Abstract
Cytoplasmic male-sterility (CMS) is important for the utilization of crop heterosis and study of the molecular mechanisms involved in CMS could improve breeding programs. In the present study, anthers of the pepper CMS line HZ1A and its maintainer line HZ1B were collected from stages S1, S2, and S3 for transcriptome sequencing. A total of 47.95 million clean reads were obtained, and the reads were assembled into 31,603 unigenes. We obtained 42 (27 up-regulated and 15 down-regulated), 691 (346 up-regulated and 345 down-regulated), and 709 (281 up-regulated and 428 down-regulated) differentially expressed genes (DEGs) in stages S1, S2, and S3, respectively. Through Gene Ontology (GO) analysis, the DEGs were found to be composed of 46 functional groups. Two GO terms involved in photosynthesis, photosynthesis (GO:0015986) and photosystem I (GO:0009522), may be related to CMS. Through Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, oxidative phosphorylation (ko00190) and phenylpropanoid biosynthesis (ko00940) were significantly enriched in the S1 and S2 stages, respectively. Through the analysis of 104 lipid metabolism-related DEGs, four significantly enriched KEGG pathways may help to regulate male sterility during anther development. The mitochondrial genes orf470 and atp6 were identified as candidate genes of male sterility for the CMS line HZ1A. Overall, the results will provide insights into the molecular mechanisms of pepper CMS.
1. Introduction
Pepper (Capsicum annuum L.) is an important global vegetable crop, and most of the varieties grown are hybrids [1]. Utilization of the maximum heterosis was responsible for approximately 163.8% [2] or 331.11% [3] of fruit pepper yield. The three-line hybrid breeding system (cytoplasmic male sterility (CMS) line, maintainer line, and restorer line) is critical and widely used for the efficient production of pepper hybrids. In this system, CMS plants must be maintained and restored for seed production. Improved knowledge of the molecular mechanisms governing CMS could help improve breeding programs in the future.
CMS is a common phenomenon that is observed in a diverse array of plant species, such as rice, maize, cotton, and soybean [4,5,6]. It occurs via interactions between the mitochondrial and nuclear genes. There are two types of genes associated with CMS: the restorer of male fertility (Rf) genes in the nucleus and the CMS genes in the mitochondria. To date, more than ten Rf genes have been isolated [6]. Approximately half of these genes contain pentatricopeptide repeat domain [7,8], such as Rf1a and Rf1b (Rf5) in Chinsurah Boro II/Taichung 65-type CMS rice [9,10], Rf5 (Rf1b) and Rf6 in Honglian-type CMS rice [11,12], Rf4 in wild-abortive-type CMS rice, Rf1 and Rf2 in sorghum CMS-A1 [13], Rfk1 from radish CMS-Kos [14], Rf0 and Rfp in rapeseed CMS Ogu and CMS Polima [15,16], and RFL79 and RFL29a in wheat [17]. Several Rf genes contain other protein characteristics, such as Rf2, which has aldehyde dehydrogenase characteristics in maize [18] and a glycine-rich domain in rice [19], Rf17 which contains an acyl-carrier protein synthase in rice [20], and Rf1 (bvORF20), which contains peptidase-like proteins in sugar beet [21]. In contrast, more than 20 CMS genes have been identified in the mitochondria from several species [6]. The reported CMS genes encode small cytotoxic transmembrane proteins that frequently cause altered mitochondrial function, leading to male sterility [6,22]. For example, WA352 triggers premature tapetal programmed cell death and pollen abortion in rice [23]. ORFH79 protein can bind to complex III and decrease its enzymatic activity through interaction with P61 and this defect results in energy production dysfunction and oxidative stress in the mitochondria, which leads to abnormal pollen development in rice [24]. Pollen fertility is influenced by gene orf79 as it induces the ORF79 protein in Lead rice-type CMS and Chinsurah Boro II/Taichung 65-type CMS rice [25], while RNA editing of the atp9 and atp6 genes is correlated with the male sterile phenotype in tobacco [26] and rice [27,28], respectively.
Male fertility is related to anther and pollen development in plants. The developmental process is complex and may cause dysplasia and ultimately male sterility. In addition to CMS-associated genes, several other genes related to male sterility have been isolated and identified, including lipid metabolism-related genes, sucrose transporter, phytohormone gibberellin-related genes, Gly-Asp-Ser-Leu (GDSL) esterase/lipases-related genes, endosomal sorting complex required for transport (ESCRT)-related genes, and transcription factors (TFs). Lipid metabolism-related genes can cause male sterility through regulating the biosynthesis and formation of the anther cuticle and pollen wall in plants [29]. The sucrose transporter CsSUT1 can cause male sterility by altering the carbohydrate supply in cucumber [30], whereas the phytohormone gibberellin-related gene, RGAT1, can cause male sterility in Arabidopsis anther development [31]. GAMYB encodes a TF associated with the gibberellin pathway and regulates anther development. Mutations in GAMYB cause an expanded and undifferentiated tapetum, leading to abnormal meiosis in rice [32,33]. GDSL esterase/lipases OsGELP34 and OsGELP110/OsGELP115 can alter the exine structure and render the plant male sterile in rice [34]. Similarly, IPE2 encodes an endoplasmic-reticulum-localized GDSL lipase, which can cause defective formation of the anther cuticle and pollen exine, and complete male sterility in maize [35]. Two ESCRT components, ISTL1 and LIP5, contribute to pollen wall development and tryptophan deposition in Arabidopsis [36]. Glycerol-3-phosphate acyltransferase 6 (GPAT6) reduces endoplasmic reticulum profiles in the tapetum and leads to pollen abortion [37]. OsTDF1, a R2R3 MYB TF, plays a crucial role in tapetum development and male fertility in rice [38]. The UDT1 gene encodes a bHLH TF that is involved in triggering tapetum differentiation in rice [39].
The Rf gene (s) and CMS gene (s) have not yet been clearly defined in pepper, but some genes related to CMS have been reported. Furthermore, with the BSA-AFLP, SLAF-sequencing, and BSA-sequencing methods, at least six genes on chromosome 6 have been considered as Rf candidate genes: CaPPR6 [40], Capana06g003028 [41], CaRf032 (CA00g82510) [42], NEDD8 (Capana06g002866) [43], Capana06g002967, and Capana06g002969 [44]. Several CMS-related mitochondrial genes have also been reported, such as atp6 [45], orf507 [46], orf456 [47], orf300a, and orf314a [48] which have been identified as candidates for CMS in pepper.
To elucidate the molecular mechanisms and genes related to male fertility in pepper during anther development, in the present study, a comparative transcriptome analysis of the anthers was conducted in a CMS line of pepper, namely HZ1A, and its maintainer line, HZ1B, during three developmental stages. By performing Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes (DEGs), few GO terms related to CMS were obtained. Simultaneously, one mitochondrial gene was identified as a candidate gene for CMS. Overall, our results will provide insights into the molecular mechanisms of pepper cytoplasmic male sterility.
2. Materials and Methods
2.1. Plant Sample Collection and Preparation
The CMS line HZ1A was completely sterile and backcrossed for at least 15 generations with its maintainer line HZ1B. The HZ1B fruits were shaped like long fingers. It was difficult to collect anther samples separately during all the developmental stages. Therefore, we collected pepper anthers at three stages according to the flower bud size for transcriptome sequencing: stage 1 (S1), sporogenous tissue to meiotic stage, the length of buds <3.5 mm; stage 2 (S2), tetrad to mononuclear stage, the length of buds between 3.5–4.5 mm; and stage 3 (S3), mature pollen stage, the length of buds about 6.5 mm. There was no difference between the HZ1A and HZ1B flower buds at each stage. The anthers were isolated from the flower buds, immediately placed into an RNA stabilization solution (RNAlater, Qiagen, Hilden, Germany), and stored at −20 °C for RNA extraction. Three biological replicates were collected per stage. All plants were grown in a greenhouse in Hangzhou, China.
