Genome-Scale Computational Identification and Characterization of UTR Introns in Atalantia buxifolia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Preparation
2.2. Genome-Wide Identification of A. buxifolia UIs
2.3. Gene Pathway-Enrichment Analysis of UI-Ts
2.4. Cis-Acting Element and Transcription Factor Binding Sites (TFBS) Prediction Analysis of 5UI Sequences
3. Results
3.1. Identification of Introns in A. buxifolia CDSs and 5′ and 3′ Untranslated Regions (UTRs)
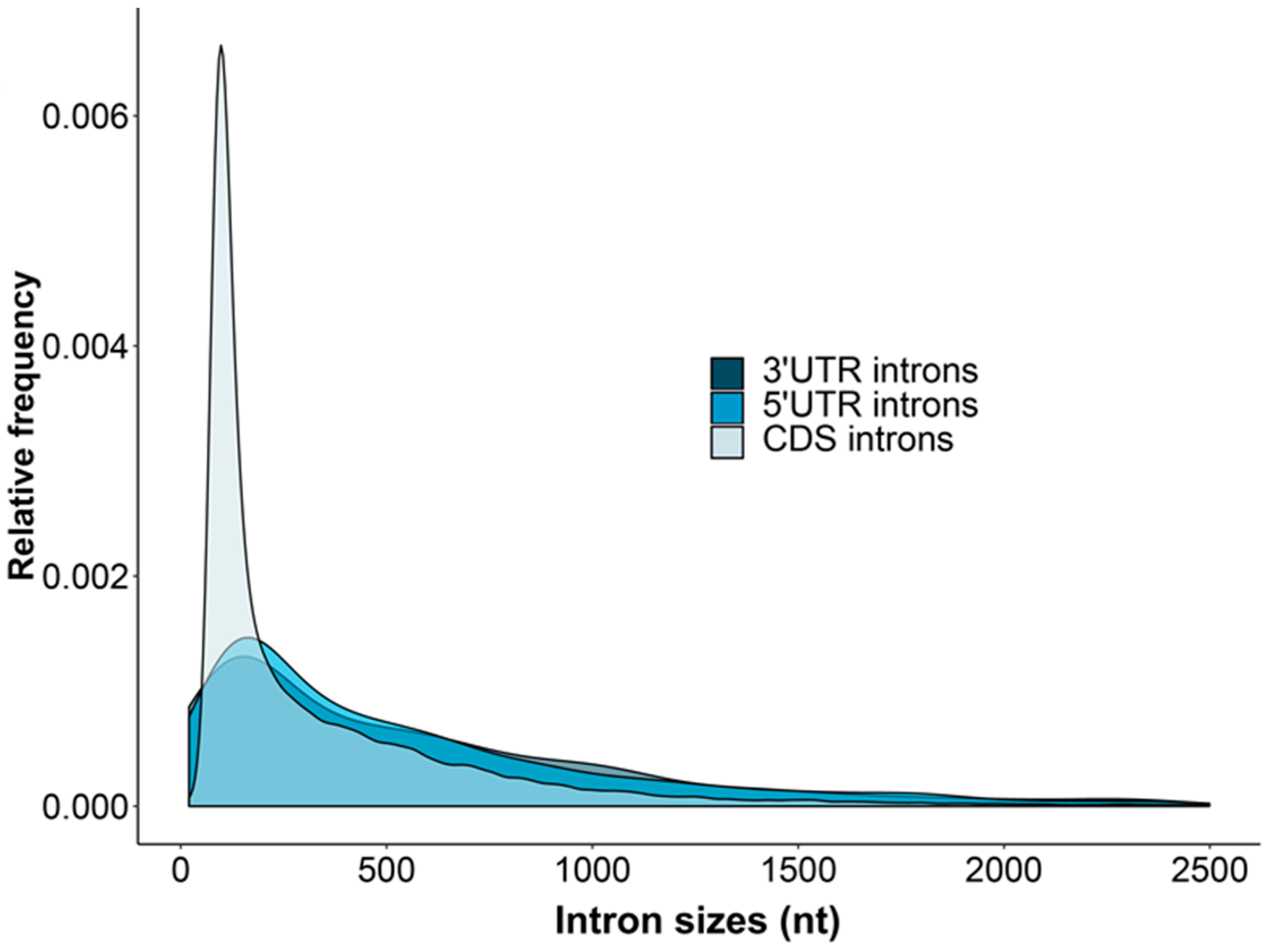
3.2. Intron Sizes and Distributions within UTRs and CDSs
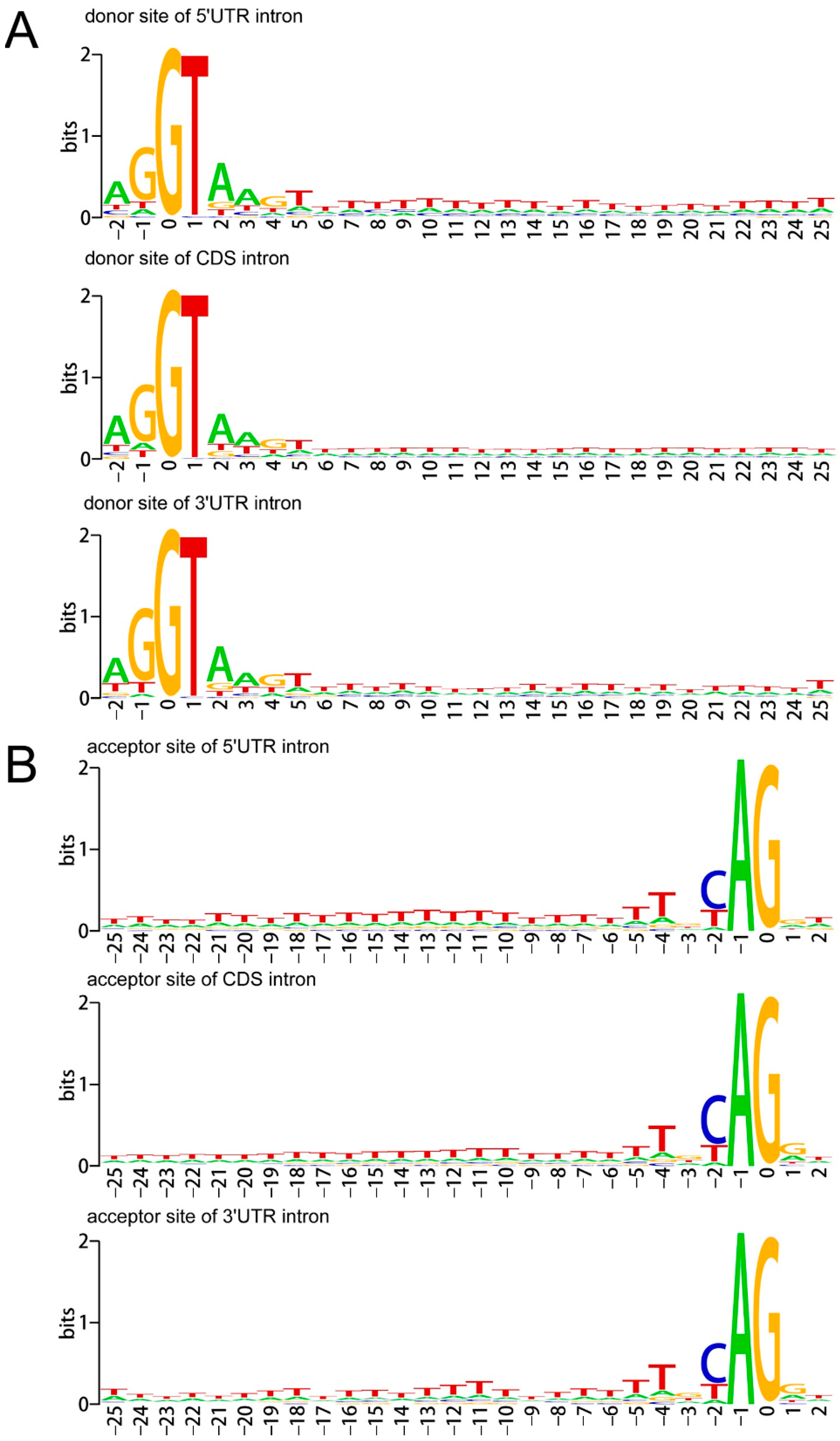
3.3. Nucleotide Conservation around the Splice Junctions
3.4. Cis-Acting Elements and TFBS Prediction Analysis of 5UIs
3.5. Gene Pathway-Enrichment Analysis of UI-Containing Transcripts (UI-Ts)
3.6. ‘RNA’ Related UI-Ts
4. Discussion
4.1. The Lengths of A. buxifolia UIs Were Less Conserved Than CIs, and Most UI-Ts Contain Only One UI
4.2. A/T-Rich Elements around Both Donor Sites and Receptor Sites of A. buxifolia UTRs Are Important for UI Recognition and Removal
4.3. A. buxifolia 5UIs Were Rich of Gene-Expression-Enhancement-Related Elements and TFBSs, Indicating That They Might Contribute Greatly to Gene Expression Regulation
4.4. Many UI-Ts Are Involved in RNA Metabolism
4.5. Most A. buxifolia 3UI-Ts Are Members of PPRP Gene Family, and Many UI-Ts Are Stress-Response-Related or with Unknown Function
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Roy, S.W.; Gilbert, W. The evolution of spliceosomal introns: Patterns, puzzles and progress. Nat. Rev. Genet. 2006, 7, 211–221. [Google Scholar]
- Sambrook, J. Adenovirus amazes at Cold Spring Harbor. Nature 1977, 268, 101–104. [Google Scholar] [CrossRef]
- Bonen, L.; Vogel, J. The ins and outs of group II introns. Trends Genet. 2001, 17, 322–331. [Google Scholar] [CrossRef]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M.; et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar]
- Wahl, M.C.; Will, C.L.; Lührmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar] [CrossRef] [Green Version]
- Le Hir, H.; Nott, A.; Moore, M.J. How introns influence and enhance eukaryotic gene expression. Trends Biochem. Sci. 2003, 28, 215–220. [Google Scholar] [CrossRef]
- Moore, M.J.; Proudfoot, N.J. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell 2009, 136, 688–700. [Google Scholar] [CrossRef] [Green Version]
- Chorev, M.; Carmel, L. The function of introns. Front. Genet. 2012, 3, 55. [Google Scholar] [CrossRef] [Green Version]
- Clark, A.J.; Archibald, A.L.; McClenaghan, M.; Simons, J.P.; Wallace, R.; Whitelaw, C.B. Enhancing the efficiency of transgene expression. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1993, 339, 225–232. [Google Scholar]
- Laxa, M. Intron-Mediated Enhancement: A Tool for Heterologous Gene Expression in Plants? Front. Plant Sci. 2016, 7, 1977. [Google Scholar] [CrossRef] [Green Version]
- Mascarenhas, D.; Mettler, I.J.; Pierce, D.A.; Lowe, H.W. Intron-mediated enhancement of heterologous gene expression in maize. Plant Mol. Biol. 1990, 15, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Callis, J.; Fromm, M.; Walbot, V. Introns increase gene expression in cultured maize cells. Genes Dev. 1987, 1, 1183–1200. [Google Scholar] [CrossRef] [Green Version]
