Using a Hybrid Neural Network Model DCNN–LSTM for Image-Based Nitrogen Nutrition Diagnosis in Muskmelon
Abstract
1. Introduction
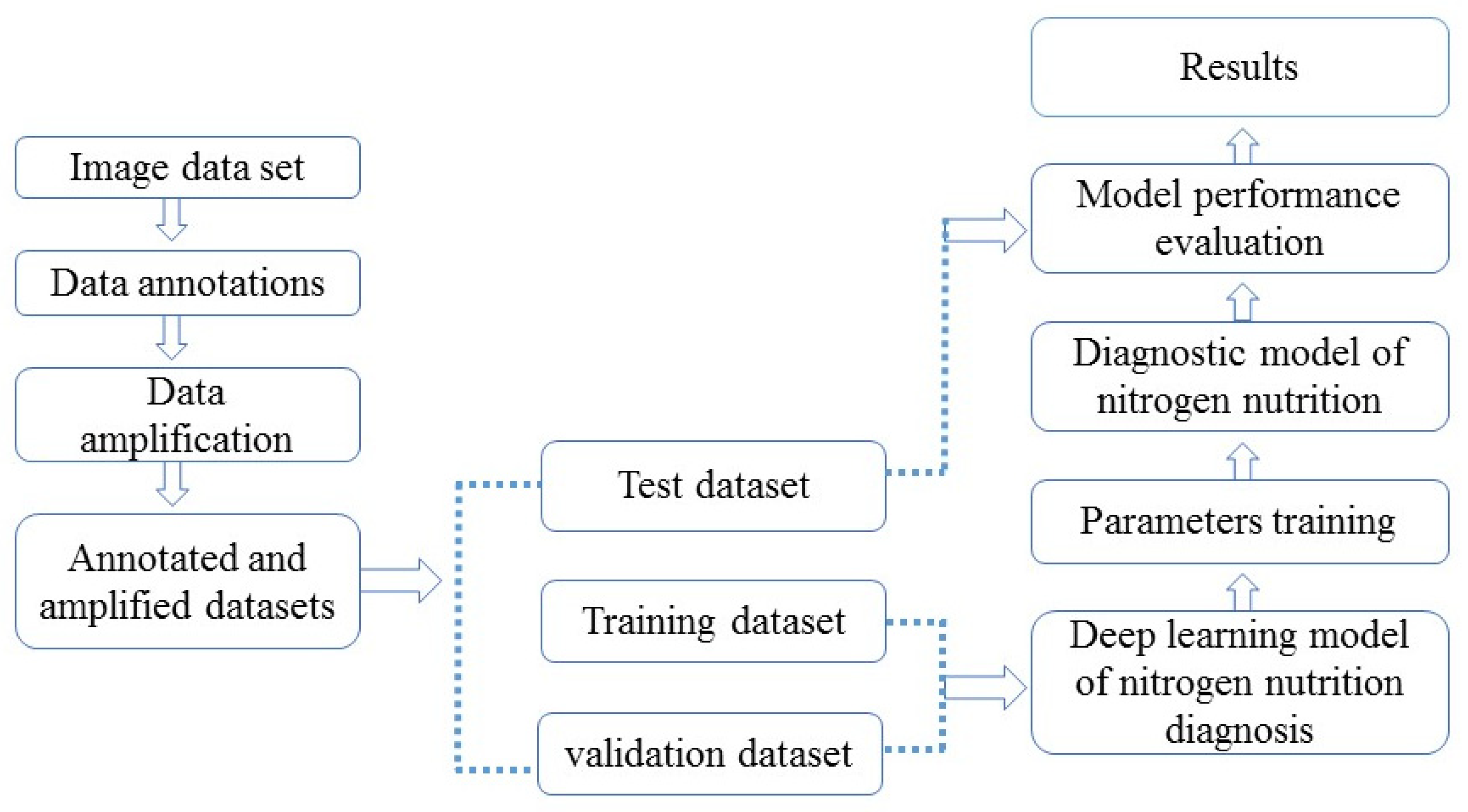
2. Materials and Methods
3. Data Collection
3.1. Measurement of Nitrogen Concentration in Plants
3.2. Leaf Image Acquisition
3.3. Collecting Meteorological Data of Greenhouse
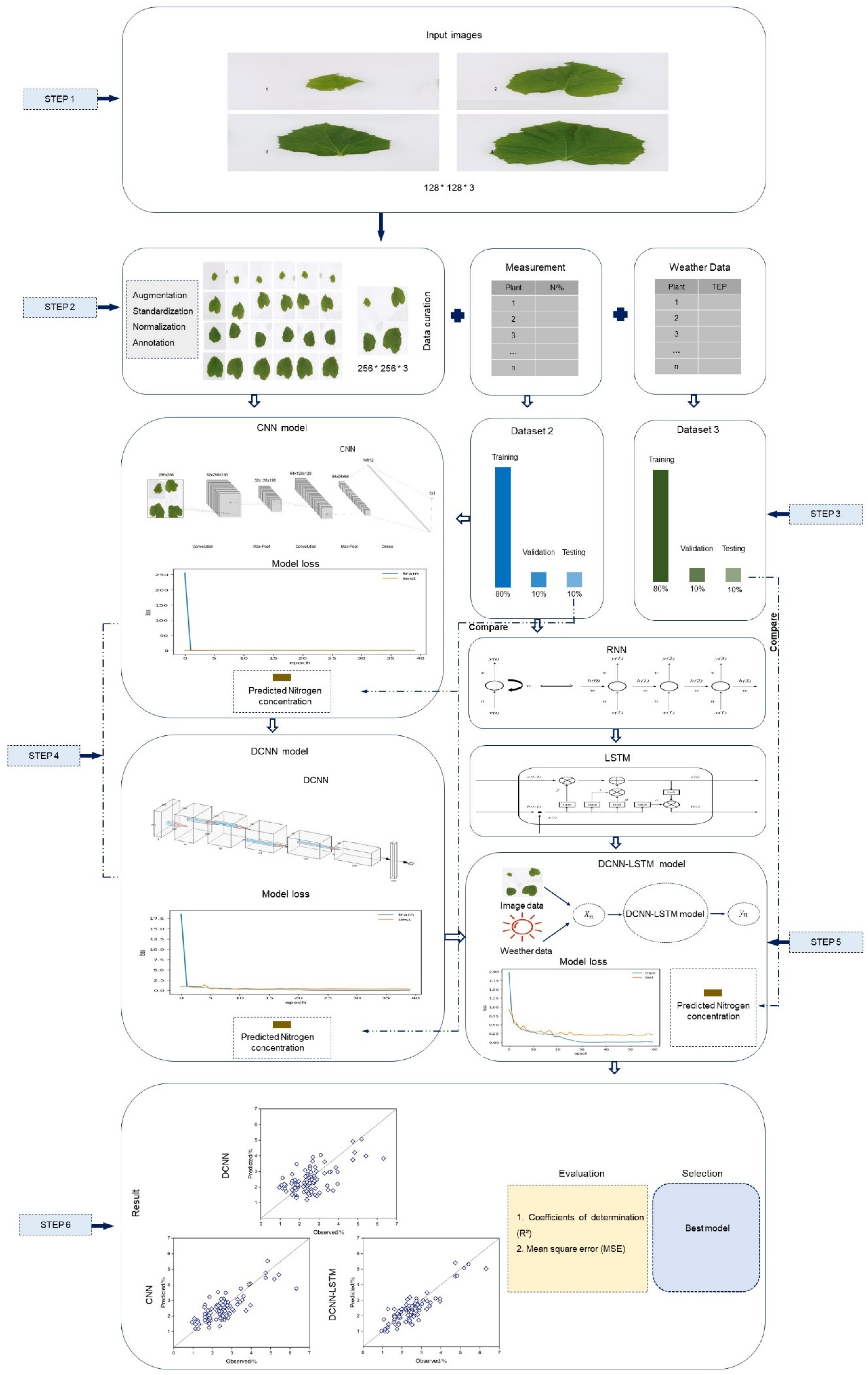
3.4. Establishment of Machine Learning (ML) Model
Extraction of Phenotypical Features
4. Result
4.1. Phenotypical Feature Parameters Screening
4.2. Establishment of Backpropagation Neural Network (BPNN)
4.3. Establishment of Deep Learning Models
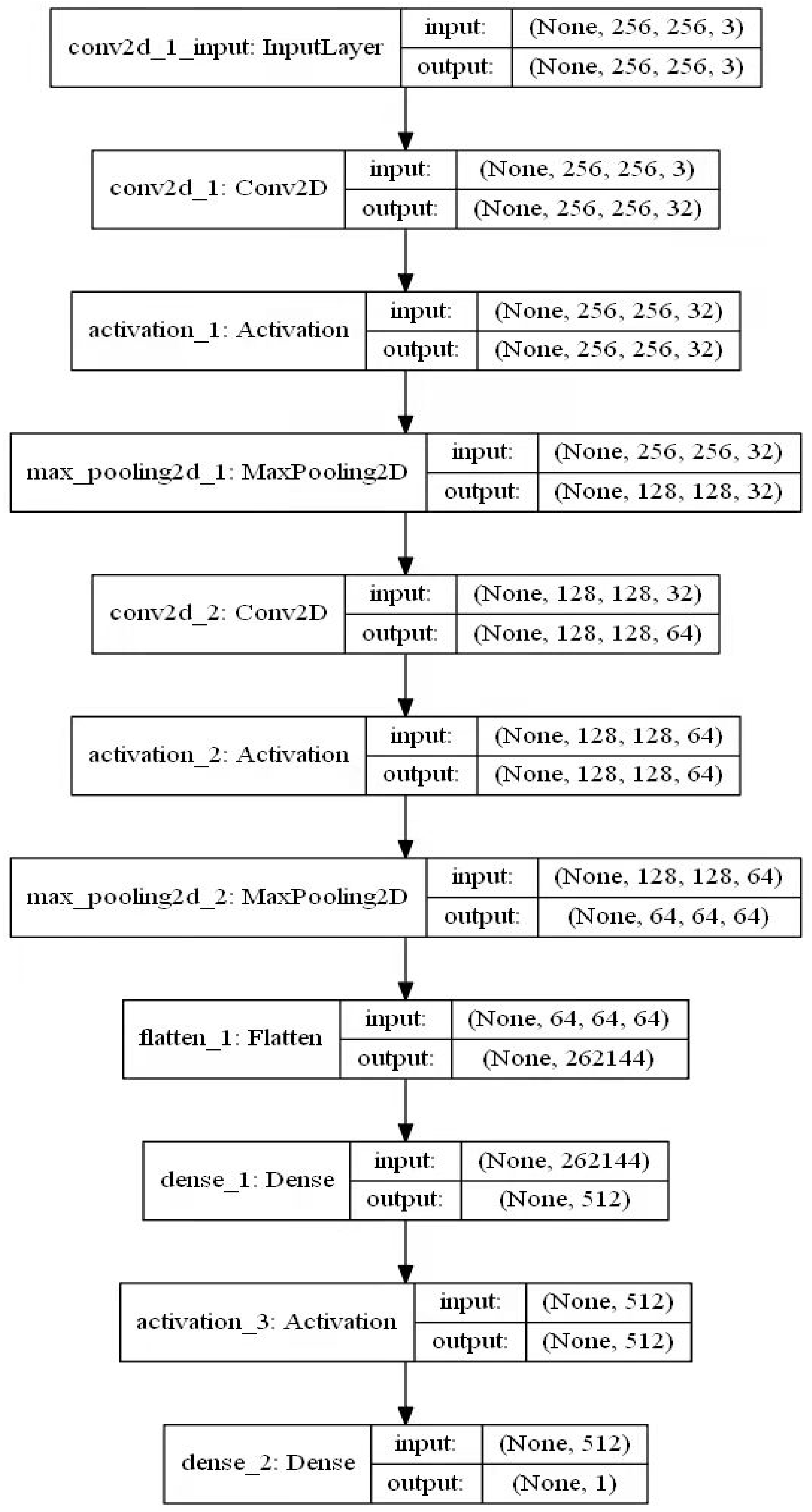
4.3.1. Image Preprocessing
4.3.2. Data Preprocessing of Environmental Factors
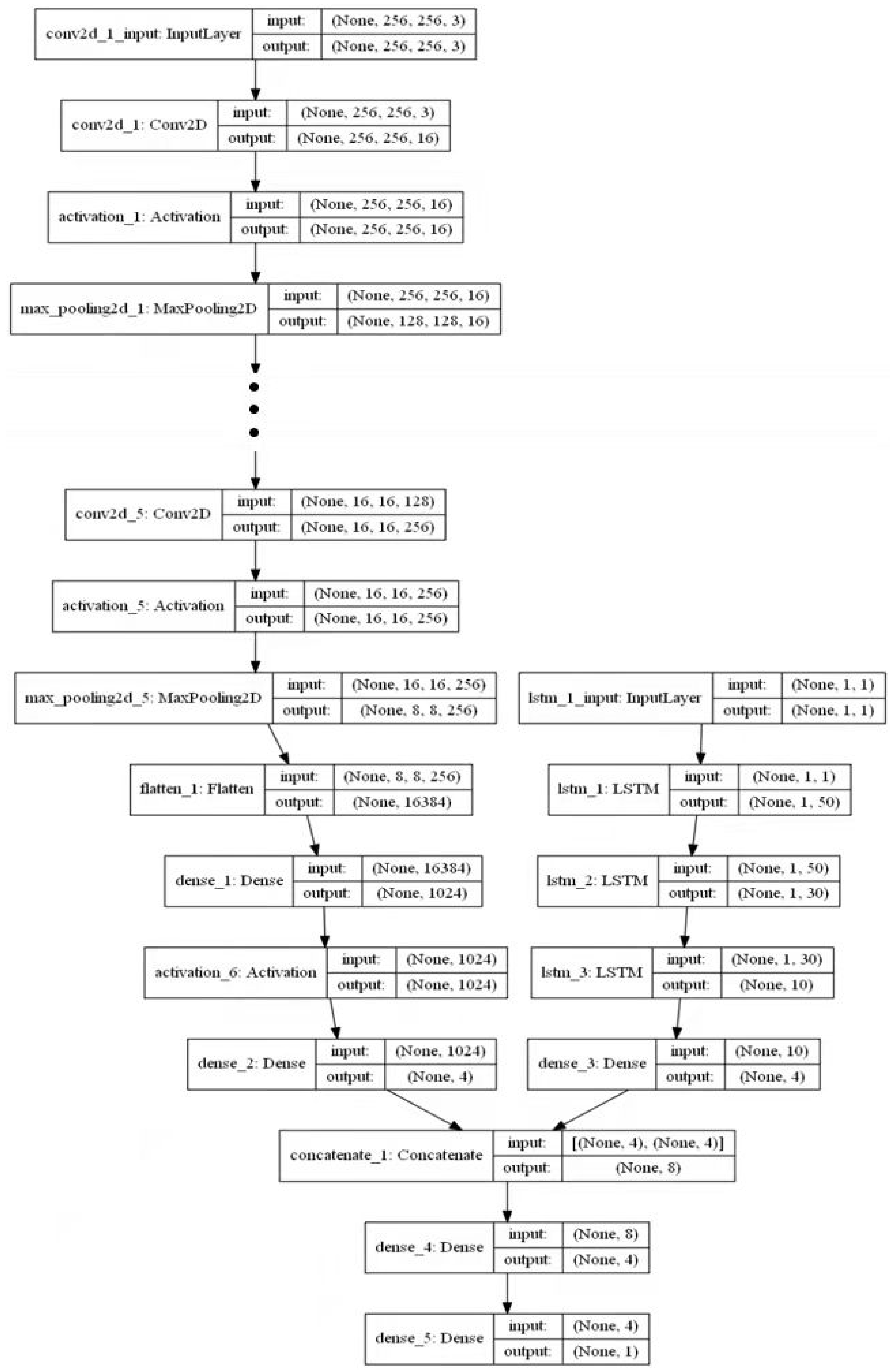
4.4. Establishment of CNN Model
4.5. Establishment of DCNN Model
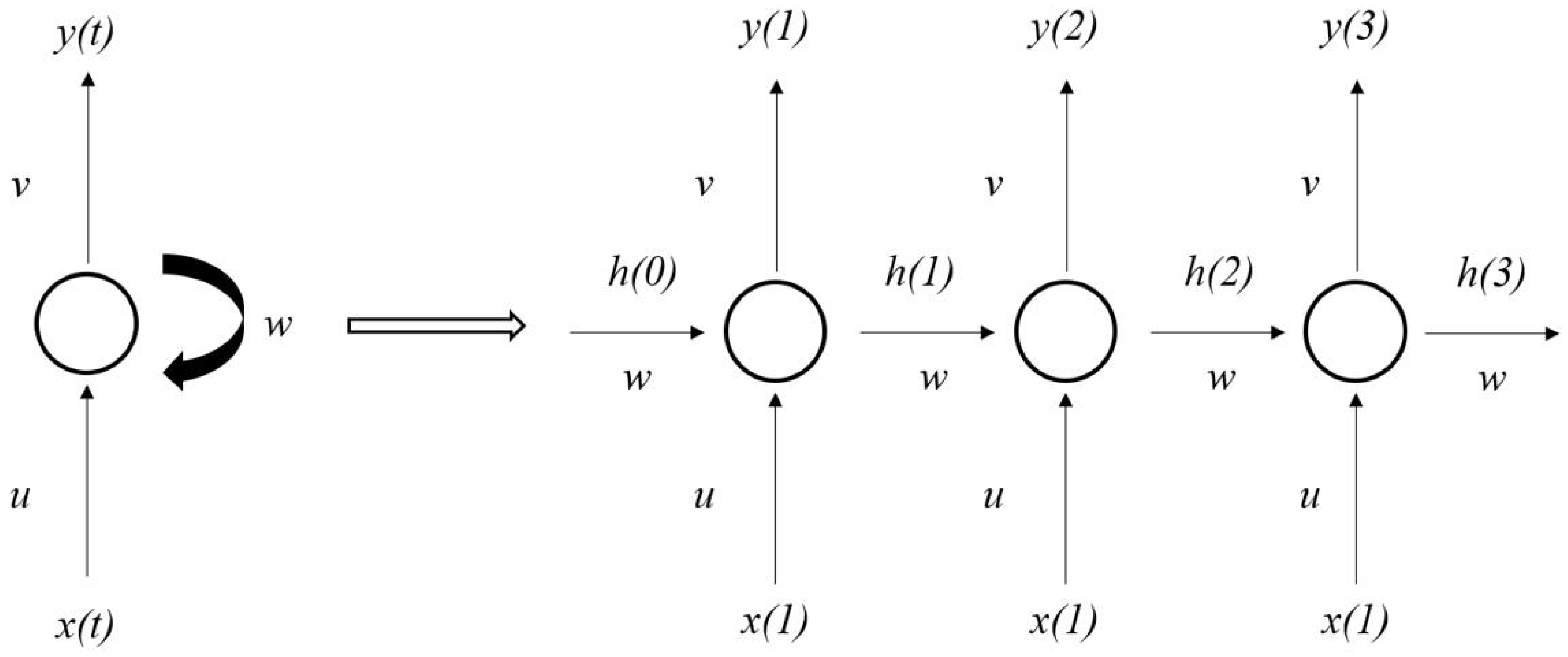
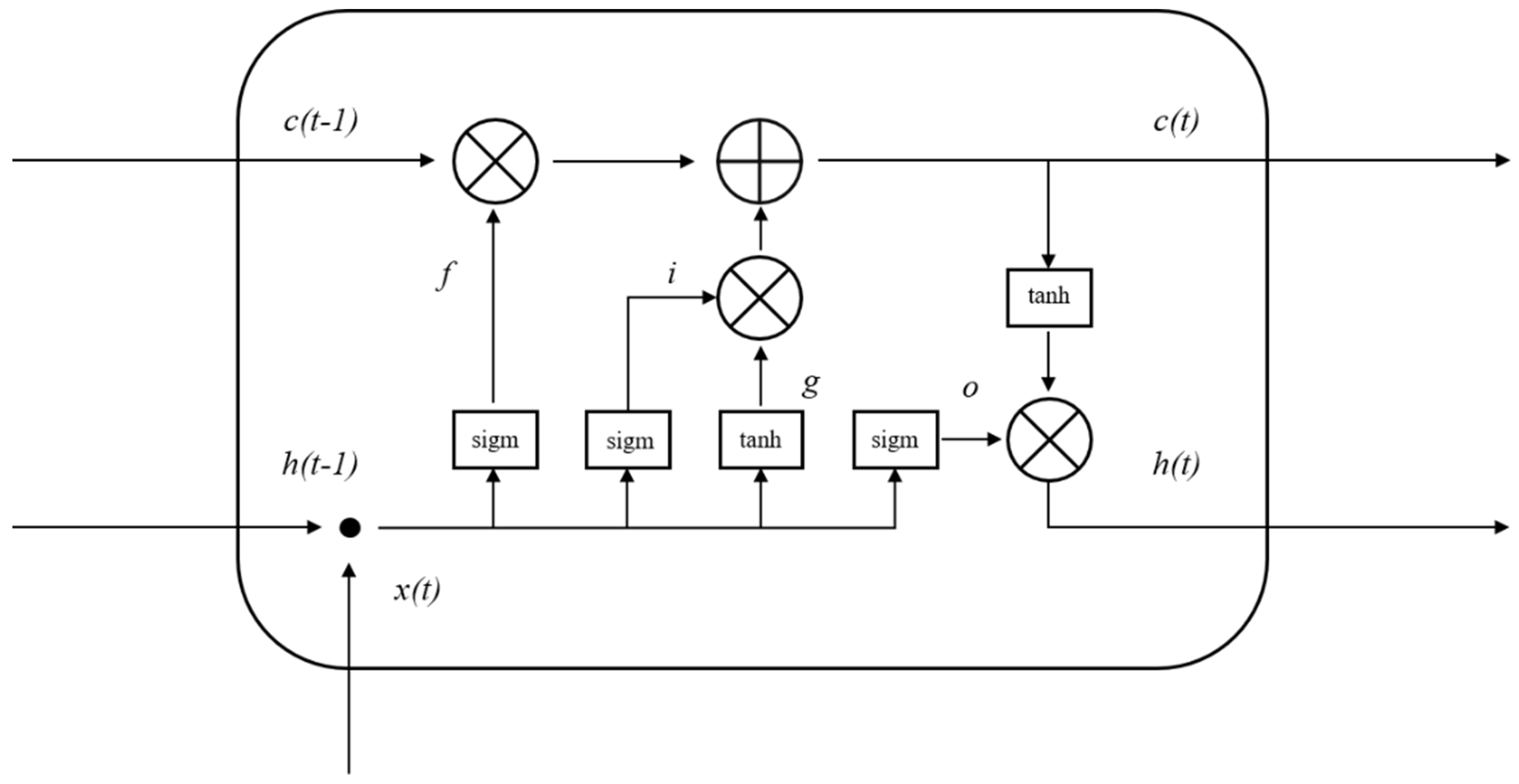
4.6. Establishment of DCNN–LSTM Model
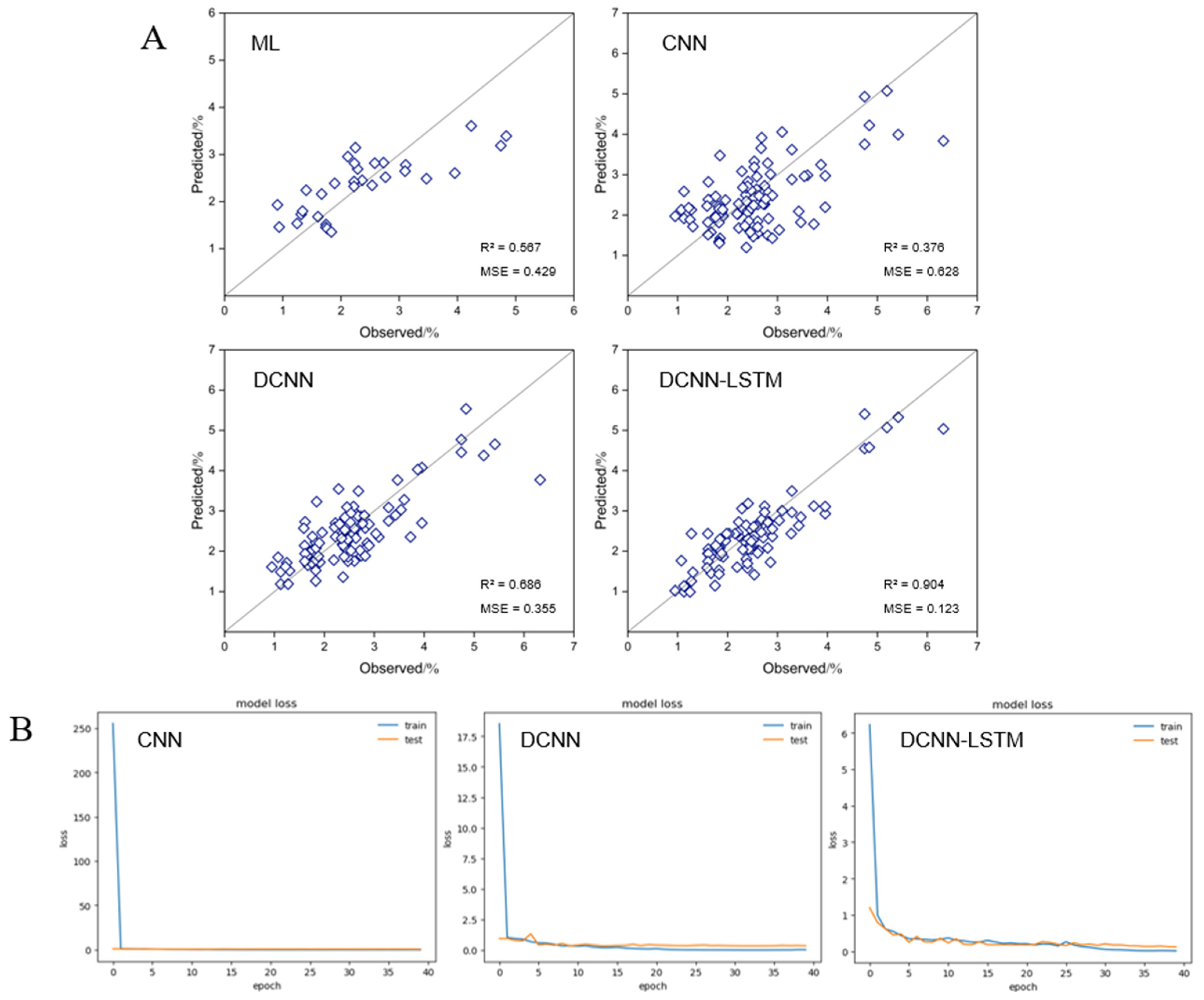
4.7. Evaluation of Models
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gallardo, M.; Gimenez, C.; Martinez-Gaitan, C.; Stoeckle, C.O.; Thompson, R.B.; Granados, M.R. Evaluation of the VegSyst model with muskmelon to simulate crop growth, nitrogen uptake and evapotranspiration. Agric. Water Manag. 2011, 101, 107–117. [Google Scholar] [CrossRef]
- Kirnak, H.; Higgs, D.; Kaya, C.; Tas, I. Effects of irrigation and nitrogen rates on growth, yield, and quality of muskmelon in semiarid regions. J. Plant Nutr. 2005, 28, 621–638. [Google Scholar] [CrossRef]
- Li, D.; Li, C.; Yao, Y.; Li, M.; Liu, L. Modern imaging techniques in plant nutrition analysis: A review. Comput. Electron. Agric. 2020, 174, 105459. [Google Scholar] [CrossRef]
- Fredes, A.; Sales, C.; Barreda, M.; Valcarcel, M.; Rosello, S.; Beltran, J. Quantification of prominent volatile compounds responsible for muskmelon and watermelon aroma by purge and trap extraction followed by gas chromatography-mass spectrometry determination. Food Chem. 2016, 190, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Lehne, P.; Le, J.; Ge, T.; Huang, D. Yield, fruit quality and nitrogen uptake of organically and conventionally grown muskmelon with different inputs of nitrogen, phosphorus, and potassium. J. Plant Nutr. 2010, 33, 130–141. [Google Scholar] [CrossRef]
- Li, X.; Hu, C.; Delgado, J.A.; Zhang, Y.; Ouyang, Z. Increased nitrogen use efficiencies as a key mitigation alternative to reduce nitrate leaching in north China plain. Agric. Water Manag. 2007, 89, 137–147. [Google Scholar] [CrossRef]
