Enzymatic Activities and Gene Transcript Levels Associated with the Augmentation of Antioxidant Constituents during Drought Stress in Lettuce
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Hydroponics and Drought Stress Treatment
2.3. Assay of ORAC
2.4. Determination of Antioxidative Enzyme Activities
2.5. RNA Extraction
2.6. RNA-Seq Analysis
2.7. Real-Time PCR
3. Results and Discussion
3.1. Effect of Drought Stress on the Biosynthesis of Antioxidants
3.2. Effect of Drought Stress O Then Transcription of Genes Related to Antioxidant Mechanisms
3.3. Differentially Expressed Genes under Drought Conditions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takatsuji, M. Present status of completely-controlled plant factories. Shokubutsu Kankyo Kogaku 2010, 22, 2–7. [Google Scholar] [CrossRef]
- Blokhina, O.; Virolainen, E.; Fagerstedt, K.V. Antioxidants, oxidative damage and oxygen deprivation stress: A review. Ann. Bot. Lond. 2003, 91, 179–194. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.M.; Fonseca, J.M. Effect of saline irrigation water on antioxidants in three hydroponically grown leafy vegetables: Diplotaxis tenuifolia, Eruca sativa, and Lepidium sativum. Hortscience 2010, 45, 546–552. [Google Scholar] [CrossRef]
- Johkan, M.; Shoji, K.; Goto, F.; Hashida, S.; Yoshihara, T. Blue light-emitting diode light irradiation of seedlings improves seedling quality and growth after transplanting in red leaf lettuce. Hortscience 2010, 45, 1809–1814. [Google Scholar] [CrossRef]
- Romani, A.; Pinelli, P.; Galardi, C.; Sani, G.; Cimato, A.; Heimler, D. Polyphenols in greenhouse and open-air-grown lettuce. Food Chem. 2002, 79, 337–342. [Google Scholar] [CrossRef]
- Sorice, A.; Guerriero, E.; Capone, F.; Colonna, G.; Castello, G.; Costantini, S. Ascorbic acid: Its role in immune system and chronic inflammation diseases. Mini-Rev. Med. Chem. 2014, 14, 444–452. [Google Scholar] [CrossRef]
- Sudha, J.D. Vitamin C: Sources, functions, sensing and analysis. In Vitamin C; Reshma Lali Raveendran, E.D.A.H.H., Ed.; IntechOpen: Rijeka, Croatia, 2017; Chapter 1. [Google Scholar]
- Andarwulan, N.; Kurniasih, D.; Apriady, R.A.; Rahmat, H.; Roto, A.V.; Bolling, B.W. Polyphenols, carotenoids, and ascorbic acid in underutilized medicinal vegetables. J. Funct. Foods 2012, 4, 339–347. [Google Scholar] [CrossRef]
- Rasouli, H.; Farzaei, M.H.; Khodarahmi, R. Polyphenols and their benefits: A review. Int. J. Food Prop. 2017, 20, 1700–1741. [Google Scholar] [CrossRef]
- The Commission of the European Communities. Commission Regulation (EC) No 1881/2006 of 19 December 2006 Setting Maximum Levels for Certain Contaminants in Foodstuffs. 2006; pp. 5–24. Available online: https://www.legislation.gov.uk/eur/2006/1881 (accessed on 21 October 2021).
- Dudley, N. Nitrates: The Threat to Food and Water; Green Print: London, UK, 1990; p. 118. [Google Scholar]
- Comly, H.H. Cyanosis in infants caused by nitrates in well water. J. Am. Med. Assoc. 1945, 129, 112–116. [Google Scholar] [CrossRef]
- Kratky, B.A. A Capillary, Noncirculating hydroponic method for leaf and semi-head lettuce. Hort Technol. Horttech 1993, 3, 206. [Google Scholar] [CrossRef]
- Koyama, R.; Itoh, H.; Kimura, S.; Morioka, A.; Uno, Y. Augmentation of antioxidant constituents by drought stress to roots in leafy vegetables. Horttechnology 2012, 22, 121–125. [Google Scholar] [CrossRef]
- Reyes-Chin-Wo, S.; Wang, Z.W.; Yang, X.H.; Kozik, A.; Arikit, S.; Song, C.; Xia, L.F.; Froenicke, L.; Lavelle, D.O.; Truco, M.J.; et al. Genome assembly with in vitro proximity ligation data and whole-genome triplication in lettuce. Nat. Commun. 2017, 8, 14953. [Google Scholar] [CrossRef] [PubMed]
- Dennis, R.J.; Micossi, E.; McCarthy, J.; Moe, E.; Gordon, E.J.; Kozielski-Stuhrmann, S.; Leonard, G.A.; McSweeney, S. Structure of the manganese superoxide dismutase from Deinococcus radiodurans in two crystal forms. Acta Crystallogr. F 2006, 62, 325–329. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Asada, K. Hydrogen-peroxide is scavenged by ascorbate-specific peroxidase in spinach-chloroplasts. Plant Cell Physiol. 1981, 22, 867–880. [Google Scholar]