- Maas, C.; Laufs, J.; Grant, S.; Korfhage, C.; Werr, W. The combination of a novel stimulatory element in the first exon of the maize Shrunken-1 gene with the following intron 1 enhances reporter gene expression up to 1000-fold. Plant Mol. Biol. 1991, 16, 199–207. [Google Scholar] [CrossRef]
- Dean, C.; Favreau, M.; Bond-Nutter, D.; Bedbrook, J.; Dunsmuir, P. Sequences downstream of translation start regulate quantitative expression of two petunia rbcS genes. Plant Cell 1989, 1, 201–208. [Google Scholar] [PubMed] [Green Version]
- Gidekel, M.; Jimenez, B.; Herrera-Estrella, L. The first intron of the Arabidopsis thaliana gene coding for elongation factor 1 beta contains an enhancer-like element. Gene 1996, 170, 201–206. [Google Scholar] [CrossRef]
- Pesole, G.; Mignone, F.; Gissi, C.; Grillo, G.; Licciulli, F.; Liuni, S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 2001, 276, 73–81. [Google Scholar] [CrossRef]
- Masuda, S.; Das, R.; Cheng, H.; Hurt, E.; Dorman, N.; Reed, R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005, 19, 1512–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nott, A.; Meislin, S.H.; Moore, M.J. A quantitative analysis of intron effects on mammalian gene expression. RNA 2003, 9, 607–617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Majewski, J.; Ott, J. Distribution and characterization of regulatory elements in the human genome. Genome Res. 2002, 12, 1827–1836. [Google Scholar] [CrossRef] [Green Version]
- Furger, A.; O’Sullivan, J.M.; Binnie, A.; Lee, B.A.; Proudfoot, N.J. Promoter proximal splice sites enhance transcription. Genes Dev. 2002, 16, 2792–2799. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, K.; Wassarman, K.M.; Wolffe, A.P. Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J. 1998, 17, 2107–2121. [Google Scholar] [CrossRef] [Green Version]
- Grant, T.N.L.; De La Torre, C.M.; Zhang, N.; Finer, J.J. Synthetic introns help identify sequences in the 5′UTR intron of the Glycine max polyubiquitin (Gmubi) promoter that give increased promoter activity. Planta 2017, 245, 849–860. [Google Scholar] [CrossRef]
- Laxa, M.; Müller, K.; Lange, N.; Doering, L.; Pruscha, J.T.; Peterhänsel, C. The 5′UTR Intron of Arabidopsis GGT1 Aminotransferase Enhances Promoter Activity by Recruiting RNA Polymerase II. Plant Physiol. 2016, 172, 313–327. [Google Scholar] [CrossRef] [Green Version]
- Kamo, K.; Kim, A.Y.; Park, S.H.; Joung, Y.H. The 5′UTR-intron of the Gladiolus polyubiquitin promoter GUBQ1 enhances translation efficiency in Gladiolus and Arabidopsis. BMC Plant Biol. 2012, 12, 79. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Kim, H.; Shin, J.S.; Chung, C.H.; Ohlrogge, J.B.; Suh, M.C. Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol. Genet. Genom. 2006, 276, 351–368. [Google Scholar] [CrossRef] [PubMed]