- Galloway, J.N.; Dentener, F.J.; Capone, D.G.; Boyer, E.W.; Howarth, R.W.; Seitzinger, S.P.; Asner, G.P.; Cleveland, C.C.; Green, P.A.; Holland, E.A.; et al. Nitrogen cycles: Past, present, and future. Biogeochemistry 2004, 70, 153–226. [Google Scholar] [CrossRef]
- Shi, Y.; Zhu, Y.; Wang, X.; Sun, X.; Ding, Y.; Cao, W.; Hu, Z. Progress and development on biological information of crop phenotype research applied to real-time variable-rate fertilization. Plant Methods 2020, 16, 11. [Google Scholar] [CrossRef]
- Li, D.; Wang, X.; Zheng, H.; Zhou, K.; Yao, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cheng, T. Estimation of area and mass-based leaf nitrogen contents of wheat and rice crops from water-removed spectra using continuous wavelet analysis. Plant Methods 2018, 14, 76. [Google Scholar] [CrossRef]
- Padilla, F.M.; Peña-Fleitas, M.T.; Gallardo, M.; Thompson, R.B. Proximal optical sensing of cucumber crop N status using chlorophyll fluorescence indices. Eur. J. Agron. 2016, 73, 83–97. [Google Scholar] [CrossRef]
- Pandey, P.; Ge, Y.; Stoerger, V.; Schnable, J.C. High Throughput In vivo Analysis of Plant Leaf Chemical Properties Using Hyperspectral Imaging. Front. Plant Sci. 2017, 8, 1348. [Google Scholar] [CrossRef]
- Agati, G.; Foschi, L.; Grossi, N.; Volterrani, M. In field non-invasive sensing of the nitrogen status in hybrid bermudagrass (Cynodon dactylon × C. transvaalensis Burtt Davy) by a fluorescence-based method. Eur. J. Agron. 2015, 63, 89–96. [Google Scholar] [CrossRef]
- Chen, D.; Shi, R.; Pape, J.M.; Neumann, K.; Arend, D.; Graner, A.; Chen, M.; Klukas, C. Predicting plant biomass accumulation from image-derived parameters. Gigascience 2018, 7, 1–13. [Google Scholar] [CrossRef]
- Fernández-Pacheco, D.G.; Escarabajal-Henarejos, D.; Ruiz-Canales, A.; Conesa, J.; Molina-Martínez, J.M. A digital image-processing-based method for determining the crop coefficient of lettuce crops in the southeast of Spain. Biosyst. Eng. 2014, 117, 23–34. [Google Scholar] [CrossRef]
- Guo, D.; Juan, J.; Chang, L.; Zhang, J.; Huang, D. Discrimination of plant root zone water status in greenhouse production based on phenotyping and machine learning techniques. Sci. Rep. 2017, 7, 8303. [Google Scholar] [CrossRef]
- Neilson, E.H.; Edwards, A.M.; Blomstedt, C.K.; Berger, B.; Moller, B.L.; Gleadow, R.M. Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C-4 cereal crop plant to nitrogen and water deficiency over time. J. Exp. Bot. 2015, 66, 1817–1832. [Google Scholar] [CrossRef]