2.2. Cytological Examination
In the morning, anthers of different-sized buds from the CMS line HZ1A and its maintainer line HZ1B were dissected. For identifying the fertility of HZ1A and HZ1B, the pollen grains of fully bloomed flowers were tested using Alexander’s stain [49]. The fertile pollen grains were stained magenta red, and the sterile pollen grains were stained blue-green. For observing the corresponding relationship between the development of flower buds and pollen, microspores were stained with hematoxylin solution and observed under an optical microscope.
For scanning electron microscopy (SEM) observations, the specimens were prepared according to Nie et al. [50]. Briefly, anthers with mature pollen of HZ1A or HZ1B were treated as follows: fixed with 4% glutaraldehyde (0.1 M sodium phosphate buffer, pH 7.2), overnight at 4 °C, rinsed with 0.1 M sodium phosphate buffer (pH 7.2), substituted in 1% OsO4 for 2 h at 4 °C, rinsed again, dehydrated in a graded ethanol series, sputter coated with gold, and then observed under a SEM (S3000N, Hitachi, Krefeld, Germany).
2.3. cDNA Preparation and RNA Sequencing
Total RNA was isolated from the anthers of each specimen (S1_HZ1A, S1_HZ1B, S2_HZ1A, S2_HZ1B, S3_HZ1A, and S3_HZ1B) using an RNAprep Pure Plant Plus Kit (DP441, Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. The quantity and quality of the mRNA were assayed using Nanodrop 2000 (NanoDrop Technologies, Wilmington, DE, USA) and LabChip GX Touch (Perkin Elmer, Boston, MA, USA), respectively. Then, 1 µg total RNA was used to construct cDNA libraries by a PCR-cDNA kit (SQK-PCS109) following the manufacturer’s instructions. Reverse transcriptase was used to enrich full-length cDNAs. Thereafter, PCR adapters were added directly to both ends of the first-strand cDNA following 14 cycles of cDNA PCR with the LongAmp Taq PCR Kit (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions. The PCR products were subjected to Oxford Nanopore Technologies (ONT, Oxford, UK) adaptor ligation using T4 DNA ligase (New England Biolabs, Ipswich, MA, USA). Thereafter, the products were purified using Agencourt AMPure XP beads (Beckman Coulter, Indianapolis, IN, USA). Finally, the cDNA libraries were added to FLO-MIN109 flow cells and run on the PromethION sequencing platform (ONT, Oxford, UK).
2.4. Bioinformatic Analysis of the Sequencing Data
Initially, raw reads were filtered with a minimum average read quality score of 6 and a minimum read length of 500 bp using the NanoFilt package, and the ribosomal RNA reads were discarded. The Phred scores (Q) were calculated from the formula: Q = −10 × log10 P, where P refers to the probability of base recognition error. Then, full-length, non-chimeric (FLNC) transcripts were determined by searching for primers at both ends of reads. Clusters of FLNC transcripts were obtained after mapping to the reference genome with mimimap2 [51]. Clusters of full-length, non-chimeric transcripts and consensus isoforms were obtained. Consensus sequences were mapped to the reference genome using the minimap2. Mapped reads were further collapsed using the cDNA_Cupcake package with min-coverage = 85% and min-identity = 90%. A 5′ difference was not considered when the redundant transcripts collapsed [52].
Clean reads were mapped onto the nuclear reference genome (C. annuum. L Zunla-1, v2.0; http://peppersequence.genomics.cn/, accessed on 1 September 2021) [53], the mitochondrional reference genome (National Center for Biotechnology Information (NCBI) RefSeq: NC_024624.1) [54], and the chloroplast reference genome (NCBI RefSeq: NC_018552.1) [55] using the mimimap2 [51]. Reads with a match quality above 5 were used for further quantification. Expression levels were estimated by reads per gene/transcript per 10,000 mapped reads. The names of newly identified genes begin with ‘ONT’. The raw data were deposited in the Short Read Archive database (https://www.ncbi.nlm.nih.gov/sra, accessed on 22 October 2021) with the accession number: PRJNA772539.
2.5. Differential Gene Expression Analysis
Differential expression analysis was performed using the DESeq2 R package (1.6.3) [56]. The resultant p values were then adjusted using Benjamini and Hochberg’s approach [57] to control the false discovery rate. Both |log2 (HZ1A vs. HZ1B)| and the p value were used to determine the threshold for the DEGs. Genes with |log2 (HZ1A vs. HZ1B)| ≥ 1.0 and p value < 0.01 were defined as DEGs. The TFs were identified by predicting all the DEGs using iTAK (Plant Transcription factor & Protein Kinase Identifier and Classifier; http://itak.feilab.net/cgi-bin/itak/index.cgi, accessed on 30 September 2021) [58] based on the databases PlnTFDB and PlantTFDB.
2.6. Functional Enrichment Analysis
Functional annotation of the DEGs was performed based on the GO [59], and the enrichment analysis of the DEGs was implemented using the GOseq R packages based on Wallenius non-central hyper-geometric distribution [60], which can adjust for gene length bias in DEGs. KEGG pathway annotation was carried out using a BLAST search against the KEGG database (http://www.genome.jp/kegg/, accessed on 22 September 2021), and pathway enrichment analysis of the DEGs was performed using the KEGG Orthology Based Annotation System [61]. GO terms and KEGG pathways with a corrected p < 0.05 were defined as significantly enriched.
2.7. Quantitative Real-Time PCR and Candidate Genes Amplification
All primers used in the present study were designed using Primer-Blast software (NCBI, Bethesda, MD, USA) and synthesized by Sangon Biotech (Shanghai, China). The pepper β-actin gene [62] was used as an internal control to normalize the expression data. The primer pairs used are listed in Table S1. Quantitative real-time PCR (qRT-PCR) was conducted according to the instructions of 2 × Hieff UNICON® Universal Blue qPCR SYBR Green Master Mix (YEASEN, Shanghai, China), and analyzed using the Quant Studio 5 real-time PCR system (Thermo Fisher Scientific, Inc., Waltham, MA, USA). The 2−ΔΔCt method was used for calculating the relative mRNA expression. Three biological replicates and three technical replicates were used for each experiment. The reaction mixture (50 μL) for the PCR amplification contained 25 μL of 2 × Taq Plus Master Mix II (Dye Plus) (Vazyme Biotech Co., Ltd., Nanjing, China), 2 μL each of 10 μmol/L forward and reverse primers, 17 μL of ddH2O, and 4 μL of DNA (30 ng). The PCR products were sequenced by Sangon Biotech (Shanghai, China).
3. Results
3.1. Morphological Characteristics of Male Fertility and Sterility
Male-fertility phenotypes of the CMS line HZ1A and its maintainer line HZ1B were observed during their flowering (Figure 1A,B). HZ1A had the typical features of male-sterility, including no pollen dissemination around its anthers. The pollen grains of the HZ1A plants were shriveled, less in number, and stained blue-green with Alexander’s stain. The pollen grains of the HZ1B plants were plump and stained magenta red with Alexander’s stain.
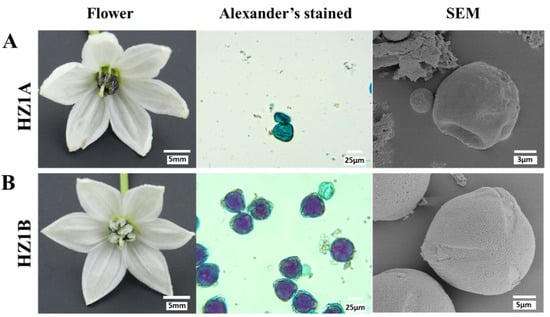
Figure 1.
Phenotypes of flowers and pollen grains of the pepper cytoplasmic male-sterility (CMS) line HZ1A and its maintainer line HZ1B. (A,B) HZ1A and HZ1B, respectively. There was no pollen dissemination around the HZ1A anthers. Pollen grains of the HZ1A plants were fewer, shriveled, and stained blue-green.