- Weischenfeldt, J.; Damgaard, I.; Bryder, D.; Theilgaard-Mönch, K.; Thoren, L.A.; Nielsen, F.C.; Jacobsen, S.E.W.; Nerlov, C.; Porse, B.T. NMD is essential for hematopoietic stem and progenitor cells and for eliminating by-products of programmed DNA rearrangements. Genes Dev. 2008, 22, 1381–1396. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.Z.; Grate, L.; Donohue, J.P.; Preston, C.; Nobida, N.; O’Brien, G.; Shiue, L.; Clark, T.A.; Blume, J.E.; Ares, M., Jr. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007, 21, 708–718. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007, 76, 51–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mendell, J.T.; Sharifi, N.A.; Meyers, J.L.; Martinez-Murillo, F.; Dietz, H.C. Nonsense surveillance regulates expression of diverse classes of mammalian transcripts and mutes genomic noise. Nat. Genet. 2004, 36, 1073–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Sun, X.; Qian, Y.; Maquat, L.E. Intron function in the nonsense-mediated decay of beta-globin mRNA: Indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA 1998, 4, 801–815. [Google Scholar] [CrossRef]
- Yepiskoposyan, H.; Aeschimann, F.; Nilsson, D.; Okoniewski, M.; Mühlemann, O. Autoregulation of the nonsense-mediated mRNA decay pathway in human cells. RNA 2011, 17, 2108–2118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rüegsegger, U.; Leber, J.H.; Walter, P. Block of HAC1 mRNA translation by long-range base pairing is released by cytoplasmic splicing upon induction of the unfolded protein response. Cell 2001, 107, 103–114. [Google Scholar] [CrossRef] [Green Version]
- Bruno, I.G.; Karam, R.; Huang, L.; Bhardwaj, A.; Lou, C.H.; Shum, E.Y.; Song, H.W.; Corbett, M.A.; Gifford, W.D.; Gecz, J.; et al. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 2011, 42, 500–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McIlwain, D.R.; Pan, Q.; Reilly, P.T.; Elia, A.J.; McCracken, S.; Wakeham, A.C.; Itie-Youten, A.; Blencowe, B.J.; Mak, T.W. Smg1 is required for embryogenesis and regulates diverse genes via alternative splicing coupled to nonsense-mediated mRNA decay. Proc. Natl. Acad. Sci. USA 2010, 107, 12186–12191. [Google Scholar] [CrossRef] [Green Version]
- Wittmann, J.; Hol, E.M.; Jäck, H.M. hUPF2 silencing identifies physiologic substrates of mammalian nonsense-mediated mRNA decay. Mol. Cell Biol. 2006, 26, 1272–1287. [Google Scholar] [CrossRef] [Green Version]
- Saltzman, A.L.; Pan, Q.; Blencowe, B.J. Regulation of alternative splicing by the core spliceosomal machinery. Genes Dev. 2011, 25, 373–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, Q.; Saltzman, A.L.; Yoon, K.K.; Misquitta, C.; Shai, O.; Maquat, L.E.; Frey, B.J.; Blencowe, B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes Dev. 2006, 20, 153–158. [Google Scholar] [CrossRef] [Green Version]
- Hong, X.; Scofield, D.G.; Lynch, M. Intron size, abundance, and distribution within untranslated regions of genes. Mol. Biol. Evol. 2006, 23, 2392–2404. [Google Scholar] [CrossRef] [Green Version]
- Chung, B.Y.W.; Simons, C.; Firth, A.E.; Brown, C.M.; Hellens, R.P. Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genom. 2006, 7, 120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, S.W.; Penny, D.; Neafsey, D.E. Evolutionary conservation of UTR intron boundaries in Cryptococcus. Mol. Biol. Evol. 2007, 24, 1140–1148. [Google Scholar] [CrossRef] [Green Version]