- Baresel, J.P.; Rischbeck, P.; Hu, Y.; Kipp, S.; Hu, Y.; Barmeier, G.; Mistele, B.; Schmidhalter, U. Use of a digital camera as alternative method for non-destructive detection of the leaf chlorophyll content and the nitrogen nutrition status in wheat. Comput. Electron. Agric. 2017, 140, 25–33. [Google Scholar] [CrossRef]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Nitrogen deficiency prediction of rice crop based on convolutional neural network. J. Ambient Intell. Humaniz. Comput. 2020, 11, 5703–5711. [Google Scholar] [CrossRef]
- Schmidhuber, J. Deep learning in neural networks: An overview. Neural Netw. 2015, 61, 85–117. [Google Scholar] [CrossRef]
- Chen, J.D.; Chen, J.X.; Zhang, D.F.; Sun, Y.D.; Nanehkaran, Y.A. Using deep transfer learning for image-based plant disease identification. Comput. Electron. Agric. 2020, 173, 11. [Google Scholar] [CrossRef]
- Dyrmann, M.; Karstoft, H.; Midtiby, H.S. Plant species classification using deep convolutional neural network. Biosyst. Eng. 2016, 151, 72–80. [Google Scholar] [CrossRef]
- Grinblat, G.L.; Uzal, L.C.; Larese, M.G.; Granitto, P.M. Deep learning for plant identification using vein morphological patterns. Comput. Electron. Agric. 2016, 127, 418–424. [Google Scholar] [CrossRef]
- Kawasaki, Y.; Uga, H.; Kagiwada, S.; Iyatomi, H. Basic study of automated diagnosis of viral plant diseases using convolutional neural networks. In International Symposium on Visual Computing; Springer: Cham, Switzerland, 2015; pp. 638–645. [Google Scholar]
- Ma, J.; Du, K.; Zheng, F.; Zhang, L.; Gong, Z.; Sun, Z. A recognition method for cucumber diseases using leaf symptom images based on deep convolutional neural network. Comput. Electron. Agric. 2018, 154, 18–24. [Google Scholar] [CrossRef]
- Khaki, S.; Wang, L. Crop Yield Prediction Using Deep Neural Networks. Front. Plant Sci. 2019, 10, 621. [Google Scholar] [CrossRef]
- Madec, S.; Jin, X.; Lu, H.; De-Solan, B.; Liu, S.; Duyme, F.; Heritier, E.; Baret, F. Ear density estimation from high resolution RGB imagery using deep learning technique. Agric. For. Meteorol. 2019, 264, 225–234. [Google Scholar] [CrossRef]
- Rahnemoonfar, M.; Sheppard, C. Deep Count: Fruit Counting Based on Deep Simulated Learning. Sensors 2017, 17, 905. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Do, D.T.; Hung, T.N.K.; Lam, L.H.T.; Huynh, T.T.; Nguyen, N.T.K. A computational framework based on ensemble deep neural networks for essential genes identification. Int. J. Mol. Sci. 2020, 21, 9070. [Google Scholar] [CrossRef]
- Le, N.Q.K.; Nguyen, V.N. SNARE-CNN: A 2D convolutional neural network architecture to identify SNARE proteins from high-throughput sequencing data. PeerJ Comput. Sci. 2019, 5, 177. [Google Scholar] [CrossRef]