3.2. The Cytological Characteristics of CMS Line HZ1A and Its Maintainer Line HZ1B Microspores during the Five Developmental Events
The length of flower buds showed a significant correlation with different stages of pollen development in pepper. The corresponding relationship in the HZ1B was as follows: about 1.5 mm, the sporogenous tissue stage; about 2.3 mm, the meiotic stage; about 3.5 mm, the tetrad period; about 4.5 mm, the mononuclear stage; and about 6.5 mm, the pollen mature stage (Figure 2A).
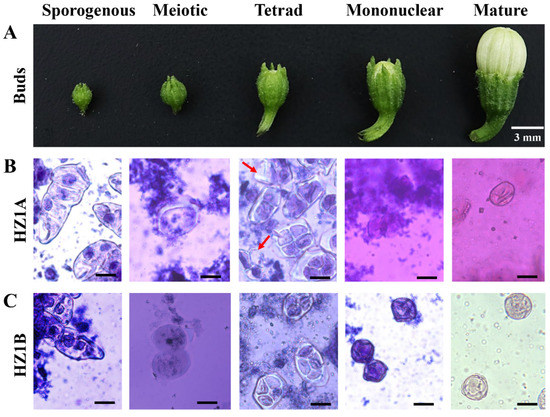
Figure 2.
Flower buds and corresponding microspores during the five developmental stages of flowers in CMS line HZ1A and its maintainer line HZ1B. (A) HZ1B flower buds at different developmental stages. No difference between HZ1A and HZ1B flower buds was observed in all developmental stages; (B) HZ1A microspores at different developmental stages of pollen; (C) HZ1B microspores at different developmental stages of pollen. Left-right: sporogenous tissue stage, meiotic stage, tetrad stage, mononuclear stage, and mature pollen stage. The male sterility in HZ1A begins at the tetrad stage of microspore development. Scale bar = 25 μm. The male sterility in HZ1A begins at the tetrad stage of microspore development.
There was no differences between the microspores of the CMS line HZ1A and its maintainer line HZ1B during the sporogenous tissue stage and meiotic stages (Figure 2B,C). In the tetrad stage, few microspores in the HZ1A were similar to that in the HZ1B in terms of their cytological features; however, few microspores (indicated by red arrows) in the HZ1A were anomalous and irregular. It was difficult to find microspores in the mononuclear stage of the HZ1A as the number of microspores was less. These results indicate that the male sterility in HZ1A to begin at the tetrad stage of microspore development.
3.3. RNA Sequencing and DEG Identification
To clarify the basic molecular mechanism regulating male sterility in the CMS line HZ1A, gene expression levels in the HZ1A and HZ1B anthers during three developmental stages (S1, S2, and S3) were compared using RNA-Seq with the PromethION platform (ONT, Oxford, UK). RNA sequencing of S1_HZ1A, S2_HZ1A, S3_HZ1A, S1_HZ1B, S2_HZ1B, and S3_HZ1B, resulted in a total of 47.95 million clean reads being obtained from the raw reads (Table 1). The number of clean reads from the six samples ranged from 7.29 to 8.52 million reads. These reads had a mean length of 711–923 bp. The data for the six samples were on an average 84.04–87.91% that of the full-length reads. The mapped rate ratios of the six samples to the pepper reference genome were 76.83–87.45% on an average. This indicated that the aligned data were suitable for subsequent analyses.

Table 1.
Statistical analysis of the RNA-Seq read quality in cytoplasmic male-sterility line HZ1A and its maintainer line HZ1B.
There was a total of 59,468 functionally annotated transcripts. The reads were assembled into 31,603 unigenes, including 35,336 genes that were matched to nuclear reference sequences, 190 genes that were matched to mitochondrial reference genome sequences, 84 genes to chloroplast reference genome sequences, and 4958 new genes. The number of genes ranged from 25,144–25,814 across all six samples (Figure 3A). The gene expression levels were compared at each developmental stage of the anthers between the CMS line HZ1A and its maintainer line HZ1B, and a total of 1377 genes across all stages were identified as being differentially expressed (Table S2). A total of 42 (27 up-regulated and 15 down-regulated), 691 (346 up-regulated and 345 down-regulated), and 709 (281 up-regulated and 428 down-regulated) DEGs were obtained from stages S1, S2, and S3 for the CMS line HZ1A, respectively (Figure 3C). Only four DEGs were common in all three stages (Figure 3D). More up-regulated DEGs were detected in the CMS line HZ1A at the S1 stage. In contrast, more down-regulated DEGs were detected in the CMS line HZ1A at the S2 and S3 stages.
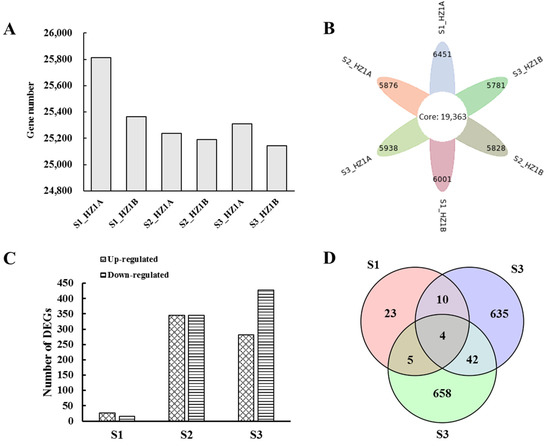
Figure 3.
The profiles of differentially expressed genes (DEGs) in anthers of the CMS line HZ1A and its maintainer line HZ1B during the three developmental stages. (A,C) Histogram of the unigenes (A) and DEGs (C) in the six samples; (B,D) Venn diagram of the unigenes (B) and DEGs (D) in the six samples. More up-regulated DEGs were detected in the CMS line HZ1A at the S1 stage. Only four DEGs were common in all three stages. More down-regulated DEGs were detected in the CMS line HZ1A at the S2 and S3 stages.
3.4. Functional Annotation by GO
A total of 1377 DEGs from S1, S2, and S3 were classified into three main categories: ‘biological process (BP)’, ‘cellular component (CC)’, and ‘molecular function (MF)’. These three categories were composed of 46 functional groups based on GO assignments (Figure 4). In the ‘BP’ category, ‘metabolic process’, ‘cellular process’ and ‘single-organism process’ were the major functional groups, including a total of 277, 276, and 223 genes in the three stages, respectively. The top six significant GO terms were different in the three stages. Photosynthesis (GO:0015986) was significantly enriched in the S1 and S3 stages (Table S3). In the ‘CC’ category, ‘membrane’, ‘membrane part’, ‘cell part’, ‘cell’ and ‘organelle’ were the dominant functional types, including a total of 308, 283, 272, 272, and 210 genes in three stages, respectively. The top six significant GO terms in the three stages are shown in Table S4. Photosystem I (GO:0009522), endomembrane system (GO:0012505), and endoplasmic reticulum (GO:0005783) were significantly enriched in the S2 and S3 stages. In the ‘MF’ category, ‘binding’ and ‘catalytic activity’ were the dominant functional types, including a total of 453 and 449 genes in the three stages, respectively. The top six significant GO terms in the three stages are shown in Table S5. Hydrogen ion transmembrane transporter activity (GO:0015078), serine-type endopeptidase activity (GO:0004252), and O-methyltransferase activity (GO:0008171) were significantly enriched in S1, S2, and S3. Interestingly, the photosynthesis (GO:0015986) term in the ‘BP’ category and photosystem I (GO:0009522) term in the ‘CC’ category were both involved in photosynthesis. Therefore, the photosynthesis process may be related to CMS.
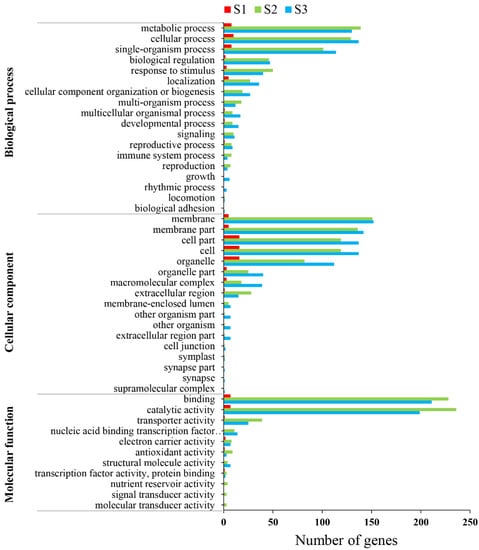
Figure 4.