- Cenik, C.; Derti, A.; Mellor, J.C.; Berriz, G.F.; Roth, F.P. Genome-wide functional analysis of human 5′ untranslated region introns. Genome Biol. 2010, 11, R29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, X.; Wu, J.; Mensah, R.A.; Tian, N.; Liu, J.; Liu, F.; Chen, J.; Che, J.; Guo, Y.; Wu, B.; et al. Genome-wide identification and characterization of UTR-introns of Citrus sinensis. Int. J. Mol. Sci. 2020, 21, 3088. [Google Scholar] [CrossRef] [PubMed]
- Liang, H.X.; Sun, J.J.; Shen, Z.B.; Yu, B.W.; Cui, H.H.; Yin, Y.Q. A novel alkaloid glycoside isolated from Atalantia buxifolia. Nat. Prod. Res. 2020, 34, 3042–3047. [Google Scholar] [CrossRef]
- Chang, F.R.; Li, P.S.; Huang Liu, R.; Hu, H.C.; Hwang, T.L.; Lee, J.C.; Chen, S.L.; Wu, Y.C.; Cheng, Y.B. Bioactive Phenolic Components from the Twigs of Atalantia buxifolia. J. Nat. Prod. 2018, 81, 1534–1539. [Google Scholar] [CrossRef]
- Yang, Y.Y.; Yang, W.; Zuo, W.J.; Zeng, Y.B.; Liu, S.B.; Mei, W.L.; Dai, H.F. Two new acridone alkaloids from the branch of Atalantia buxifolia and their biological activity. J. Asian Nat. Prod. Res. 2013, 15, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Nat. Biotechnol. 2014, 32, 656–662. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. MAPMAN: A user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lurin, C.; Andrés, C.; Aubourg, S.; Bellaoui, M.; Bitton, F.; Bruyère, C.; Caboche, M.; Debast, C.; Gualberto, J.; Hoffmann, B.; et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 2004, 16, 2089–2103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, J.W.; Simpson, C.G.; Thow, G.; Clark, G.P.; Jennings, S.N.; Medina-Escobar, N.; Haupt, S.; Chapman, S.C.; Oparka, K.J. Splicing signals and factors in plant intron removal. Biochem. Soc. Trans. 2002, 30, 146–149. [Google Scholar] [CrossRef]
- Goguel, V.; Rosbash, M. Splice site choice and splicing efficiency are positively influenced by pre-mRNA intramolecular base pairing in yeast. Cell 1993, 72, 893–901. [Google Scholar]
- Rohs, R.; West, S.M.; Sosinsky, A.; Liu, P.; Mann, R.S.; Honig, B. The role of DNA shape in protein-DNA recognition. Nature 2009, 461, 1248–1253. [Google Scholar] [CrossRef] [Green Version]
- Badis, G.; Berger, M.F.; Philippakis, A.A.; Talukder, S.; Gehrke, A.R.; Jaeger, S.A.; Chan, E.T.; Metzler, G.; Vedenko, A.; Chen, X.; et al. Diversity and complexity in DNA recognition by transcription factors. Science 2009, 324, 1720–1723. [Google Scholar] [CrossRef] [Green Version]
- Hecker, A.; Brand, L.H.; Peter, S.; Simoncello, N.; Kilian, J.; Harter, K.; Gaudin, V.; Wanke, D. The Arabidopsis GAGA-Binding Factor Basic Pentacysteine6 Recruits the Polycomb-Repressive Complex1 Component Like Heterochromatin Protein1 to GAGA DNA Motifs. Plant Physiol. 2015, 168, 1013–1024. [Google Scholar] [CrossRef]
- Simonini, S.; Kater, M.M. Class I Basic Pentacysteine factors regulate Homeobox genes involved in meristem size maintenance. J. Exp. Bot. 2014, 65, 1455–1465. [Google Scholar] [CrossRef] [Green Version]