- Namin, S.T.; Esmaeilzadeh, M.; Najafi, M.; Brown, T.B.; Borevitz, J.O. Deep phenotyping: Deep learning for temporal phenotype/genotype classification. Plant Methods 2018, 14, 66. [Google Scholar] [CrossRef]
- Condori, R.H.M.; Romualdo, L.M.; Bruno, O.M.; de Cerqueira-Luz, P.H. Comparison between traditional texture methods and deep learning descriptors for detection of nitrogen deficiency in maize crops. In Proceedings of the 2017 Workshop of Computer Vision (WVC), Natal, Brazil, 30 October–1 November 2017; pp. 7–12. [Google Scholar]
- Yu, X.; Lu, H.; Liu, Q. Deep-learning-based regression model and hyper-spectral imaging for rapid detection of nitrogen concentration in oilseed rape (Brassica napus L.) leaf. Chemom. Intell. Lab. Syst. 2018, 172, 188–193. [Google Scholar] [CrossRef]
- Ni, C.; Wang, D.; Tao, Y. Variable weighted convolutional neural network for the nitrogen content quantization of Masson pine seedling leaves with near-infrared spectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 209, 32–39. [Google Scholar] [CrossRef]
- Mistele, B.; Schmidhalter, U. Estimating the nitrogen nutrition index using spectral canopy reflectance measurements. Eur. J. Agron. 2008, 29, 184–190. [Google Scholar] [CrossRef]
- Padilla, F.M.; Teresa, P.F.M.; Gallardo, M.; Thompson, R.B. Evaluation of optical sensor measurements of canopy reflectance and of leaf flavonols and chlorophyll contents to assess crop nitrogen status of muskmelon. Eur. J. Agron. 2014, 58, 39–52. [Google Scholar] [CrossRef]
- Csurka, G.; Dance, C.; Fan, L.; Willamowski, J.; Bray, C. Visual categorization with bags of keypoints. In Workshop on Statistical Learning in Computer Vision, ECCV; 2004; Volume 1, pp. 1–2. [Google Scholar]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Yang, B.; Xu, Y. Applications of deep-learning approaches in horticultural research: A review. Hortic. Res. 2021, 8, 123. [Google Scholar] [CrossRef] [PubMed]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Lin, K.; Gong, L.; Huang, Y.; Liu, C.; Pan, J. Deep learning-based segmentation and quantification of cucumber powdery mildew using convolutional neural network. Front. Plant Sci. 2019, 10, 155. [Google Scholar] [CrossRef]
- Pascanu, R.; Gulcehre, C.; Cho, K.; Bengio, Y. How to construct deep recurrent neural networks? arXiv 2013, arXiv:1312.6026. [Google Scholar]
- Graves, A.; Mohamed, A.R.; Hinton, G. Speech recognition with deep recurrent neural networks. In Proceedings of the 2013 IEEE International Conference on Acoustics, Speech and Signal Processing, Vancouver, Canada, 26–31 May 2013; pp. 6645–6649. [Google Scholar]