Gene Ontology (GO) enrichment analysis results for the differentially expressed genes in anthers of the cytoplasmic male-sterility line HZ1A and its maintainer line HZ1B during S1, S2, and S3 stages. There were 46 functional groups in three categories.
3.5. Pathway Mapping by KEGG
Functional annotation and pathway enrichment analysis of the DEGs was performed using the KEGG database. DEGs were enriched in 13, 100, and 97 (Tables S6–S8) pathways in the three stages, respectively, as observed by the KEGG pathway analysis, respectively. Only two pathways, oxidative phosphorylation (ko00190) (Figure 5A) and phenylpropanoid biosynthesis (ko00940) (Figure 5B) were significantly enriched in the S1 and S2 stages, respectively. Although the numbers of DEGs in the S2 and S3 stages were almost the same, there was no significantly enriched KEGG pathway. There were four DEGs in the oxidative phosphorylation pathway and 24 DEGs in the phenylpropanoid biosynthesis pathway (ko00940). In the latter pathway, almost all the DEGs (20) were up-regulated in the CMS anthers. This indicated that the up-regulation of phenylpropanoid biosynthesis might cause sterility in the pepper CMS line, HZ1A.
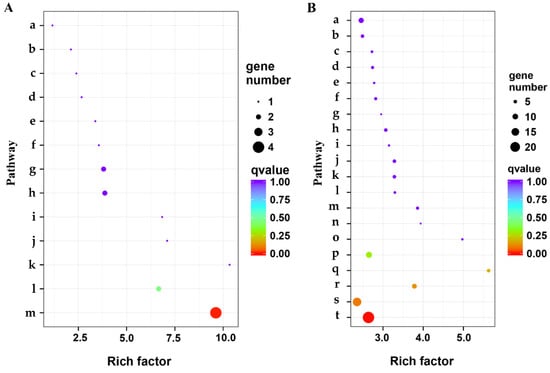
Figure 5.
Scatterplot of the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment results for the DEGs in anthers of the cytoplasmic male-sterility line HZ1A and its maintainer line HZ1B during S1 (A) and S2 (B) stages of development. Oxidative phosphorylation (ko00190) and phenylpropanoid biosynthesis (ko00940) were significantly enriched in the S1 and S2 stages, respectively. Note: The small letter indicates the KEGG pathway. In part A, a: MAPK signaling pathway–plant; b: Phenylpropanoid biosynthesis; c: Biosynthesis of amino acids; d: Ubiquitin mediated proteolysis; e: Photosynthesis; f: Glycolysis/Gluconeogenesis; g: Protein processing in endoplasmic reticulum; h: Carbon metabolism; i: Glycine, serine and threonine metabolism; j: Zeatin biosynthesis; k: Citrate cycle (TCA cycle); l: Amino sugar and nucleotide sugar metabolism; m: Oxidative phosphorylation. In part B, a: Cysteine and methionine metabolism; b: Tyrosine metabolism; c: Selenocompound metabolism; d: Ubiquinone and other terpenoid−quinone biosynthesis; e: Histidine metabolism; f: Diterpenoid biosynthesis; g: Limonene and pinene degradation; h: Alanine, aspartate and glutamate metabolism; i: Benzoxazinoid biosynthesis; j: Tropane, piperidine and pyridine alkaloid biosynthesis; k: Isoquinoline alkaloid biosynthesis; l: Glycosylphosphatidylinositol (GPI)−anchor biosynthesis; m: Isoflavonoid biosynthesis; n: Glycosphingolipid biosynthesis−globo and isoglobo series; o: Betalain biosynthesis; p: Flavonoid biosynthesis; q: Biosynthesis of unsaturated fatty acids; r: Cyanoamino acid metabolism; s: Ubiquitin mediated proteolysis; t: Phenylpropanoid biosynthesis. Rich factor indicates the ratio of frequency of DEGs annotated to the pathway to that of all genes annotated to the pathway.
3.6. Identification of Differentially Expressed TFs
The TFs of the DEGs were analyzed at the S1, S2, and S3 stages of the CMS line HZ1A and its maintainer line HZ1B. A total of 72 TFs were identified by predicting all the DEGs, and 1 (1 up-regulated), 52 (34 up-regulated and 18 down-regulated), and 21 (15 up-regulated and 6 down-regulated) TFs were identified in S1, S2, and S3 stages of the CMS line HZ1A, respectively (Table S9). These TFs were classified into 30 families, including NAC (9), LOB (8), bZIP (5), MADS-M-type (4), MYB (4), and 25 other TFs (Figure 6). Only three TFs (Capana02g000496, Capana02g002747, and Capana06g001532) were up-regulated in the S2, and S3 stages simultaneously. There were 45 TFs that were classified into 25 families that were up-regulated in the CMS line, whereas 27 TFs that were classified into 12 families that were down-regulated in the CMS line. In the S1 stage, only one TF (Capana02g000758) was up-regulated in the CMS line. Most of the TFs were up-regulated or down-regulated during the S2 and S3 developmental stages. These results suggested that there is a complex transcriptional network was involved in the anther development.
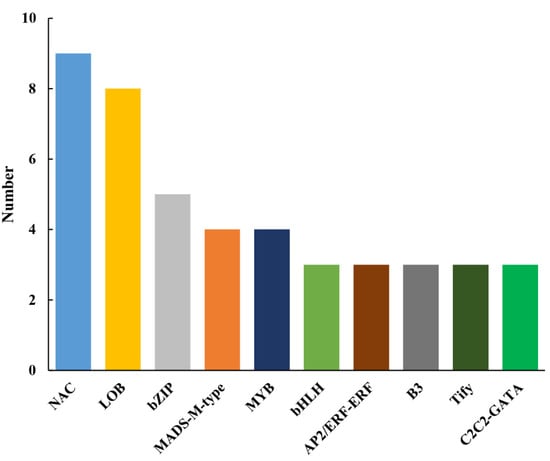
Figure 6.
Top 10 transcription factors (TFs) identified from the differentially expressed genes in the S1, S2, and S3 developmental stages of the cytoplasmic male-sterility line HZ1A and its maintainer line HZ1B. There is a complex transcriptional network involved in the anther development.
3.7. Validation of the DEGs Using Quantitative Real-Time PCR
Lipid metabolism and endoplasmic reticulum-related genes are associated with male sterility [29,37]. To validate the reliability of the RNA-Seq, nine DEGs related to lipid metabolism (Capana00g002971, Capana04g001148, Capana07g002334, Capana08g001266, and Capana12g002508), and endoplasmic reticulum (Capana01g000067, Capana06g000385, Capana06g002873, and Capana09g001928) were randomly selected and used for qRT-PCR analysis. The expression levels of the nine DEGs showed similar trends in both RNA-Seq analysis and qRT-PCR results (Figure 7). These results indicate that the RNA-Seq results were reliable.
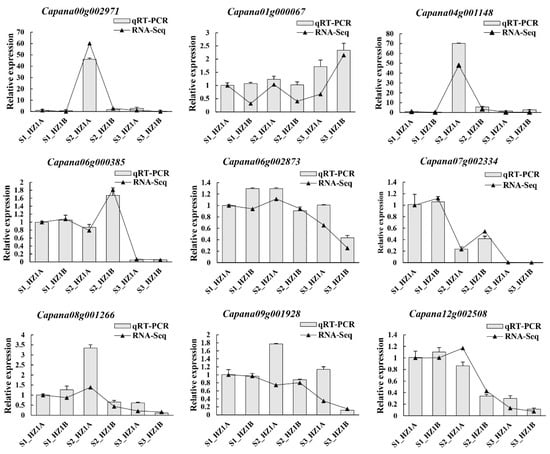
Figure 7.
