrés, O.; Selena, G.I.; Nathalie, L.; Roberto, S. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol. J. 2018, 17, 665–673. [Google Scholar]
- Haque, E.; Taniguchi, H.; Hassan, M.M.; Bhowmik, P.; Karim, M.R.; Śmiech, M.; Zhao, K.; Rahman, M.; Islam, T. Application of CRISPR/Cas9 Genome Editing Technology for the Improvement of Crops Cultivated in Tropical Climates: Recent Progress, Prospects, and Challenges. Front. Plant Sci. 2018, 9, 617. [Google Scholar] [CrossRef]
- Yin, Y.; Qin, K.; Song, X.; Zhang, Q.; Zhou, Y.; Xia, X.; Yu, J. BZR1 Transcription Factor Regulates Heat Stress Tolerance Through FERONIA Receptor-Like Kinase-Mediated Reactive Oxygen Species Signaling in Tomato. Plant Cell Physiol. 2018, 59, 2239–2254. [Google Scholar] [CrossRef]
- Li, R.; Zhang, L.; Wang, L.; Chen, L.; Zhao, R.; Sheng, J.; Shen, L. Reduction of Tomato-Plant Chilling Tolerance by CRISPR-Cas9-Mediated SlCBF1 Mutagenesis. J. Agric. Food Chem. 2018, 66, 9042–9051. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, L.; Li, R.; Zhao, R.; Yang, M.; Sheng, J.; Shen, L. Reduced drought tolerance by CRISPR/Cas9-mediated SlMAPK3 mutagenesis in tomato plants. J. Agric. Food Chem. 2017, 65, 8674–8682. [Google Scholar] [CrossRef] [PubMed]
- Tian, S.; Jiang, L.; Cui, X.; Zhang, J.; Guo, S.; Li, M.; Zhang, H.; Ren, Y.; Gong, G.; Zong, M. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018, 37, 1353–1356. [Google Scholar] [CrossRef]
- Butler, N.M.; Atkins, P.A.; Voytas, D.F.; Douches, D.S. Generation and Inheritance of Targeted Mutations in Potato (Solanum tuberosum L.) Using the CRISPR/Cas System. PLoS ONE 2015, 10, e0144591. [Google Scholar] [CrossRef] [Green Version]
- Bari, V.K.; Nassar, J.A.; Aly, R. CRISPR/Cas9 mediated mutagenesis of MORE AXILLARY GROWTH 1 in tomato confers resistance to root parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2021, 11, 3905. [Google Scholar] [CrossRef] [PubMed]
- Bari, V.K.; Nassar, J.A.; Kheredin, S.M.; Gal-On, A.; Ron, M.; Britt, A.; Steele, D.; Yoder, J.; Aly, R. CRISPR/Cas9-mediated mutagenesis of CAROTENOID CLEAVAGE DIOXYGENASE 8 in tomato provides resistance against the parasitic weed Phelipanche aegyptiaca. Sci. Rep. 2019, 9, 11438. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Qi, M.; Sun, M.; Liu, Y.; Liu, Y.; Xu, T.; Li, Y.; Li, T. Tomato Transcription Factor SlWUS Plays an Important Role in Tomato Flower and Locule Development. Front. Plant Sci. 2017, 8, 457. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Yang, X.; Yu, Y.; Si, X.; Zhai, X.; Zhang, H.; Dong, W.; Gao, C.; Xu, C. Domestication of wild tomato is accelerated by genome editing. Nat. Biotechnol. 2018, 36, 1160–1163. [Google Scholar] [CrossRef]
- Rodríguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Molthoff, J.; de Vos, R.; Hekkert, B.T.L.; Orzaez, D.; Fernaݩndez-Moreno, J.P.; Tripodi, P.; Gran-dillo, S.; Martin, C.; Heldens, J.; et al. Biochemical and Molecular Analysis of Pink Tomatoes: Deregulated Expression of the Gene Encoding Transcription Factors SlMYB12 Leads to Pink Tomato Fruit Color. Plant Physiol. 2010, 152, 71–84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Čermák, T.; Baltes, N.J.; Čegan, R.; Zhang, Y.; Voytas, D.F. High-frequency, precise modification of the tomato genome. Genome Biol. 2015, 16, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, L.; Wang, H.; Sun, C.; Li, Q.; Jiang, H.; Du, M.; Li, C.B.; Li, C. Efficient generation of pink-fruited tomatoes using CRISPR/Cas9 system. J. Genet. Genom. 2018, 45, 51–54. [Google Scholar] [CrossRef]