- Santi, L.; Wang, Y.; Stile, M.R.; Berendzen, K.; Wanke, D.; Roig, C.; Pozzi, C.; Müller, K.; Müller, J.; Rohde, W.; et al. The GA octodinucleotide repeat binding factor BBR participates in the transcriptional regulation of the homeobox gene Bkn3. Plant J. 2003, 34, 813–826. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Sivamani, E.; Azhakanandam, K.; Samadder, P.; Li, X.; Qu, R. Gene expression enhancement mediated by the 5′UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol. Genet. Genom. 2008, 279, 563–572. [Google Scholar] [CrossRef]
- Wang, J.; Oard, J.H. Rice ubiquitin promoters: Deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep. 2003, 22, 129–134. [Google Scholar] [CrossRef] [PubMed]
- McElroy, D.; Zhang, W.; Cao, J.; Wu, R. Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 1990, 2, 163–171. [Google Scholar]
- Rose, A.B.; Elfersi, T.; Parra, G.; Korf, I. Promoter-proximal introns in Arabidopsis thaliana are enriched in dispersed signals that elevate gene expression. Plant Cell 2008, 20, 543–551. [Google Scholar] [CrossRef] [Green Version]
- Rose, A.B. The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J. 2004, 40, 744–751. [Google Scholar] [CrossRef]
- Samadder, P.; Sivamani, E.; Lu, J.; Li, X.; Qu, R. Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol. Genet. Genom. 2008, 279, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Bellini, C. The plant-specific dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [CrossRef] [Green Version]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A.K. Overexpression of OsiSAP8, a member of stress associated protein (SAP) gene family of rice confers tolerance to salt, drought and cold stress in transgenic tobacco and rice. Plant Mol. Biol. 2008, 66, 445–462. [Google Scholar] [CrossRef]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Bicknell, A.A.; Cenik, C.; Chua, H.N.; Roth, F.P.; Moore, M.J. Introns in UTRs: Why we should stop ignoring them. Bioessays 2012, 34, 1025–1034. [Google Scholar] [CrossRef]
- Lorković, Z.J. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci. 2009, 14, 229–236. [Google Scholar] [CrossRef]
- Lambermon, M.H.L.; Fu, Y.; Kirk, D.A.W.; Dupasquier, M.; Filipowicz, W.; Lorković, Z.J. UBA1 and UBA2, two proteins that interact with UBP1, a multifunctional effector of pre-mRNA maturation in plants. Mol. Cell Biol. 2002, 22, 4346–4357. [Google Scholar] [CrossRef] [Green Version]
- Xing, H.; Fu, X.; Yang, C.; Tang, X.; Guo, L.; Li, C.; Xu, C.; Luo, K. Genome-wide investigation of pentatricopeptide repeat gene family in poplar and their expression analysis in response to biotic and abiotic stresses. Sci. Rep. 2018, 8, 2817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmitz-Linneweber, C.; Small, I. Pentatricopeptide repeat proteins: A socket set for organelle gene expression. Trends Plant Sci. 2008, 13, 663–670. [Google Scholar] [CrossRef]