Expression levels for nine differentially expressed genes in the cytoplasmic male-sterility line HZ1A and its maintainer line HZ1B during the S1, S2, and S3 developmental stages by qRT-PCR and the RNA-seq results. The expression levels of the nine DEGs showed similar trends in both RNA-Seq analysis and qRT-PCR results.
3.8. Mitochondrial Genes Involved in Energy Production Are Related to CMS
Mitochondria play an important role in energy production. Mitochondrial deficiencies may lead to failure in acquiring sufficient energy for normal pollen development [62]. In some studies, the sterility-related genes in plant mitochondria, such as orf138, orf79, atp4, and atp6 [8], cause abnormal energy supplies and eventually leading CMS. In the present study, RNA sequencing data were compared to the mitochondrial reference genome and the chloroplast reference genome. In total, 12 mitochondrial DEGs were obtained. Of these DEGs, of which three (orf470, atp6 and cox1) were associated with energy production. Comparison of the gene expression levels from the RNA-Seq analysis suggested that the genes orf470 and cox1 in the CMS line HZ1A showed lower expression than those in the HZ1B during the S1 and S3 stages, respectively; however, the atp6 gene showed higher expression levels in HZ1A than in HZ1B during the S1 and S2 stages. Primers (Table S1) for these three DEGs were designed to amplify in the CMS line HZ1A and its maintainer line HZ1B. The results showed that orf470 was amplified only in the HZ1B; however, the other two DEGs were effectively amplified (Figure 8A). According to the sequencing results of the PCR products, there was no difference in the sequences of cox1 between the HZ1A and HZ1B; however, several nucleotide differences in the sequence of atp6 were observed between the HZ1A and HZ1B, which lead to changes in the amino acid sequences of corresponding proteins (Figure 8B). These results indicate that orf470 and atp6 may be related to male sterility and were thus considered the candidate genes for male sterility in the CMS line, HZ1A. This will require further verification through functional characterization and other methods in the future.
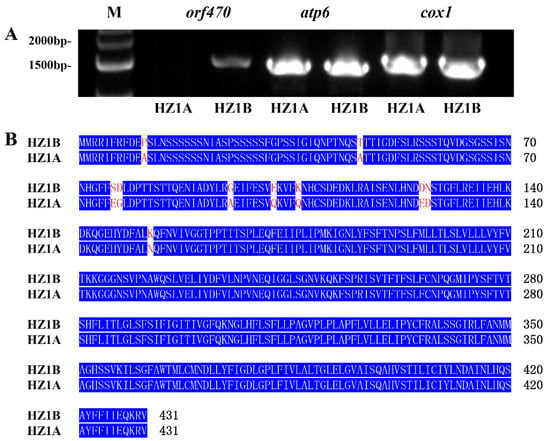
Figure 8.
Expression levels of the DEGs of mitochondria that are involved in energy production. (A) Agarose gel electrophoresis of three mitochondrial DEGs in cytoplasmic male-sterility line HZ1A and its maintainer line HZ1B. M Marker lane (1 Kb DNA ladder). Each amplified DEG of HZ1A and HZ1B was loaded in two lanes. The amplified product from HZ1A was loaded on the left and that of HZ1B was loaded on the right. (B) Amino acid sequences of atp6 in HZ1A and HZ1B.
4. Discussion
4.1. The Anther Was the Best Organ for Studying Male Sterility in Pepper
In plants, the phenomenon of male fertility is reflected in the anthers and pollen grains, and the development of pollen grains is generally related to the anthers. The anthers are consequently the best organs in which to study male sterility. To study gene expression related to male fertility in pepper, directly study of the anther and pollen grains would be the best practice.
Peppers have small anthers, especially in their early stages of development, which makes it difficult to obtain the anther samples separately for RNA extraction across all the developmental stages. Therefore, the pepper floral buds have been used in previous studies for transcriptome analysis [62,63]. In wheat and rice, RNA stabilization solutions have been used to collect anthers and pollen grains, and qualified RNA has been extracted [64]. Therefore, the RNA stabilization solution (RNALater) was used to collect the pepper anthers and extract the RNA successfully in the present study. On the other hand, according to the results of the expressions of genes in pepper floral buds and anthers using qRT-PCR (not published data), the expressions of some genes were different in the flower buds and anthers. This result was the same as in soybean [65]. Consequently, the anther was considered the best organ to study male sterility.
4.2. Male Sterility Occurs alongside Gene Expression Level Changes in the Middle and Late Stages of Anther Development
In each of the anther development stages assessed (S1, S2, and S3), the number of detected genes was more than 25,000, and there was little difference between the samples. This shows that a large number of genes are involved in the anther development of pepper at different developmental stages, however, the number of DEGs was significantly different at different developmental stages. In the S1 stage, only 42 DEGs were identified. While in the S2 and S3 stages, the number of DEGs (691 and 709), respectively, increased rapidly, which was similar to previous results for eggplant anthers [66] and pepper buds [62]. This shows that the change in gene expression is not obviously caused by male sterility in the early stage of the anther development, and that male sterility mainly affects the gene expression level in the S2 and S3 stages. These results were in-line with the male sterility beginning at the tetrad stage (S2) of the CMS line HZ1A microspore development. This suggests that male sterility causes changes in the gene expression levels in the middle and late stages of anther development in the CMS line HZ1A.
4.3. TFs Related to the CMS in Pepper Line HZ1A
TFs are associated with fertility in some plant species. GAMYB encodes a TF associated with the gibberellin pathway and leads to abnormal meiosis through regulating another development in rice [32,33]. CSA encodes an R2R3 MYB TF that leads to male sterility through reduced levels of carbohydrates in rice [67]. A barley PHD TF, MALE STERTILITY1 (MS1) is expressed in the anther tapetum and plays a critical role during pollen development [68]. The barley HvMS1 gene encodes a PHD-finger TF that is expressed in the anther tapetum, which is essential for pollen development and causes complete male sterility when overexpressed in barley [68]. The TF ZmDREB1.7 partially restores male fertility of CMS-S maize by activating the expression of the mitochondria-encoded CMS gene orf355 [69]. The SlMS10 gene (Solyc02g079810) encodes the bHLH TF that regulates meiosis and cell death of the tapetum during microsporogenesis in tomato [70].
In our study, the highest number of TFs was observed in the S2 stage, and only one TF was identified in the S1 stage. These results were in-line with the male sterility in the CMS line HZ1A beginning at the tetrad stage (S2) of microspore development. The TFs in the S2 stage are most likely related to male sterility. We analyzed the TFs that belong to the same family of the above described TFs in HZ1A and HZ1B. Only bHLH (two TFs) and MYB (one TF) families were identified in the S2 stage. These three TFs (Capana01g000416, Capana01g000418, and Capana02g000991) might regulate fertility in HZ1A.
4.4. Genes Involved in Lipid Metabolism Are Related to Male Sterility
Male sterility is a phenomenon caused by a complex process that occurs widely in flowering plants. Lipid transport and metabolism are related to male fertility development in the anther. In the present study, according to the functional annotation of DEGs, 104 lipid metabolism-related genes were found to be differentially expressed, and 1 (1 down-regulated), 68 (46 up- and 22 down-regulated), and 36 (18 up- and 18 down-regulated) genes were identified in stages S1, S2, and S3 of the CMS line HZ1A, respectively. In the S2 stage, the number of lipid metabolism-related genes was higher than that in the S1 and S3 stages. These results indicated that S2 is an important stage in the lipid metabolism pathway.
The lipid metabolism-related genes were analyzed using KEGG pathway enrichment. The significantly enriched terms were fatty acid metabolism (ko01212, 10 genes), biosynthesis of unsaturated fatty acids (ko01040, 5 genes), sphingolipid metabolism (ko00600, 5 genes), and glycerophospholipid metabolism (ko00564, 6 genes) (Figure S1). To date, many lipid metabolism-related male sterile genes have been identified and characterized, and the transcriptional regulation pathways, lipid metabolism, and control of male fertility have been well investigated in plants [29,71]. Therefore, these four pathways in our study may play important roles in regulating male sterility in the CMS line HZ1A during anther development.