- Filler Hayut, S.; Melamed Bessudo, C.; Levy, A.A. Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat. Commun. 2017, 8, 15605. [Google Scholar] [CrossRef]
- Sun, B.; Jiang, M.; Zheng, H.; Jian, Y.; Huang, W.L.; Yuan, Q.; Zheng, A.H.; Chen, Q.; Zhang, Y.T.; Lin, Y.X.; et al. Color-related chlorophyll and carotenoid concentrations of Chinese kale can be altered through CRISPR/Cas9 targeted editing of the carotenoid isomerase gene BoaCRTISO. Hortic. Res. 2020, 7, 161. [Google Scholar] [CrossRef]
- Chen, L.; Yang, D.; Zhang, Y. Evidence for a specific and critical role of mitogen-activated protein kinase 20 in uni-to-binucleate transition of microgametogenesis in tomato. Plant Biol. 2018, 219, 176–194. [Google Scholar] [CrossRef] [Green Version]
- Kris-Etherton, P.M.; Hecker, K.D.; Bonanome, A.; Coval, S.M.; Binkoski, A.E.; Hilpert, K.F.; Griel, A.E.; Etherton, T.D. Bioactive compounds in foods: Their role in the prevention of cardiovascular disease and cancer. Am. J. Med. 2002, 113, 71–88. [Google Scholar] [CrossRef]
- Meng, X.; Yang, D.; Li, X.; Zhao, S.; Sui, N.; Meng, Q. Physiological changes in fruit ripening caused by overexpression of tomato SlAN2, an R2R3-MYB factor. Plant Physiol. Biochem. 2015, 89, 24–30. [Google Scholar] [CrossRef]
- Ye, J.; Wang, X.; Hu, T.; Zhang, F.; Wang, B.; Li, C.; Yang, T.; Li, H.; Lu, Y. An InDel in the Promoter of Al-ACTIVATED MALATE TRANSPORTER9 Selected during Tomato Domestication Determines Fruit Malate Contents and Aluminum Tolerance. Plant Cell 2017, 29, 2249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nonaka, S.; Arai, C.; Takayama, M.; Matsukura, C.; Ezura, H. Efficient increase of γ-aminobutyric acid (GABA) content in tomato fruits by targeted mutagenesis. Sci. Rep. 2017, 7, 7057. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Wang, Y.; Chen, S.; Tian, H.; Fu, D.; Zhu, B.; Luo, Y.; Zhu, H. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front. Plant Sci. 2018, 9, 559. [Google Scholar] [CrossRef] [PubMed]
- Nakayasu, M.; Akiyama, R.; Lee, H.J.; Osakabe, K.; Osakabe, Y.; Watanabe, B.; Sugimoto, Y.; Umemoto, N.; Saito, K.; Muranaka, T.; et al. Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol. Biochem. 2018, 131, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Toki, S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 2015, 467, 76–82. [Google Scholar] [CrossRef]
- Lang, Z.; Wang, Y.; Tang, K.; Tang, D.; Datsenka, T.; Cheng, J.; Zhang, Y.; Handa, A.K.; Zhu, J.K. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc. Natl. Acad. Sci. USA 2017, 114, E4511–E4519. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Samsulrizal, A.; Yan, B.C.; Allcock, C.; Craigon, D.J. Characterization of CRISPR Mutants Targeting Genes Modulating Pectin Degradation in Ripening Tomato. Plant Physiol. 2019, 179, 544–557. [Google Scholar]
- Yu, Q.H.; Wang, B.; Li, N.; Tang, Y.; Yang, S.; Yang, T.; Xu, J.; Guo, C.; Yan, P.; Wang, Q.; et al. CRISPR/Cas9-induced Targeted Mutagenesis and Gene Replacement to Generate Long-shelf Life Tomato Lines. Sci. Rep. 2017, 7, 11874. [Google Scholar] [CrossRef]
- Ledford, H. Fixing the tomato: CRISPR edits correct plant-breeding snafu. Nature 2017, 545, 394–395. [Google Scholar] [CrossRef] [Green Version]
- Soyk, S.; Lemmon, Z.H.; Oved, M.; Fisher, J.; Liberatore, K.L.; Park, S.J.; Goren, A.; Jiang, K.; Ramos, A.; van der Knaap, E.; et al. Bypassing Negative Epistasis on Yield in Tomato Imposed by a Domestication Gene. Cell 2017, 169, 1142–1155. [Google Scholar] [CrossRef] [Green Version]