- Laluk, K.; Abuqamar, S.; Mengiste, T. The Arabidopsis mitochondria-localized pentatricopeptide repeat protein PGN functions in defense against necrotrophic fungi and abiotic stress tolerance. Plant Physiol. 2011, 156, 2053–2068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, D.; Prasad, A.M.; Srinivasan, R. Pentatricopeptide repeat proteins and their emerging roles in plants. Plant Physiol. Biochem. 2007, 45, 521–534. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zou, Y.; Li, X.; Zhang, Q.; Chen, L.; Wu, H.; Su, D.; Chen, Y.; Guo, J.; Luo, D.; et al. Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 2006, 18, 676–687. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; An, Y.; Li, Y.X.; Li, C.; Shi, Y.; Song, Y.; Zhang, D.; Wang, T.; Li, Y. Candidate Loci for Yield-Related Traits in Maize Revealed by a Combination of MetaQTL Analysis and Regional Association Mapping. Front. Plant Sci. 2017, 8, 2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.W.; Liu, C.; Marx, H.E.; Olmstead, R.G. The pentatricopeptide repeat (PPR) gene family, a tremendous resource for plant phylogenetic studies. New Phytol. 2009, 182, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Ner-Gaon, H.; Halachmi, R.; Savaldi-Goldstein, S.; Rubin, E.; Ophir, R.; Fluhr, R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004, 39, 877–885. [Google Scholar] [CrossRef]
- Simpson, G.G.; Filipowicz, W. Splicing of precursors to mRNA in higher plants: Mechanism, regulation and sub-nuclear organisation of the spliceosomal machinery. Plant Mol. Biol. 1996, 32, 1–41. [Google Scholar] [CrossRef]
- Luehrsen, K.R.; Taha, S.; Walbot, V. Nuclear pre-mRNA processing in higher plants. Prog. Nucleic Acid Res. Mol. Biol. 1994, 47, 149–193. [Google Scholar] [PubMed]
| No. of Sequences | Sequences with Introns | Total Bases (Genomic) | Intron/Sequence | No. of Introns/Nucleotide (mRNA) | |
|---|---|---|---|---|---|
| 5′UTR | 16,218 | 597 | 1.28 × 107 | 0.07 | 8.42 × 10−5 |
| CDS | 28,412 | 21,005 | 3.64 × 107 | 3.99 | 3.11 × 10−3 |
| 3′UTR | 16,337 | 452 | 2.81 × 107 | 0.05 | 3.08 × 10−5 |
| UI Number | 5UI-Ts Number/Percentage (Gene ID) | 3UI-Ts Number/Percentage (Gene ID) |
|---|---|---|
| 1 UI | 765/85.28% (NS) | 527/78.66% (NS) |
| 2 UIs | 106/11.82% (NS) | 90/13.43% (NS) |
| 3 UIs | 16/1.78% (sb11639.1, sb32006.1, sb10151.1, sb18930.1, sb18698.1, sb31744.1, sb37205.1, sb20998.1, sb14514.1, sb29193.1, sb21675.1, sb20177.1, sb34312.1, sb34492.1, sb36282.1, sb12383.1) | 25/3.73% (sb13064.1, sb32006.1, sb37841.1, sb18145.1, sb18385.1, sb34575.1, sb33241.1, sb37027.1, sb32422.1, sb12195.1, sb24162.1, sb10905.1, sb20663.1, sb32886.1, sb22221.1, sb35222.1, sb26419.1, sb26292.1, sb26045.1, sb22509.1, sb37886.1, sb18573.1, sb20840.1, sb15172.1, sb28503.1) |
| 4 UIs | 4/0.45% (sb19593.1, sb20478.1, sb12332.1, sb28722.1) | 4/0.60% (sb17114.1, sb31386.1, sb26143.1, sb25688.1) |
| 5 UIs | 3/0.33% (sb27690.1, sb31747.1, sb28046.1) | 4/0.60% (sb13718.1, sb31879.1, sb10627.1, sb31747.1) |
| 6 UIs | 1/0.11% (sb26751.1) | 2/0.30% (sb25651.1, sb31448.1) |
| 7 UIs | 1/0.11% (sb14804.1) | 0 |
| 8 UIs | 1/0.11% (sb36468.1) | 0 |
| Function | Elements Number | 5UI Number (Ratio) |
|---|---|---|
| core promoter element around −30 of transcription start | 12,106 | 893 (82.92%) |
| common cis-acting element in promoter and enhancer regions | 5538 | 948 (88.02%) |
| light responsive element | 1297 | 271 (25.16%) |
| part of a conserved DNA module involved in light responsiveness | 1003 | 489 (45.40%) |