There were 21 DEGs in the four KEGG pathways. Of them, 10 DEGs were down-regulated in the CMS line, HZ1A and these DEGs may be related to male sterility. Among these 10 DEGs, Capana02g003412 was down-regulated and showed the lowest expression (log2 (fold change) = −2.88) in the CMS line HZ1A. According to the KEGG pathway annotation, Capana02g003412 was a glycosphingolipid biosynthesis-related gene. Glycosphingolipids are the major constituents of the outer leaflet of the plasma membrane in eukaryotic cells [72]. On the other hand, we searched the homologous genes using the amino acid sequence of Capana02g003412 through NCBI-Blast (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 30 October 2021). One of the homologous genes, Arabidopsis thaliana gene AT5G20710, was found to be related to pollen development [73]. These results suggest that Capana02g003412 may be related to pollen development and male sterility.
5. Conclusions
In the present study, a comparative transcriptome analysis of the anthers was conducted between the CMS line HZ1A and its maintainer line HZ1B during three developmental stages. RNA-Seq analysis identified that 42, 691, and 709 DEGs were obtained in stages S1, S2, and S3, respectively. GO and KEGG analysis of the DEGs revealed that two GO terms and one KEGG pathway may be related to CMS. The mitochondrial genes orf470 and atp6 were identified as candidate genes for CMS. Therefore, our study elucidated the pathways and key genes related to the CMS of pepper, and our results provided insights into the molecular mechanisms of CMS, laying the foundation for further research in pepper.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae7120580/s1, Figure S1: Scatterplot of the KEGG pathway enrichment results for the lipid metabolism-related DEGs between HZ1A and HZ1B anthers, Table S1: Primers of qRT-PCR and mitochondrial genes in the present study, Table S2: DEGs in HZ1B vs. HZ1A anthers during three stages, Table S3: The top six significant GO terms in ‘biological process’ during three stages, Table S4: The top six significant GO terms in ‘cellular component’ during three stages, Table S5: The top six significant GO terms in ‘molecular function’ during three stages, Table S6: The pathways of the DEGs at S1 stage based on KEGG metabolic pathways, Table S7: The pathways of the DEGs at S2 stage based on KEGG metabolic pathways, Table S8: The pathways of the DEGs at S3 stage based on KEGG metabolic pathways, Table S9: The transcription factors from the DEGs in the S1, S2, and S3 development stages.
Author Contributions
Conceptualization, W.Z., Q.N. and Z.N.; methodology, J.C., H.F. and H.W.; investigation, Z.N. and J.C.; resources, J.C.; data curation, Z.N. and Y.S.; writing—original draft preparation, Z.N.; writing—2021 review and editing, W.Z. and Q.N.; supervision, W.Z. and Q.N.; funding acquisition, Z.N. and W.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the national major vegetable industry technology system construction project [grant number CARS-25], the independent declaration project of agricultural and social research in Hangzhou [grant number 20191203B58], and the science and technology project of the Hangzhou academy of agricultural sciences [grant number 2020HNCT-10].
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| Abbreviation | Full definition |
| CMS | Cytoplasmic male-sterility |
| DEGs | Differentially expressed genes |
| ESCRT | Endosomal sorting complex required for transport |
| FLNC | Full-length, non-chimeric |
| GDSL | Gly-Asp-Ser-Leu |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| NCBI | National Center for Biotechnology Information |
| ONT | Oxford Nanopore Technologies |
| Rf | Restorer of male fertility |
| SEM | Scanning electron microscopy |
| TFs | Transcription factors |
References
- Wang, L.; Zhang, B.; Zhang, Z.; Cao, Y.; Yu, H.; Feng, X. Status in breeding and production of Capsicum spp. in China during ‘the thirteenth five-year plan’ period and future prospect. China Veg. 2021, 2, 21–29. [Google Scholar]
- Marame, F.; Dessalegne, L.; Fininsa, C.; Sigvald, R. Heterosis and heritability in crosses among Asian and Ethiopian parents of hot pepper genotypes. Euphytica 2009, 168, 235–247. [Google Scholar] [CrossRef]
- Singh, P.; Cheema, D.S.; Dhaliwal, M.S.; Garg, N. Heterosis and combining ability for earliness, plant growth, yield and fruit attributes in hot pepper (Capsicum annuum L.) involving genetic and cytoplasmic-genetic male sterile lines. Sci. Hortic.-Amst. 2014, 168, 175–188. [Google Scholar] [CrossRef]
- Li, S.; Yang, D.; Zhu, Y. Characterization and use of male sterility in hybrid rice breeding. J. Integr. Plant. Biol. 2007, 49, 791–804. [Google Scholar] [CrossRef]
- Nie, Z.X.; Zhao, T.J.; Yang, S.P.; Gai, J.Y. Development of a cytoplasmic male-sterile line NJCMS4A for hybrid soybean production. Plant Breed. 2017, 136, 516–525. [Google Scholar] [CrossRef]
- Kim, Y.J.; Zhang, D. Molecular control of male fertility for crop hybrid breeding. Trends. Plant Sci. 2018, 23, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Gaborieau, L.; Brown, G.G.; Mireau, H. The propensity of pentatricopeptide repeat genes to evolve into restorers of cytoplasmic male sterility. Front. Plant Sci. 2016, 7, 1816. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, T.; Ohta, S.; Murai, N.; Takakura, Y.; Kuraya, Y.; Suzuki, S.; Hiei, Y.; Imaseki, H.; Nitta, N. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.). Plant J. 2004, 10, 1046–1056. [Google Scholar]
- Hu, J.; Wang, K.; Huang, W.; Liu, G.; Gao, Y.; Wang, J.; Huang, Q.; Ji, Y.; Qin, X.; Wan, L. The rice pentatricopeptide repeat protein RF5 restores fertility in Hong-Lian cytoplasmic male-sterile lines via a complex with the glycine-rich protein GRP162. Plant Cell 2012, 24, 109–122. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Yu, C.; Hu, J.; Wang, L.; Dan, Z.; Zhou, W.; He, C.; Zeng, Y.; Yao, G.; Qi, J. Pentatricopeptide-repeat family protein RF6 functions with hexokinase 6 to rescue rice cytoplasmic male sterility. Proc. Natl. Acad. Sci. USA 2015, 112, 14984–14989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jordan, D.; Mace, E.S.; Henzell, R.; Klein, P.; Klein, R. Molecular mapping and candidate gene identification of the Rf2 gene for pollen fertility restoration in sorghum [Sorghum bicolor (L.) Moench]. Theor. Appl. Genet. 2010, 120, 1279–1287. [Google Scholar] [CrossRef] [PubMed]
- Koizuka, N.; Imai, R.; Fujimoto, H.; Hayakawa, T.; Kimura, Y.; Kohno-Murase, J.; Sakai, T.; Kawasaki, S.; Imamura, J. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that restores fertility in the cytoplasmic male-sterile Kosena radish. Plant J. 2003, 34, 407–415. [Google Scholar] [CrossRef]