- Roldan, M.V.G.; Périlleux, C.; Morin, H.; Huerga-Fernandez, S.; Latrasse, D.; Benhamed, M.; Bendahmane, A. Natural and induced loss of function mutations in SlMBP21 MADS-box gene led to jointless-2 phenotype in tomato. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Klap, C.; Yeshayahou, E.; Bolger, A.M.; Arazi, T.; Gupta, S.K.; Shabtai, S.; Usadel, B.R.; Salts, Y.; Barg, R. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 2016, 15, 634–647. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Alon, I.; Naomi, O.; Sun, T.P. DELLA-ARF/IAA Interaction Mediates Crosstalk between Gibberellin and Auxin Signaling in Controlling Fruit Initiation in Solanum lycopersicum. Plant Cell 2018, 30, 1710–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueta, R.; Abe, C.; Watanabe, T.; Sugano, S.S.; Ishihara, R.; Ezura, H.; Osakabe, Y.; Osakabe, K. Rapid breeding of parthenocarpic tomato plants using CRISPR/Cas9. Sci. Rep. 2017, 7, 507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef] [Green Version]
- Zsögön, A.; Čermák, T.; Naves, E.R.; Notini, M.M.; Edel, K.H.; Weinl, S.; Freschi, L.; Voytas, D.F.; Kudla, J.; Peres, L.E.P. De novo domestication of wild tomato using genome editing. Nat. Biotechnol. 2018, 36, 1211–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Tavano, E.C.d.R.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; de Maagd, R.A. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 2019, 9, 1696. [Google Scholar] [CrossRef] [PubMed]
- Soyk, S.; Müller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jiménez-Gómez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Zsögön, A.; Cermak, T.; Voytas, D.; Peres, L.E. Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: Case study in tomato. Plant Sci. 2017, 256, 120–130. [Google Scholar] [CrossRef] [PubMed]
- Bisognin, D.A. Breeding vegetatively propagated horticultural crops. Crop Breed. Appl. Biotechnol. 2011, 11, 35–43. [Google Scholar] [CrossRef] [Green Version]
- Corte, E.D.; MMahmoud, L.; SMoraes, T.; Mou, Z.; WGrosser, J.; Dutt, M. Development of Improved Fruit, Vegetable, and Ornamental Crops Using the CRISPR/Cas9 Genome Editing Technique. Plants 2019, 8, 601. [Google Scholar] [CrossRef] [Green Version]
- Charrier, A.; Vergne, E.; Dousset, N.; Richer, A.; Petiteau, A.; Chevreau, E. Efficient Targeted Mutagenesis in Apple and First Time Edition of Pear Using the CRISPR-Cas9 System. Front. Plant Sci. 2019, 10, 40. [Google Scholar] [CrossRef] [Green Version]
- Malnoy, M.; Viola, R.; Jung, M.H.; Koo, O.J.; Kim, S.; Kim, J.S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.G.; Kim, S.T.; Choe, S.; Kim, J.S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef]
- Pompili, V.; Dalla Costa, L.; Piazza, S.; Pindo, M.; Malnoy, M. Reduced fire blight susceptibility in apple cultivars using a high-efficiency CRISPR/Cas9-FLP/FRT-based gene editing system. Plant Biotechnol. J. 2020, 18, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Dalla Costa, L.; Piazza, S.; Campa, M.; Flachowsky, H.; Hanke, M.-V.; Malnoy, M. Efficient heat-shock removal of the selectable marker gene in genetically modified grapevine. Plant Cell Tissue Organ Cult. 2016, 124, 471–481. [Google Scholar] [CrossRef]
- Herzog, K.; Flachowsky, H.; Deising, H.B.; Hanke, M.V. Heat-shock-mediated elimination of the nptII marker gene in transgenic apple (Malus×domestica Borkh.). Gene 2012, 498, 41–49. [Google Scholar] [CrossRef]