- Tran, T.T.; Choi, J.W.; Le, T.T.H.; Kim, J.W. A comparative study of deep CNN in forecasting and classifying the macronutrient deficiencies on development of tomato plant. Appl. Sci. 2019, 9, 1601. [Google Scholar] [CrossRef]
- Zhu, L.; Li, Z.; Li, C.; Wu, J.; Yue, J. High performance vegetable classification from images based on alexnet deep learning model. Int. J. Agric. Biol. Eng. 2018, 11, 217–223. [Google Scholar] [CrossRef]
- Jiang, Z.; Liu, C.; Hendricks, N.P.; Ganapathysubramanian, B.; Hayes, D.J.; Sarkar, S. Predicting county level corn yields using deep long short term memory models. arXiv 2018, arXiv:1805.12044. [Google Scholar]
- Haider, S.A.; Naqvi, S.R.; Akram, T.; Umar, G.A.; Shahzad, A.; Sial, M.R.; Khaliq, S.; Kamran, M. LSTM Neural Network Based Forecasting Model for Wheat Production in Pakistan. Agronomy 2019, 9, 72. [Google Scholar] [CrossRef]
- Alhnaity, B.; Pearson, S.; Leontidis, G.; Kollias, S. Using deep learning to predict plant growth and yield in greenhouse environments. In Proceedings of the International Symposium on Advanced Technologies and Management for Innovative Greenhouses: GreenSys2019, Angers, France, 16–20 June 2019; pp. 425–432. [Google Scholar]
- Gavahi, K.; Abbaszadeh, P.; Moradkhani, H. Deep Yield: A Combined Convolutional Neural Network with Long Short-Term Memory for Crop Yield Forecasting. Expert Syst. Appl. 2021, 184, 115511. [Google Scholar] [CrossRef]
- Hu, G.; Xiong, T.; Zhang, Y.; Feng, J.; Wu, H.; Li, Q. Spatial distribution and nitrogen diagnosis of SPAD value for different leaves position on main stem of muskmelon. Soil Fertil. Sci. China 2017, 80–85, 148. [Google Scholar]
- Villanueva, M.J.; Tenorio, M.D.; Esteban, M.A.; Mendoza, M.C. Compositional changes during ripening of two cultivars of muskmelon fruits. Food Chem. 2004, 87, 179–185. [Google Scholar] [CrossRef]
- Gehan, M.A.; Fahlgren, N.; Abbasi, A.; Berry, J.C.; Sax, T. PlantCV v2: Image analysis software for high-throughput plant phenotyping. PeerJ 2017, 5, e4088. [Google Scholar] [CrossRef]
- Xiong, X.; Zhang, J.; Guo, D.; Chang, L.; Huang, D. Non-Invasive Sensing of Nitrogen in Plant Using Digital Images and Machine Learning for Brassica Campestris ssp. Chinensis L. Sensors 2019, 19, 2448. [Google Scholar] [CrossRef]
- Kaiser, H.F. An index of factorial simplicity. Psychometrika 1974, 39, 31–36. [Google Scholar] [CrossRef]
- Bartlett, M.S. Tests of significance in factor analysis. Br. J. Stat. Psychol. 1950, 3, 77–85. [Google Scholar] [CrossRef]
- Macbeth, C.; Dai, H. Effects of Learning Parameters on Learning Procedure and Performance of a BPNN. Neural Netw. Off. J. Int. Neural Netw. Soc. 1997, 10, 1505–1521. [Google Scholar]
- Chollet, F. Deep Learning with Python; Manning: New York, NY, USA, 2018; Volume 361. [Google Scholar]