- Uyttewaal, M.; Arnal, N.; Quadrado, M.; Martin-Canadell, A.; Vrielynck, N.; Hiard, S.; Gherbi, H.; Bendahmane, A.; Budar, F.; Mireau, H. Characterization of Raphanus sativus pentatricopeptide repeat proteins encoded by the fertility restorer locus for Ogura cytoplasmic male sterility. Plant Cell 2008, 20, 3331–3345. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Fu, T.; Yi, B.; Hong, D. A mitochondria-targeted PPR protein restores pol cytoplasmic male sterility by reducing orf224 transcript levels in oilseed rape. Mol. Plant 2016, 9, 1082–1084. [Google Scholar]
- Melonek, J.; Duarte, J.; Martin, J.; Beuf, L.; Murigneux, A.; Varenne, P.; Comadran, J.; Specel, S.; Levadoux, S.; Bernath-Levin, K.; et al. The genetic basis of cytoplasmic male sterility and fertility restoration in wheat. Nat. Commun. 2021, 12, 1036. [Google Scholar] [CrossRef]
- Cui, X.; Wise, R.P.; Schnable, P.S. The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. Science 1996, 272, 1334–1336. [Google Scholar] [CrossRef]
- Itabashi, E.; Iwata, N.; Fujii, S.; Kazama, T.; Toriyama, K. The fertility restorer gene, Rf2, for Lead Rice-type cytoplasmic male sterility of rice encodes a mitochondrial glycine-rich protein. Plant J. 2011, 65, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Fujii, S.; Toriyama, K. Suppressed expression of RETROGRADE-REGULATED MALE STERILITY restores pollen fertility in cytoplasmic male sterile rice plants. Proc. Natl. Acad. Sci. USA 2009, 106, 9513–9518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitazaki, K.; Arakawa, T.; Matsunaga, M.; Yui-Kurino, R.; Matsuhira, H.; Mikami, T.; Kubo, T. Post-translational mechanisms are associated with fertility restoration of cytoplasmic male sterility in sugar beet (Beta vulgaris). Plant J. 2015, 83, 290–299. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.; Huang, W.; Huang, Q.; Qin, X.; Yu, C.; Wang, L.; Li, S.; Zhu, R.; Zhu, Y. Mitochondria and cytoplasmic male sterility in plants. Mitochondrion 2014, 19, 282–288. [Google Scholar] [CrossRef]
- Luo, D.P.; Xu, H.; Liu, Z.L.; Guo, J.X.; Li, H.Y.; Chen, L.T.; Fang, C.; Zhang, Q.Y.; Bai, M.; Yao, N.; et al. A detrimental mitochondrial-nuclear interaction causes cytoplasmic male sterility in rice. Nat. Genet. 2013, 45, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Gao, F.; Ji, Y.X.; Liu, Y.; Dan, Z.W.; Yang, P.F.; Zhu, Y.G.; Li, S.Q. ORFH79 impairs mitochondrial function via interaction with a subunit of electron transport chain complex III in Honglian cytoplasmic male sterile rice. New Phytol. 2013, 198, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Kazama, T.; Itabashi, E.; Fujii, S.; Nakamura, T.; Toriyama, K. Mitochondrial ORF79 levels determine pollen abortion in cytoplasmic male sterile rice. Plant J. 2016, 85, 707–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabaleta, E.; Mouras, A.; Hernould, M.; Suharsono, S.; Araya, A. Transgenic male-sterile plant induced by an unedited atp9 gene is restored to fertility by inhibiting its expression with antisense RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 11259–11263. [Google Scholar] [CrossRef] [Green Version]
- Das, S.; Sen, S.; Chakraborty, A.; Chakraborti, P.; Maiti, M.K.; Basu, A.; Basu, D.; Sen, S.K. An unedited 1.1 kb mitochondrial orfB gene transcript in the wild abortive cytoplasmic male sterility (WA-CMS) system of Oryza sativa L. subsp indica. BMC Plant Biol. 2010, 10, 39. [Google Scholar] [CrossRef] [Green Version]
- Tang, W.; Luo, C. Molecular and functional diversity of RNA editing in plant mitochondria. Mol. Biotechnol. 2018, 60, 935–945. [Google Scholar] [CrossRef]
- Wan, X.; Wu, S.; Li, Z.; An, X.; Tian, Y. Lipid metabolism: Critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 2020, 13, 955–983. [Google Scholar] [CrossRef]
- Sun, L.; Sui, X.; Lucas, W.J.; Li, Y.; Feng, S.; Ma, S.; Fan, J.; Gao, L.; Zhang, Z. Down-regulation of the sucrose transporter CsSUT1 causes male sterility by altering carbohydrate supply. Plant Physiol. 2019, 180, 986–997. [Google Scholar] [CrossRef] [Green Version]
- Qian, Q.; Yang, Y.H.; Zhang, W.B.; Hu, Y.L.; Li, Y.G.; Yu, H.; Hou, X.L. A novel Arabidopsis gene RGAT1 is required for GA-mediated tapetum and pollen development. New Phytol. 2021, 231, 137–151. [Google Scholar] [CrossRef]
- Aya, K.; Ueguchi-Tanaka, M.; Kondo, M.; Hamada, K.; Yano, K.; Nishimura, M.; Matsuoka, M. Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 2009, 21, 1453–1472. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Bao, W.; Liang, W.; Yin, J.; Zhang, D. Identification of gamyb-4 and analysis of the regulatory role of GAMYB in rice anther development. J. Integr. Plant Biol. 2010, 52, 670–678. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, M.; Li, Y.; Yan, W.; Chang, Z.; Ni, H.; Chen, Z.; Wu, J.; Xu, C.; Deng, X.W.; et al. GDSL esterase/lipases OsGELP34 and OsGELP110/OsGELP115 are essential for rice pollen development. J. Integr. Plant Biol. 2020, 62, 1574–1593. [Google Scholar] [CrossRef] [Green Version]
- Huo, Y.; Pei, Y.; Tian, Y.; Zhang, Z.; Li, K.; Liu, J.; Xiao, S.; Chen, H.; Liu, J. IRREGULAR POLLEN EXINE2 encodes a GDSL lipase essential for male fertility in maize. Plant Physiol. 2020, 184, 1438–1454. [Google Scholar] [CrossRef] [PubMed]
- Goodman, K.; Paez-Valencia, J.; Pennington, J.; Sonntag, A.; Ding, X.; Lee, H.N.; Ahlquist, P.G.; Molina, I.; Otegui, M.S. ESCRT components ISTL1 and LIP5 are required for tapetal function and pollen viability. Plant Cell 2021, 33, 2850–2868. [Google Scholar] [CrossRef]
- Li, X.-C.; Zhu, J.; Yang, J.; Zhang, G.R.; Xing, W.F.; Zhang, S.; Yang, Z.N. Glycerol-3-phosphate acyltransferase 6 (GPAT6) is important for tapetum development in Arabidopsis and plays multiple roles in plant fertility. Mol. Plant 2012, 5, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Cai, C.F.; Zhu, J.; Lou, Y.; Guo, Z.L.; Xiong, S.X.; Wang, K.; Yang, Z.N. The functional analysis of OsTDF1 reveals a conserved genetic pathway for tapetal development between rice and Arabidopsis. Sci. Bull. 2015, 60, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- Jung, K.H.; Han, M.J.; Lee, Y.S.; Kim, Y.W.; Hwang, I.W.; Kim, M.J.; Kim, Y.K.; Nahm, B.H.; An, G.H. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 2005, 17, 2705–2722. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.D.; Ha, Y.; Lee, J.H.; Park, M.; Bergsma, A.C.; Choi, H.I.; Goritschnig, S.; Kloosterman, B.; van Dijk, P.J.; Choi, D.; et al. Fine mapping of Restorer-of-fertility in pepper (Capsicum annuum L.) identified a candidate gene encoding a pentatricopeptide repeat (PPR)-containing protein. Theor. Appl. Genet. 2016, 129, 2003–2017. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.W.; Chen, Y.J.; Hu, Y.F.; Zhou, Z.Y.; Hu, F.; Dong, J.C.; Chen, W.L.; Cui, J.J.; Wu, Z.M.; Hu, K.L. Fine mapping of restorer-of-fertility gene based on high-density genetic mapping and collinearity analysis in pepper (Capsicum annuum L.). Theor. Appl. Genet. 2020, 133, 889–902. [Google Scholar] [CrossRef]