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020, 10, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Akihiro, Y.; Takashi, I.; Mika, Y.; Yuri, K.; Shinichiro, S. Developing Heritable Mutations in Arabidopsis thaliana Using a Modified CRISPR/Cas9 Toolkit Comprising PAM-Altered Cas9 Variants and gRNAs. Plant Cell Physiol. 2019, 60, 2255–2262. [Google Scholar]
- Hu, X.; Meng, X.; Liu, Q.; Li, J.; Wang, K. Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice. Plant Biotechnol. J. 2018, 16, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, C.; Fu, Y.; Liu, Q.; Jiao, X.; Wang, K. Expanding the Range of CRISPR/Cas9 Genome Editing in Rice. Mol. Plant 2016, 9, 943–945. [Google Scholar] [CrossRef] [Green Version]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.W.; Li, Z.; Peterson, R.T.; Yeh, J.-R.J.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.K. Genome Engineering in Rice Using Cas9 Variants that Recognize NG PAM Sequences. Mol. Plant 2019, 12, 1003–1014. [Google Scholar] [CrossRef]
- Endo, M.; Mikami, M.; Endo, A.; Kaya, H.; Itoh, T.; Nishimasu, H.; Nureki, O.; Toki, S. Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat. Plants 2019, 5, 14–17. [Google Scholar] [CrossRef]
- Zhong, Z.; Sretenovic, S.; Ren, Q.; Yang, L.; Bao, Y.; Qi, C.; Yuan, M.; He, Y.; Liu, S.; Liu, X.; et al. Improving Plant Genome Editing with High-Fidelity xCas9 and Non-canonical PAM-Targeting Cas9-NG. Mol. Plant 2019, 12, 1027–1036. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Pattanayak, V.; Prew, M.S.; Tsai, S.Q.; Nguyen, N.T.; Zheng, Z.; Joung, J.K. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 2016, 529, 490–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slaymaker, I.M.; Gao, L.; Zetsche, B.; Scott, D.A.; Yan, W.X.; Zhang, F. Rationally engineered Cas9 nucleases with improved specificity. Science 2016, 351, 84–88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Zhang, H.; Li, T.; Chen, K.; Qiu, J.L.; Gao, C. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017, 18, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Casini, A.; Olivieri, M.; Petris, G.; Bianchi, A.; Montagna, C.; Reginato, G.; Maule, G.; Lorenzin, F.; Prandi, D.; Romanel, A.; et al. Evocas9, a highly specific SpCas9 variant from a yeast in vivo screening. Nat. Biotechnol. 2018, 36, 265–271. [Google Scholar] [CrossRef]
- Lee, J.K.; Jeong, E.; Lee, J.; Jung, M.; Shin, E.; Kim, Y.-H.; Lee, K.; Jung, I.; Kim, D.; Kim, S.; et al. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat. Commun. 2018, 9, 3048. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Meng, X.; Hu, X.; Sun, T.; Li, J.; Wang, K.; Yu, H. xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol. J. 2019, 17, 709–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Xu, W.; Wang, F.; Zhao, S.; Yang, J. Increasing Cytosine Base Editing Scope and Efficiency With Engineered Cas9-PmCDA1 Fusions and the Modified sgRNA in RiceData_Sheet_1.docx. Front. Genet. 2019, 10, 379. [Google Scholar] [CrossRef] [Green Version]
- Hou, Z.; Zhang, Y.; Propson, N.E.; Howden, S.E.; Chu, L.F.; Sontheimer, E.J.; Thomson, J.A. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc. Natl. Acad. Sci. USA 2013, 110, 15644–15649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Müller, M.; Lee, C.M.; Gasiunas, G.; Davis, T.H.; Cradick, T.J.; Siksnys, V.; Bao, G.; Cathomen, T.; Mussolino, C. Streptococcus thermophilus CRISPR-Cas9 Systems Enable Specific Editing of the Human Genome. Mol. Ther. 2016, 24, 636–644. [Google Scholar] [CrossRef] [Green Version]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F. Structure and Engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [Green Version]