| part of a light responsive element | 904 | 491 (45.59%) |
| cis-acting regulatory element essential for the anaerobic induction | 768 | 426 (39.55%) |
| cis-acting regulatory element involved in the MeJA-responsiveness | 678 | 252 (23.40%) |
| cis-acting regulatory element involved in light responsiveness | 486 | 270 (25.7%) |
| cis-acting element involved in the abscisic acid responsiveness | 430 | 246 (22.84%) |
| light responsive element | 392 | 271 (25.16%) |
| Family | Number (Ratio) |
|---|---|
| BARLEY B RECOMBINANT/BASIC PENTACYSTEINE (BBR-BPC) | 268 (24.54%) |
| MADS intervening keratin-like and C-terminal -type MADS (MIKC_MADS) | 266 (24.35%) |
| APETALA2/Ethylene-Responsive factor (AP2) | 176 (16.12%) |
| DNA binding with one finger (Dof) | 149 (13.64%) |
| The three-amino-acid-loop-extension (TALE) | 53 (4.85%) |
| GIBBERELLIC-ACID INSENSITIVE, REPRESSOR of GAI and SCARECROW (GRAS) | 50 (4.58%) |
| GATA | 25 (2.29%) |
| The Cys 2 His 2-like fold group (C2H2) | 24 (2.20%) |
| NODULE-INCEPTION-like (Nin-like) | 22 (2.01%) |
| Leafy (LFY) | 10 (0.92%) |
| BIN | Pathway Name | Number | p-Value |
|---|---|---|---|
| 35.1.5 | not assigned. no ontology. pentatricopeptide (PPR) repeat-containing protein | 139 | 2.45 × 10−22 |
| 35.1 | not assigned. no ontology | 217 | 3.40 × 10−16 |
| 35 | not assigned | 733 | 2.57 × 10−4 |
| 27.3 | RNA. regulation of transcription | 199 | 2.77 × 10−4 |
| 27.4 | RNA. RNA binding | 11 | 4.14 × 10−3 |
| BIN | Pathway Name | 5UI Number | BIN | Pathway Name | 3UI Number |
|---|---|---|---|---|---|
| 35.2 | not assigned. unknown | 328 | 35.2 | not assigned. unknown | 221 |
| 27 | RNA | 160 | 35.1 | not assigned. no ontology | 157 |
| 27.3 | RNA. regulation of transcription | 151 | 35.1.5 | not assigned. no ontology. pentatricopeptide (PPR) repeat-containing protein | 119 |
| 29 | protein | 120 | 27 | RNA | 85 |
| 35.1 | not assigned. no ontology | 77 | 27.3 | RNA. regulation of transcription | 62 |
| 29.5 | protein. degradation | 60 | 29 | protein | 57 |
| 29.5.11 | protein. degradation. ubiquitin | 55 | 30 | signaling | 32 |
| 30 | signaling | 48 | 20 | stress | 31 |
| 29.5.11.4 | protein. degradation. ubiquitin. E3 | 45 | 29.5 | protein. degradation | 30 |
| 20 | stress | 43 | 29.5.11 | protein. degradation. ubiquitin | 25 |
| BIN | Pathway Name | 5UI-Ts | 3UI-Ts | 5+3UI-Ts |
|---|---|---|---|---|
| 27.3.99 | RNA. regulation of transcription. unclassified | 30 | 12 | 2 |
| 27.3.6 | RNA. regulation of transcription. bHLH, basic Helix-Loop-Helix family | 15 | 4 | 1 |
| 27.3.69 | RNA. regulation of transcription. SET-domain transcriptional regulator family | 12 | 8 | 3 |
| 27.3.29 | RNA. regulation of transcription. TCP transcription factor family | 11 | 6 | 2 |
| 27.3.11 | RNA. regulation of transcription. C2H2 zinc finger family | 11 | 3 | 1 |
| 27.3.21 | RNA. regulation of transcription. GRAS transcription factor family | 10 | 3 | 1 |
| 27.4 | RNA. RNA binding | 0 | 11 | 0 |
| 27.3.67 | RNA. regulation of transcription. putative transcription regulator | 9 | 3 | 2 |
| 27.3.30 | RNA. regulation of transcription. Trihelix, Triple-Helix transcription factor family | 4 | 6 | 0 |