- Zhang, Z.H.; Zhu, Y.S.; Cao, Y.C.; Yu, H.L.; Bai, R.Q.; Zhao, H.; Zhang, B.X.; Wang, L.H. Fine mapping of the male fertility restoration gene CaRf032 in Capsicum annuum L. Theor. Appl. Genet. 2020, 133, 1177–1187. [Google Scholar] [CrossRef]
- Wei, B.Q.; Bosland, P.W.; Zhang, Z.H.; Wang, Y.F.; Zhang, G.Y.; Wang, L.L.; Yu, J.H. A predicted NEDD8 conjugating enzyme gene identified as a Capsicum candidate Rf gene using bulk segregant RNA sequencing. Hortic. Res.-Engl. 2020, 7, 210. [Google Scholar] [CrossRef]
- Wu, L.; Wang, P.; Wang, Y.H.; Cheng, Q.; Lu, Q.H.; Liu, J.Q.; Li, T.; Ai, Y.X.; Yang, W.C.; Sun, L.; et al. Genome-wide correlation of 36 agronomic traits in the 287 pepper (Capsicum) accessions obtained from the SLAF-seq-based GWAS. Int. J. Mol. Sci. 2019, 20, 5675. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.J.; Huang, W.; Li, D.W.; Yin, Y.X.; Chai, W.G.; Gong, Z.H. A CMS-related gene, ψatp6-2, causes increased ATP hydrolysis activity of the mitochondrial F1Fo-ATP synthase and induces male sterility in pepper (Capsicum annuum L.). Plant Mol. Biol. Rep. 2014, 32, 888–899. [Google Scholar] [CrossRef]
- Li, J.; Pandeya, D.; Jo, Y.D.; Liu, W.Y.; Kang, B.C. Reduced activity of ATP synthase in mitochondria causes cytoplasmic male sterility in chili pepper. Planta 2013, 237, 1097–1109. [Google Scholar] [CrossRef]
- Kim, D.H.; Kang, J.G.; Kim, B.D. Isolation and characterization of the cytoplasmic male sterility-associated orf456 gene of chili pepper (Capsicum annuum L.). Plant Mol. Biol. 2007, 63, 519–532. [Google Scholar] [CrossRef]
- Wang, P.; Lu, Q.; Ai, Y.; Wang, Y.; Li, T.; Wu, L.; Liu, J.; Cheng, Q.; Sun, L.; Shen, H. Candidate gene selection for cytoplasmic male sterility in pepper (Capsicum annuum L.) through whole mitochondrial genome sequencing. Int. J. Mol. Sci. 2019, 20, 578. [Google Scholar] [CrossRef] [Green Version]
- Peterson, R.; Slovin, J.P.; Chen, C. A simplified method for differential staining of aborted and non-aborted pollen grains. Int. J. Plant Biol. 2010, 1, e13. [Google Scholar] [CrossRef] [Green Version]
- Nie, Z.; Zhao, T.; Liu, M.; Dai, J.; He, T.; Lyu, D.; Zhao, J.; Yang, S.; Gai, J. Molecular mapping of a novel male-sterile gene msNJ in soybean [Glycine max (L.) Merr.]. Plant Reprod. 2019, 32, 371–380. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Teng, L.; Feng, Y.C.; Guo, S.T.; Wang, P.L.; Qi, T.F.; Yue, Y.M.; Wang, S.X.; Zhang, S.N.; Tang, C.X.; La, T.; et al. The pan-cancer lncRNA PLANE regulates an alternative splicing program to promote cancer pathogenesis. Nat. Commun. 2021, 12, 3734. [Google Scholar] [CrossRef]
- Qin, C.; Yu, C.; Shen, Y.; Fang, X.; Chen, L.; Min, J.; Cheng, J.; Zhao, S.; Xu, M.; Luo, Y.; et al. Whole-genome sequencing of cultivated and wild peppers provides insights into Capsicum domestication and specialization. Proc. Natl. Acad. Sci. USA 2014, 111, 5135–5140. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.D.; Choi, Y.; Kim, D.H.; Kim, B.D.; Kang, B.C. Extensive structural variations between mitochondrial genomes of CMS and normal peppers (Capsicum annuum L.) revealed by complete nucleotide sequencing. BMC Genom. 2014, 15, 561. [Google Scholar] [CrossRef] [Green Version]
- Jo, Y.D.; Park, J.; Kim, J.; Song, W.; Hur, C.G.; Lee, Y.H.; Kang, B.C. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 2011, 30, 217–229. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yi, Z.; Chen, J.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Goseq: Gene ontology testing for RNA-seq datasets. R. Bioconduct. 2012, 8, 1–25. [Google Scholar]
- Mao, X.Z.; Cai, T.; Olyarchuk, J.G.; Wei, L.P. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Lv, J.H.; Liu, Z.B.; Liu, Y.H.; Ou, L.J.; Deng, M.H.; Wang, J.; Song, J.S.; Ma, Y.Q.; Chen, W.C.; Zhang, Z.Q.; et al. Comparative transcriptome analysis between cytoplasmic male-sterile line and its maintainer during the floral bud development of pepper. Hortic. Plant J. 2020, 6, 89–98. [Google Scholar] [CrossRef]
- Qiu, Y.; Liao, L.; Jin, X.; Mao, D.; Liu, R. Analysis of the meiotic transcriptome reveals the genes related to the regulation of pollen abortion in cytoplasmic male-sterile pepper (Capsicum annuum L.). Gene 2018, 641, 8–17. [Google Scholar] [CrossRef]
- Browne, R.G.; Li, S.F.; Iacuone, S.; Dolferus, R.; Parish, R.W. Differential responses of anthers of stress tolerant and sensitive wheat cultivars to high temperature stress. Planta 2021, 254, 4. [Google Scholar] [CrossRef]
- Nie, Z. Discovery of a New Cytoplasmic Male-Sterile Source, Detection of a Nuclear Male-Sterile Gene and QTL-Allele Constitutions of Heterosis in Soybean [Glycine max (L.) Merr.]. Ph.D. Thesis, Nanjing Agricultural University, Nanjing, China, 2017. [Google Scholar]
- Yang, Y.; Bao, S.Y.; Zhou, X.H.; Liu, J.; Zhuang, Y. The key genes and pathways related to male sterility of eggplant revealed by comparative transcriptome analysis. BMC Plant Biol. 2018, 18, 209. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.J.; Park, S.W.; Hong, W.J.; Silva, J.; Liang, W.Q.; Zhang, D.B.; Jung, K.H.; Kim, Y.J. Genome-wide analysis of RopGEF gene family to identify genes contributing to pollen tube growth in rice (Oryza sativa). BMC Plant Biol. 2020, 20, 95. [Google Scholar] [CrossRef] [Green Version]
- Fernández Gómez, J.; Wilson, Z.A. A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 2014, 12, 765–777. [Google Scholar] [CrossRef]
- Xiao, S.; Zang, J.; Pei, Y.; Liu, J.; Liu, J.; Song, W.; Shi, Z.; Su, A.; Zhao, J.; Chen, H. Activation of mitochondrial orf355 gene expression by a nuclear-encoded DREB transcription factor causes cytoplasmic male sterility in maize. Mol. Plant 2020, 13, 1270–1283. [Google Scholar] [CrossRef]
- Jung, Y.J.; Kim, D.H.; Lee, H.J.; Nam, K.H.; Bae, S.; Nou, I.S.; Cho, Y.G.; Kim, M.K.; Kang, K.K. Knockout of SlMS10 Gene (Solyc02g079810) encoding bHLH transcription factor using CRISPR/Cas9 system confers male sterility phenotype in tomato. Plants 2020, 9, 1189. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.M.; Kong, X.D.; Chen, F.; Huang, J.X.; Lou, X.Y.; Zhao, J.Y. Transcriptome analysis of Brassica napus pod using RNA-Seq and identification of lipid-related candidate genes. BMC Genom. 2015, 16, 858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lannert, H.; Gorgas, K.; Meißner, I.; Wieland, F.T.; Jeckel, D. Functional organization of the Golgi apparatus in glycosphingolipid biosynthesis: Lactosylceramide and subsequent glycosphingolipids are formed in the lumen of the late Golgi. J. Biol. Chem. 1998, 273, 2939–2946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobsen, M.K. Pollen development and fertilization in arabidopsis is dependent on the male gametogenesis impaired anthers gene encoding a type v p-type ATPase. Genes Dev. 2005, 19, 2757–2769. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).