- Steinert, J.; Schiml, S.; Fauser, F.; Puchta, H. Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J. 2015, 84, 1295–1305. [Google Scholar] [CrossRef]
- Abudayyeh, O.O.; Gootenberg, J.S.; Konermann, S.; Joung, J.; Slaymaker, I.M.; Cox, D.B.; Shmakov, S.; Makarova, K.S.; Semenova, E.; Minakhin, L.; et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 2016, 353, aaf5573. [Google Scholar] [CrossRef] [Green Version]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Harrington, L.B.; Burstein, D.; Chen, J.S.; Paez-Espino, D.; Ma, E.; Witte, I.P.; Cofsky, J.C.; Kyrpides, N.C.; Banfield, J.F.; Doudna, J.A. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science 2018, 362, 839–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, W.X.; Hunnewell, P.; Alfonse, L.E.; Carte, J.M.; Keston-Smith, E.; Sothiselvam, S.; Garrity, A.J.; Chong, S.; Makarova, K.S.; Koonin, E.V.; et al. Functionally diverse type V CRISPR-Cas systems. Science 2019, 363, 88. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Kim, J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef] [PubMed]
- Globus, R.; Qimron, U. A technological and regulatory outlook on CRISPR crop editing. J. Cell Biochem. 2018, 119, 1291–1298. [Google Scholar] [CrossRef]
- USDA Re: Confirmation of Regulatory Status of Waxy Com Developed by CRISPR-Cas Technology. Available online: https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/15-352-01_air_response_signed.pdf (accessed on 2 October 2020).
- USDA Re: Request for Confirmation that Transgene-Free, CRISPR-Edited Mushroom Is Not a Regulated Article. Available online: https://www.aphis.usda.gov/biotechnology/downloads/reg_loi/15-321-01_air_response_signed.pdf (accessed on 2 October 2020).
- Callaway, E. CRISPR plants now subject to tough GM laws in European Union. Nature 2018, 560, 16. [Google Scholar] [CrossRef]
- Mallapaty, S. Australian gene-editing rules adopt ‘middle ground’. Nature 2019. [Google Scholar] [CrossRef]
- Metje-Sprink, J.; Sprink, T.; Hartung, F. Genome-edited plants in the field. Curr. Opin. Biotechnol. 2020, 61, 1–6. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Li, Q.; Sapkota, M.; van der Knaap, E. Perspectives of CRISPR/Cas-mediated cis-engineering in horticulture: Unlocking the neglected potential for crop improvement. Hortic. Res. 2020, 7, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz, D.J.; Ali, Z.; Wang, C.; Aljedaani, F.; Hooykaas, P.J.J.; Mahfouz, M.; de Pater, S. CRISPR/Cas9 Mutagenesis by Translocation of Cas9 Protein Into Plant Cells via the Agrobacterium Type IV Secretion System. Front. Genome Ed. 2020, 2, 6. [Google Scholar] [CrossRef]
- Schmidt, C.; Pacher, M.; Puchta, H. Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J. 2019, 98, 577–589. [Google Scholar] [CrossRef] [Green Version]
- Beying, N.; Schmidt, C.; Pacher, M.; Houben, A.; Puchta, H. CRISPR–Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat. Plants 2020, 6, 638–645. [Google Scholar] [CrossRef]
- Chuang, Y.F.; Phipps, A.J.; Lin, F.L.; Hecht, V.; Hewitt, A.W.; Wang, P.Y.; Liu, G.S. Approach for in vivo delivery of CRISPR/Cas system: A recent update and future prospect. Cell Mol. Life Sci. 2021, 78, 2683–2708. [Google Scholar] [CrossRef]
- Lowe, K.; Wu, E.; Wang, N.; Hoerster, G.; Hastings, C.; Cho, M.J.; Scelonge, C.; Lenderts, B.; Chamberlin, M.; Cushatt, J.; et al. Morphogenic Regulators Baby boom and Wuschel Improve Monocot Transformation. Plant Cell 2016, 28, 1998–2015. [Google Scholar] [CrossRef] [Green Version]