| 27.3.3 | RNA. regulation of transcription. AP2/EREBP, APETALA2/Ethylene-responsive element binding protein family | 5 | 3 | 0 |
| 27.3.12 | RNA. regulation of transcription. C3H zinc finger family | 6 | 1 | 1 |
| 27.1 | RNA. processing | 4 | 3 | 0 |
| 27.1.1 | RNA. processing. splicing | 0 | 5 | 0 |
| 27.3.25 | RNA. regulation of transcription. MYB domain transcription factor family | 5 | 0 | 0 |
| 27.3.37 | RNA. regulation of transcription. AS2, Lateral Organ Boundaries Gene Family | 5 | 0 | 0 |
| 27.3.16 | RNA. regulation of transcription. CCAAT box binding factor family, HAP5 | 0 | 4 | 0 |
| 27.3.19 | RNA. regulation of transcription. EIN3-like (EIL) transcription factor family | 4 | 0 | 0 |
| 27.3.68 | RNA. regulation of transcription. PWWP domain protein | 4 | 1 | 1 |
| 27.3.8 | RNA. regulation of transcription. C2C2 (Zn) DOF zinc finger family | 3 | 1 | 0 |
| 27.2 | RNA. transcription | 3 | 0 | 0 |
| 27.1.2 | RNA. processing. RNA helicase | 0 | 3 | 0 |
| 27.3.1 | RNA. regulation of transcription. ABI3/VP1-related B3-domain-containing transcription factor family | 3 | 0 | 0 |
| 27.3.20 | RNA. regulation of transcription. G2-like transcription factor family, GARP | 0 | 3 | 0 |
| 27.3.24 | RNA. regulation of transcription. MADS box transcription factor family | 3 | 0 | 0 |
| 27.3.52 | RNA. regulation of transcription. Global transcription factor group | 3 | 0 | 0 |
| 27.3.80 | RNA. regulation of transcription. zf-HD | 3 | 0 | 0 |
| 27.1.19 | RNA. processing. ribonucleases | 1 | 1 | 1 |
| 27.3.35 | RNA. regulation of transcription. bZIP transcription factor family | 2 | 0 | 0 |
| 27.3.15 | RNA. regulation of transcription. CCAAT box binding factor family, HAP3 | 0 | 1 | 0 |
| 27.3.26 | RNA. regulation of transcription. MYB-related transcription factor family | 0 | 1 | 0 |
| 27.3.49 | RNA. regulation of transcription. GeBP like | 0 | 1 | 0 |
| 27.3.73 | RNA. regulation of transcription. Zn-finger (CCHC) | 1 | 0 | 0 |
| 27.3.75 | RNA. regulation of transcription. GRP | 0 | 1 | 0 |
| 27.3.84 | RNA. regulation of transcription. BBR/BPC | 1 | 0 | 0 |
| 27.3.89 | RNA. regulation of transcription. ovate family OFP | 1 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.; Shi, X.; Wu, J.; Zhang, Y.; Lü, P. Genome-Scale Computational Identification and Characterization of UTR Introns in Atalantia buxifolia. Horticulturae 2021, 7, 556. https://doi.org/10.3390/horticulturae7120556
Cheng C, Shi X, Wu J, Zhang Y, Lü P. Genome-Scale Computational Identification and Characterization of UTR Introns in Atalantia buxifolia. Horticulturae. 2021; 7(12):556. https://doi.org/10.3390/horticulturae7120556
Chicago/Turabian StyleCheng, Chunzhen, Xiaobao Shi, Junwei Wu, Yongyan Zhang, and Peitao Lü. 2021. "Genome-Scale Computational Identification and Characterization of UTR Introns in Atalantia buxifolia" Horticulturae 7, no. 12: 556. https://doi.org/10.3390/horticulturae7120556
APA StyleCheng, C., Shi, X., Wu, J., Zhang, Y., & Lü, P. (2021). Genome-Scale Computational Identification and Characterization of UTR Introns in Atalantia buxifolia. Horticulturae, 7(12), 556. https://doi.org/10.3390/horticulturae7120556