- Abadi, M.; Barham, P.; Chen, J.; Chen, Z.; Davis, A.; Dean, J.; Devin, M.; Ghemawat, S.; Irving, G.; Isard, M.; et al. Tensorflow: A system for large-scale machine learning. In Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI ’16), Savannah, GA, USA, 2–4 November 2016. [Google Scholar]
- Chang, L.Y.; He, S.P.; Qian, L.I.U.; Xiang, J.L.; Huang, D.F. Quantifying muskmelon fruit attributes with A-TEP-based model and machine vision measurement. J. Integr. Agric. 2018, 17, 1369–1379. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A.; Bengio, Y. Deep Learning; MIT Press: Cambridge, UK, 2016. [Google Scholar]
- Graves, A. Connectionist temporal classification. In Supervised Sequence Labelling with Recurrent Neural Networks; Springer: Berlin/Heidelberg, Germany, 2012; pp. 61–93. [Google Scholar]
- Lee, K.J.; Lee, B.W. Estimation of rice growth and nitrogen nutrition status using colour digital camera image analysis. Eur. J. Agron. 2013, 48, 57–65. [Google Scholar] [CrossRef]
- Wu, K.; Du, C.; Ma, F.; Shen, Y.; Zhou, J. Rapid diagnosis of nitrogen status in rice based on Fourier transform infrared photoacoustic spectroscopy (FTIR-PAS). Plant Methods 2019, 15, 94. [Google Scholar] [CrossRef] [PubMed]
- Prey, L.; Schmidhalter, U. Sensitivity of Vegetation Indices for Estimating Vegetative N Status in Winter Wheat. Sensors 2019, 19, 3712. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zhao, J.; Xu, X.; Liang, D.; Yang, G.; Feng, H.; Yang, H.; Wang, Y.; Chen, G.; Wei, P. Hyperspectral-Based Estimation of Leaf Nitrogen Content in Corn Using Optimal Selection of Multiple Spectral Variables. Sensors 2019, 19, 2898. [Google Scholar] [CrossRef] [PubMed]
- Nguy-Robertson, A.L.; Peng, Y.; Gitelson, A.A.; Arkebauer, T.J.; Pimstein, A.; Herrmann, I.; Karnieli, A.; Rundquist, D.C.; Bonfil, D.J. Estimating green LAI in four crops: Potential of determining optimal spectral bands for a universal algorithm. Agric. For. Meteorol. 2014, 192, 140–148. [Google Scholar] [CrossRef]
- Li, H.; Zhao, C.; Huang, W.; Yang, G. Non-uniform vertical nitrogen distribution within plant canopy and its estimation by remote sensing: A review. Field Crop. Res. 2013, 142, 75–84. [Google Scholar] [CrossRef]
- Ma, J.F.; Yan, Z.; Xia, Y.; Tian, Y.C.; Liu, X.J.; Cao, W.X. Relationship between leaf nitrogen content and fluorescence parameters in rice. Zhongguo Shuidao Kexue 2007, 21, 65–70. [Google Scholar]
- De-Freitas, F.M.A.; Andriolo, J.L.; Godoi, R.D.S.; Peixoto-de-Barros, C.A.; Janisch, D.I.; Braz-Vaz, M.A. Nitrogen critical dilution curve for the muskmelon crop. Cienc. Rural 2008, 38, 345–350. [Google Scholar] [CrossRef][Green Version]