- Ali, Z.; Abul-faraj, A.; Li, L.; Ghosh, N.; Piatek, M.; Mahjoub, A.; Aouida, M.; Piatek, A.; Baltes, N.J.; Voytas, D.F.; et al. Efficient Virus-Mediated Genome Editing in Plants Using the CRISPR/Cas9 System. Mol. Plant 2015, 8, 1288–1291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Crop Species | Target | Mutation | Transformation Method | Trait Modification | References |
|---|---|---|---|---|---|
| Tomato | SlAGO7 | Loss of function | Agrobacterium-mediated transformation | Wiry phenotype | [33] |
| Cabbage | BoPDS | Loss of function | Agrobacterium-mediated hypocotyl transformation | Albino phenotype | [35] |
| Watermelon | ClPDS | Loss of function | Agrobacterium-mediated cotyledon transformation | Albino phenotype | [36] |
| Chinese kale | BaPDS1BaPDS2 | Loss of function | Agrobacterium-mediated cotyledon transformation | Albino phenotype | [37] |
| Cucumber | elF4E | Loss of function | Agrobacterium-mediated cotyledon transformation | Resistance against cucumber vein yellowing virus, zucchini yellow mosaic virus, and papaya ring spot mosaic virus | [38] |
| Tomato | ClDMR6 | Loss of function | Agrobacterium-mediated cotyledon transformation | Resistance against downy mildew | [44] |
| Tomato | ClMlo1 | Loss of function | Agrobacterium-mediated cotyledon transformation | Resistance against downy mildew | [45] |
| Tomato | Solyc08g075770 | Loss of function | Agrobacterium-mediated cotyledon transformation | Fusarium wilt susceptibility | [47] |
| Watermelon | ClPSK1 | Loss of function | Agrobacterium-mediated cotyledon transformation | Resistance to Fusarium oxysporum f.sp. niveum | [48] |
| Tomato | MAPK3 | Loss of function | Agrobacterium-mediated cotyledon transformation | Resistance to Botrytis cinerea | [49] |
| Tomato | BZR1 | Loss of function | Agrobacterium-mediated cotyledon transformation | Decrease in heat stress tolerance | [52] |
| Tomato | CBF1 | Loss of function | Agrobacterium-mediated cotyledon transformation | Decrease in chilling stress tolerance | [53] |
| Tomato | SlMAPK3 | Loss of function | Agrobacterium-mediated cotyledon transformation | Decrease in drought stress tolerance | [54] |
| Watermelon | ClALS | Site-directed mutagenesis | Agrobacterium-mediated cotyledon transformation | Herbicide resistance | [55] |
| Tomato and Potato | StALS2 | Site-directed mutagenesis | Agrobacterium-mediated transformation | Herbicide resistance | [56] |
| Tomato | Carotenoid cleavage dioxygenase8 (CCD8) | Loss of function | Agrobacterium-mediated transformation | Resistance against Phelipanche aegytiaca | [57] |
| Tomato | More Axillary Growth1 (MAX1) | Loss of function | Agrobacterium-mediated transformation | Resistance against Phelipanche aegytiaca | [58] |
| Tomato | SP,SP5G,CLV3, WUS, GGP1 | Cis-regulatory variation and loss of function | Agrobacterium-mediated transformation | Introduction of desirable traits with morphology, flower number, fruit size and number, and ascorbic acid synthesis | [60] |
| Tomato | SlWUS CarG element, SlCLV3 promoter | Cis-regulatory variation | Agrobacterium-mediated transformation | Fruit size, inflorescence branching, and plant architecture | [61] |
| Tomato | PSY1 | Different mutations in alleles | Agrobacterium-mediated cotyledon transformation | Yellow-coloured tomato | [64] |
| Tomato | MYB12 | Different mutations in alleles | Agrobacterium-mediated cotyledon transformation | Pink-coloured tomato | [65] |