- Singh, A.K.; Ganapathysubramanian, B.; Sarkar, S.; Singh, A. Deep learning for plant stress phenotyping: Trends and future perspectives. Trends Plant. Sci. 2018, 23, 883–898. [Google Scholar] [CrossRef]
- Sa, I.; Popovic, M.; Khanna, R.; Chen, Z.; Lottes, P.; Liebisch, F.; Nieto, J.; Stachniss, C.; Walter, A.; Siegwart, R. WeedMap: A Large-Scale Semantic Weed Mapping Framework Using Aerial Multispectral Imaging and Deep Neural Network for Precision Farming. Remote Sens. 2018, 10, 1423. [Google Scholar] [CrossRef]
- Agarwal, M.; Sinha, A.; Gupta, S.K.; Mishra, D.; Mishra, R.; Agarwal, M.; Sinha, A.; Gupta, S.K.; Mishra, D.; Mishra, R. Potato crop disease classification using convolutional neural network. In Smart Systems and IoT: Innovations in Computing; Springer: Singapore, 2020; pp. 391–400. [Google Scholar]
- You, J.; Li, X.; Low, M.; Lobell, D.; Ermon, S.; Aaai. Deep gaussian process for crop yield prediction based on remote sensing data. In Proceedings of the AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–9 February 2017; Volume 31, pp. 4559–4565. [Google Scholar]
- Ghazaryan, G.; Skakun, S.; Konig, S.; Rezaei, E.E.; Siebert, S.; Dubovyk, O. Crop yield estimation using multi-source satellite image series and deep learning. In Proceedings of the IGARSS 2020–2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, HI, USA, 26 September–2 October 2020; pp. 5163–5166. [Google Scholar] [CrossRef]
- Haryono; Anam, K.; Saleh, A. A novel herbal leaf identification and authentication using deep learning neural network. In Proceedings of the International Conference on Computer Engineering, Network, and Intelligent Multimedia (CENIM), Surabaya, Indonesia, 17–18 November 2020; pp. 338–342. [Google Scholar] [CrossRef]
- Baek, S.S.; Pyo, J.; Chun, J.A. Prediction of water level and water quality using a CNN-LSTM combined deep learning approach. Water 2020, 12, 3399. [Google Scholar] [CrossRef]
- Sun, J.; Di, L.P.; Sun, Z.H.; Shen, Y.L.; Lai, Z.L. County-level soybean yield prediction using deep CNN-LSTM model. Sensors 2019, 19, 4363. [Google Scholar] [CrossRef] [PubMed]

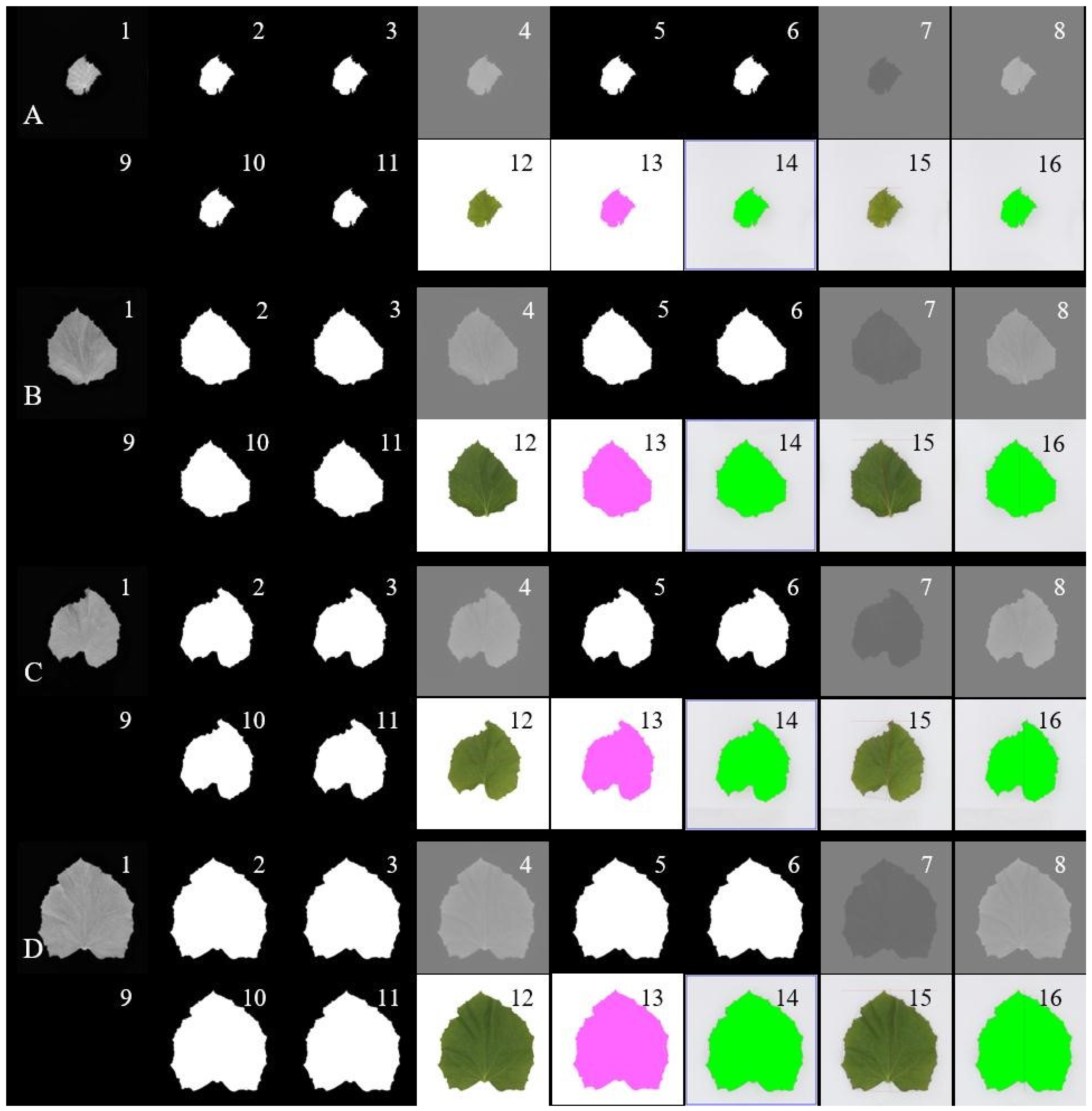
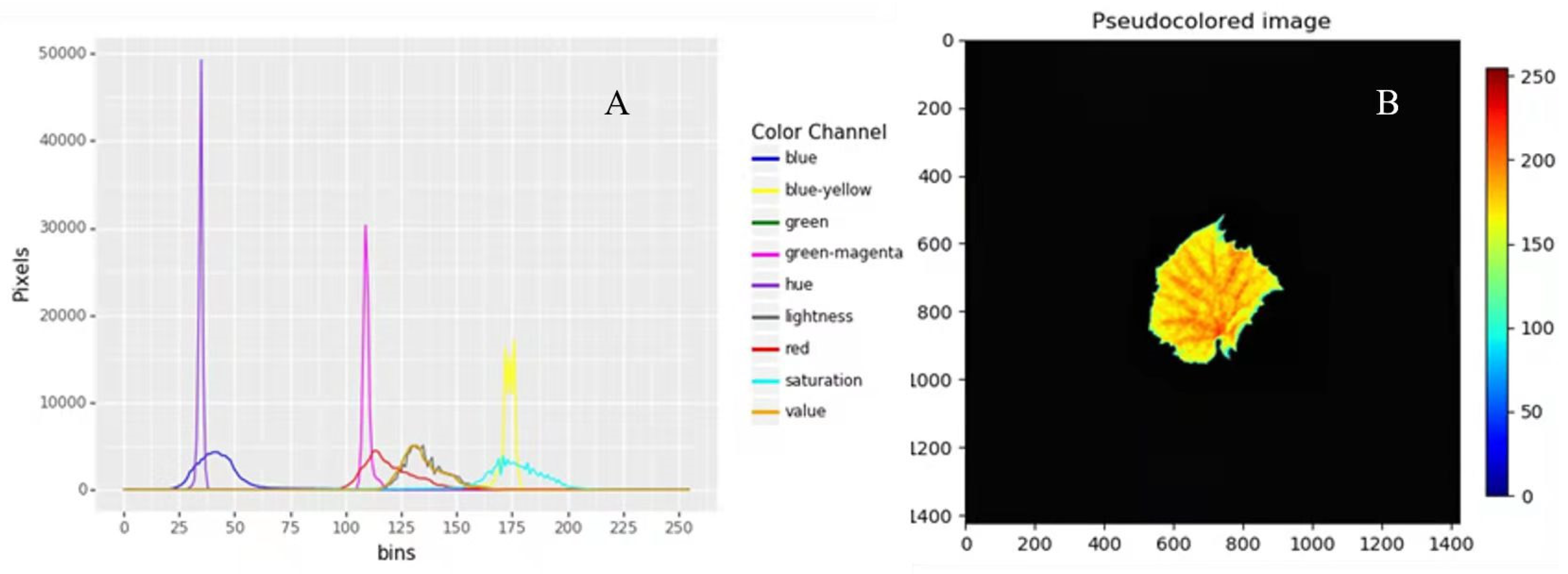
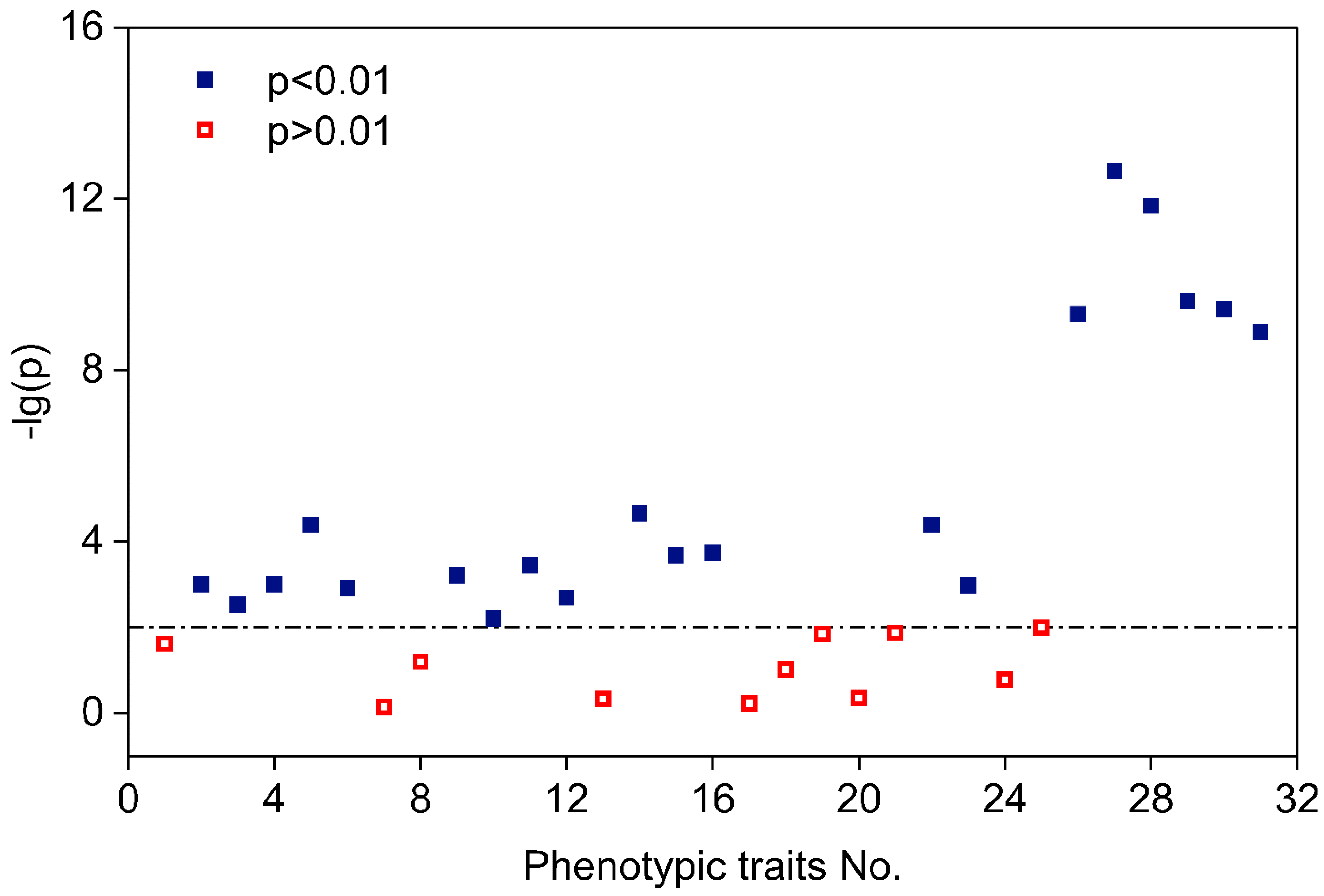
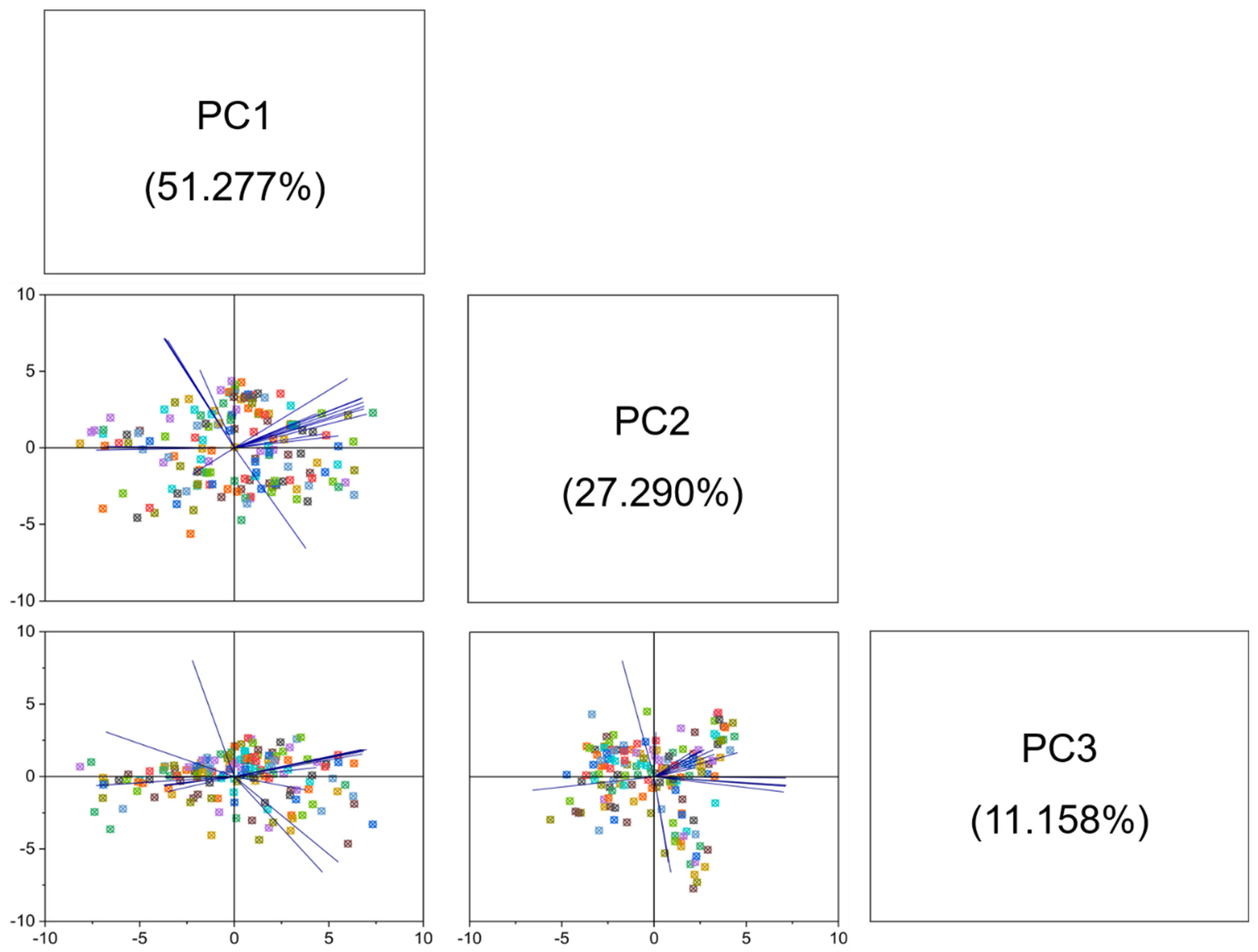
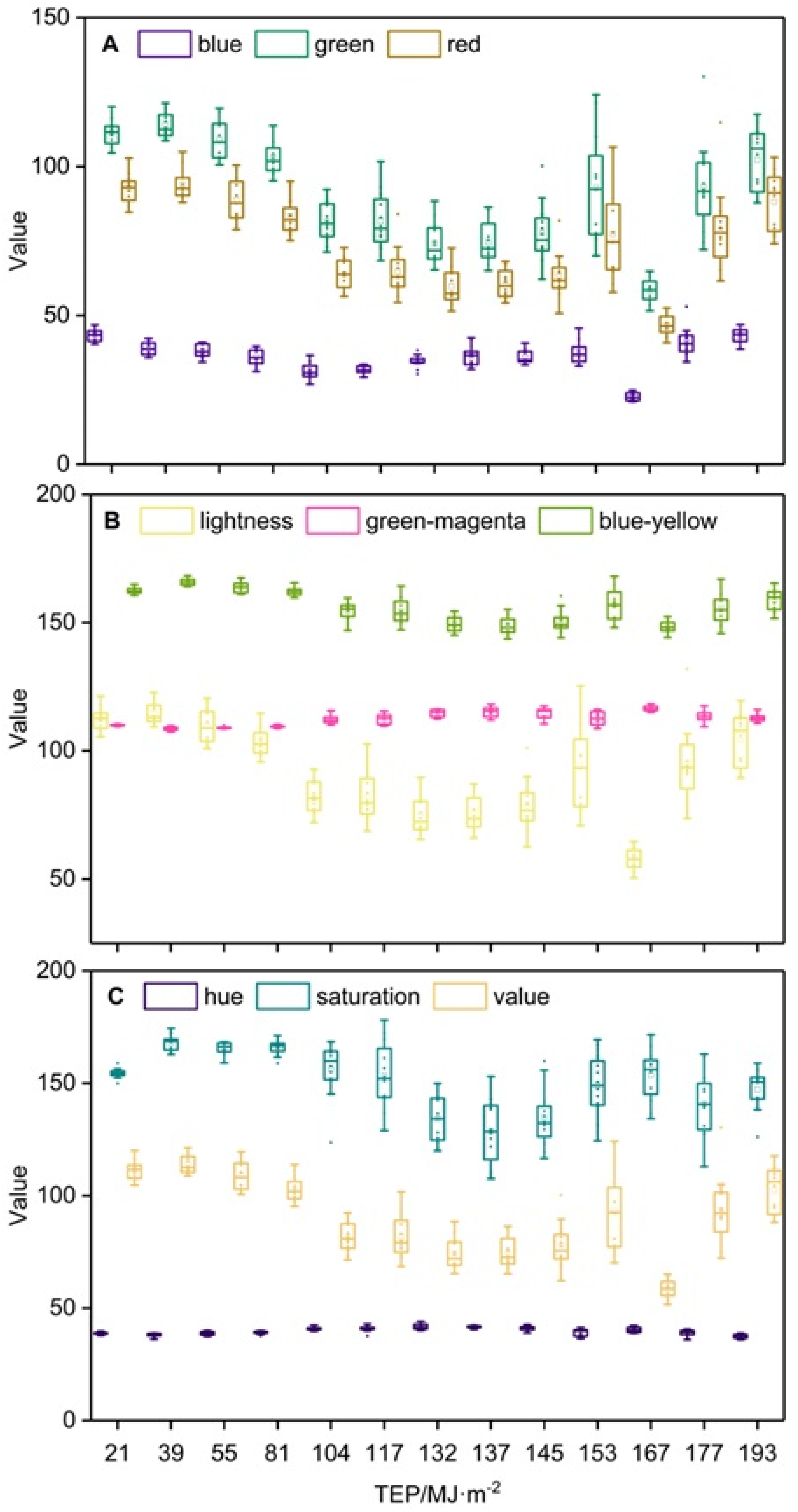

| Category | Serial No. | Extracted Index | Reference |
|---|---|---|---|
| Color | 1–3 | blue/green/red mean | |
| 4–6 | lightness/green-magenta/blue-yellow mean | ||
| 7–9 | hue/saturation/value mean | ||
| Morphology | 10 | area | |
| 11 | hull-area | ||
| 12 | solidity | ||
| 13 | perimeter | ||
| 14 | width | ||
| 15 | height | [52] | |
| 16 | longest-axis | ||
| 17 | center-of-mass-x | ||
| 18 | center-of-mass-y | ||
| 19 | hull-vertices | ||
| 20 | ellipse-center-x | ||
| 21 | ellipse-center-y | ||
| 22 | ellipse-major-axis | ||
| 23 | ellipse-minor-axis | ||
| 24 | ellipse-angle | ||
| 25 | ellipse-eccentricity | ||
| Texture | 26 | contrast | |
| 27 | dissimilarity | ||
| 28 | homogeneity | ||
| 29 | ASM | ||
| 30 | energy | ||
| 31 | correlation |
| Category | No. | Parameters Name | F1 | F2 | F3 |
|---|---|---|---|---|---|
| Yan Color Special Sign | 2 | Green | −0.488 | 0.855 | −0.067 |
| 3 | Red | −0.462 | 0.840 | −0.119 | |
| 4 | Lightness | −0.484 | 0.853 | −0.076 | |
| 5 | green-magenta | 0.496 | −0.787 | −0.107 | |
| 6 | blue-yellow | −0.480 | 0.854 | −0.008 | |
| 9 | Value | −0.486 | 0.856 | −0.068 | |
| Shape State Special Sign | 10 | Area | 0.784 | 0.540 | 0.190 |
| 11 | hull-area | 0.896 | 0.356 | 0.199 | |
| 12 | Solidity | −0.239 | 0.606 | 0.001 | |
| 14 | Width | 0.917 | 0.263 | 0.214 | |
| 15 | Height | 0.897 | 0.304 | 0.208 | |
| 16 | longest-axis | 0.900 | 0.325 | 0.214 | |
| 22 | ellipse-major-axis | 0.887 | 0.390 | 0.178 | |
| 23 | ellipse-minor-axis | 0.880 | 0.383 | 0.214 | |
| 26 | Contrast | 0.611 | 0.109 | −0.757 | |
| Pattern Reason Special Sign | 27 | dissimilarity | 0.721 | 0.093 | −0.676 |
| 28 | homogeneity | −0.889 | 0.011 | 0.353 | |
| 29 | ASM | −0.933 | 0.003 | −0.070 | |
| 30 | Energy | −0.957 | −0.018 | −0.073 | |
| 31 | correlation | −0.290 | −0.207 | 0.919 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, L.; Li, D.; Hameed, M.K.; Yin, Y.; Huang, D.; Niu, Q. Using a Hybrid Neural Network Model DCNN–LSTM for Image-Based Nitrogen Nutrition Diagnosis in Muskmelon. Horticulturae 2021, 7, 489. https://doi.org/10.3390/horticulturae7110489
Chang L, Li D, Hameed MK, Yin Y, Huang D, Niu Q. Using a Hybrid Neural Network Model DCNN–LSTM for Image-Based Nitrogen Nutrition Diagnosis in Muskmelon. Horticulturae. 2021; 7(11):489. https://doi.org/10.3390/horticulturae7110489
Chicago/Turabian StyleChang, Liying, Daren Li, Muhammad Khalid Hameed, Yilu Yin, Danfeng Huang, and Qingliang Niu. 2021. "Using a Hybrid Neural Network Model DCNN–LSTM for Image-Based Nitrogen Nutrition Diagnosis in Muskmelon" Horticulturae 7, no. 11: 489. https://doi.org/10.3390/horticulturae7110489
APA StyleChang, L., Li, D., Hameed, M. K., Yin, Y., Huang, D., & Niu, Q. (2021). Using a Hybrid Neural Network Model DCNN–LSTM for Image-Based Nitrogen Nutrition Diagnosis in Muskmelon. Horticulturae, 7(11), 489. https://doi.org/10.3390/horticulturae7110489