| Tomato | ANT2 | Gene insertion | Agrobacterium-mediated cotyledon transformation | Purple-coloured tomato | [66] |
| Chinese kale | BoaCRTISO | Gene insertion and replacement | Agrobacterium-mediated transformation | Colour of mutants changed from green to yellow | [67] |
| Tomato | MPK20 | Loss of function | Agrobacterium-mediated transformation | Repression of genes controlling sugar and auxin metabolism | [68] |
| Tomato | GAD2,GAD3,SlyGABA-TP1,SlyGABA-TP2,SlyGABA-TP3,SlyCAT9,SlySSADH | Autoinhibitory domain deletion | Agrobacterium-mediated transformation | Increase in γ-aminobutyric acid (GABA) content | [72] |
| Tomato | SGR1,lycopene ε-cyclase (LCY-E), beta-lycopene cyclase(Blc), lycopene β-cyclase1(LCY-B1) and LCY-B2 | Loss of function | Agrobacterium-mediated apical segments of hypocotyls transformation | Lycopene content | [73] |
| Potato | 16α-hydroxylation (St16DOX) | Loss of function | Agrobacterium-mediated shoot transformation | Steroidal glycoalkaloid (SGA) biosynthesis | [74] |
| Tomato | Ripening inhibitor (RIN) | Single base insertion or deletion of more than three bases | Agrobacterium-mediated transformation | MADS-box transcription factor regulating fruit ripening | [75] |
| Tomato | DNA demethylases (SlDML2) | Loss of function | Agrobacterium-mediated cotyledon transformation | Activation and inhibition of fruit ripening | [76] |
| Tomato | Enzymes pectate lyase (PL), Polygalacturonase 2a (PG2a), and β-galactanase (TBG4) | Generation of a range of CRISPR alleles | Agrobacterium-mediated transformation | Pectin degradation control | [77] |
| Tomato | Alcobaca (SLALC) | Loss of function | Agrobacterium-mediated hypocotyls transformation | Long shelf-life | [78] |
| Tomato | SlMBP21 | Loss of function | Agrobacterium-mediated transformation | Jointless fruit stem | [81] |
| Tomato | SlAGAMOUS-LIKE 6 (Sl AGL6) | Loss of function | Agrobacterium-mediated transformation | Parthenocarpic | [82] |
| Tomato | ARF7 | Loss of function | Agrobacterium-mediated transformation | Parthenocarpic | [83] |
| Tomato | SlIAA9 | Loss of function | Agrobacterium-mediated leaf disk transformation | Parthenocarpic | [84] |
| Tomato | Blade-on-petiole (SlBOP) | Loss of function | Agrobacterium-mediated cotyledon segments transformation | Early flowering with simplified inflorescence architecture | [85] |
| Tomato | Self-pruning 5G(SlSP5G) | cis-regulatory variation | Agrobacterium-mediated transformation | Day-length-sensitive flowering | [88] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wan, L.; Wang, Z.; Tang, M.; Hong, D.; Sun, Y.; Ren, J.; Zhang, N.; Zeng, H. CRISPR-Cas9 Gene Editing for Fruit and Vegetable Crops: Strategies and Prospects. Horticulturae 2021, 7, 193. https://doi.org/10.3390/horticulturae7070193
Wan L, Wang Z, Tang M, Hong D, Sun Y, Ren J, Zhang N, Zeng H. CRISPR-Cas9 Gene Editing for Fruit and Vegetable Crops: Strategies and Prospects. Horticulturae. 2021; 7(7):193. https://doi.org/10.3390/horticulturae7070193
Chicago/Turabian StyleWan, Lili, Zhuanrong Wang, Mi Tang, Dengfeng Hong, Yuhong Sun, Jian Ren, Na Zhang, and Hongxia Zeng. 2021. "CRISPR-Cas9 Gene Editing for Fruit and Vegetable Crops: Strategies and Prospects" Horticulturae 7, no. 7: 193. https://doi.org/10.3390/horticulturae7070193
APA StyleWan, L., Wang, Z., Tang, M., Hong, D., Sun, Y., Ren, J., Zhang, N., & Zeng, H. (2021). CRISPR-Cas9 Gene Editing for Fruit and Vegetable Crops: Strategies and Prospects. Horticulturae, 7(7), 193. https://doi.org/10.3390/horticulturae7070193