- Ishibashi, M.; Nabe, T.; Nitta, Y.; Uno, Y. Efficient isolation of high-quality total RNA from strawberry. Hortscience 2019, 54, 380–384. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef]
- Kitano, M.; Hidaka, K.; Zushi, K.; Araki, T. Production of value-added vegetables by applying environmental stresses to roots in soil-less culture. Shokubutsu Kankyo Kogaku 2008, 20, 210–218. [Google Scholar] [CrossRef][Green Version]
- Nicolle, C.; Carnat, A.; Fraisse, D.; Lamaison, J.L.; Rock, E.; Michel, H.; Amouroux, P.; Remesy, C. Characterisation and variation of antioxidant micronutrients in lettuce (Lactuca sativa folium). J. Sci. Food Agrc. 2004, 84, 2061–2069. [Google Scholar] [CrossRef]
- Oh, M.M.; Carey, E.E.; Rajashekar, C.B. Regulated water deficits improve phytochemical concentration in lettuce. J. Am. Soc. Hortic. Sci. 2010, 135, 223–229. [Google Scholar] [CrossRef]
- Guo, Z.; Ou, W.; Lu, S.; Zhong, Q. Differential responses of antioxidative system to chilling and drought in four rice cultivars differing in sensitivity. Plant Physiol. Bioch. 2006, 44, 828–836. [Google Scholar] [CrossRef]
- Pan, Y.; Wu, L.J.; Yu, Z.L. Effect of salt and drought stress on antioxidant enzymes activities and SOD isoenzymes of liquorice (Glycyrrhiza uralensis Fisch). Plant Growth Regul. 2006, 49, 157–165. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Riadh, K.; Gopi, R.; Manivannan, P.; Ines, J.; Al-Juburi, H.; Zhao, C.X.; Shao, H.B.; Panneerselvam, R. Antioxidant defense responses: Physiological plasticity in higher plants under abiotic constraints. Acta Physiol. Plant. 2009, 31, 427–436. [Google Scholar] [CrossRef]
- Asada, K. The water-water cycle in chloroplasts: Scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Phys. 1999, 50, 601–639. [Google Scholar] [CrossRef]
- Mahmoudi, H.; Nasri, M.; Baâtour, O.; Huang, J.; Tarchoune, I.; Nasri, N.; Rym, K.; Gruber, M.; Lachaâl, M.; Ouerghi, Z. Differential response to elevated NaCl by antioxidant enzymes and gene transcripts in two contrasting lettuce genotypes. Aust. J. Crop Sci. 2012, 6, 632–640. [Google Scholar]
- Seo, H.S.; Song, J.T.; Cheong, J.J.; Lee, Y.H.; Lee, Y.W.; Hwang, I.; Lee, J.S.; Choi, Y.D. Jasmonic acid carboxyl methyltransferase: A key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. USA 2001, 98, 4788–4793. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.H.; Kim, Y.S.; Park, S.H.; Koo, Y.J.; Do Choi, Y.; Chung, Y.Y.; Lee, I.J.; Kim, J.K. Methyl jasmonate reduces grain yield by mediating stress signals to alter spikelet development in rice. Plant Physiol. 2009, 149, 1751–1760. [Google Scholar] [CrossRef]
- Ma, H.X.; Liu, M. The microtubule cytoskeleton acts as a sensor for stress response signaling in plants. Mol. Biol. Rep. 2019, 46, 5603–5608. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Valadier, M.H.; Migge, A.; Becker, T.W. Drought-induced effects on nitrate reductase activity and mRNA and on the coordination of nitrogen and carbon metabolism in maize leaves. Plant Physiol. 1998, 117, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008, 148, 6–24. [Google Scholar] [CrossRef]
- Hara, M. The multifunctionality of dehydrins: An overview. Plant Signal. Behav. 2010, 5, 503–508. [Google Scholar] [CrossRef]
- Zhang, H.; Zheng, J.X.; Su, H.X.; Xia, K.F.; Jian, H.G.; Zhang, M. Molecular cloning and functional characterization of the dehydrin (IpDHN) gene from Ipomoea pes-caprae. Front. Plant Sci. 2018, 9, 1454. [Google Scholar] [CrossRef] [PubMed]
- Porcel, R.; Azcon, R.; Ruiz-Lozano, J.M. Evaluation of the role of genes encoding for dehydrin proteins (LEA D-11) during drought stress in arbuscular mycorrhizal Glycine max and Lactuca sativa plants. J. Exp. Bot. 2005, 56, 1933–1942. [Google Scholar] [CrossRef] [PubMed]
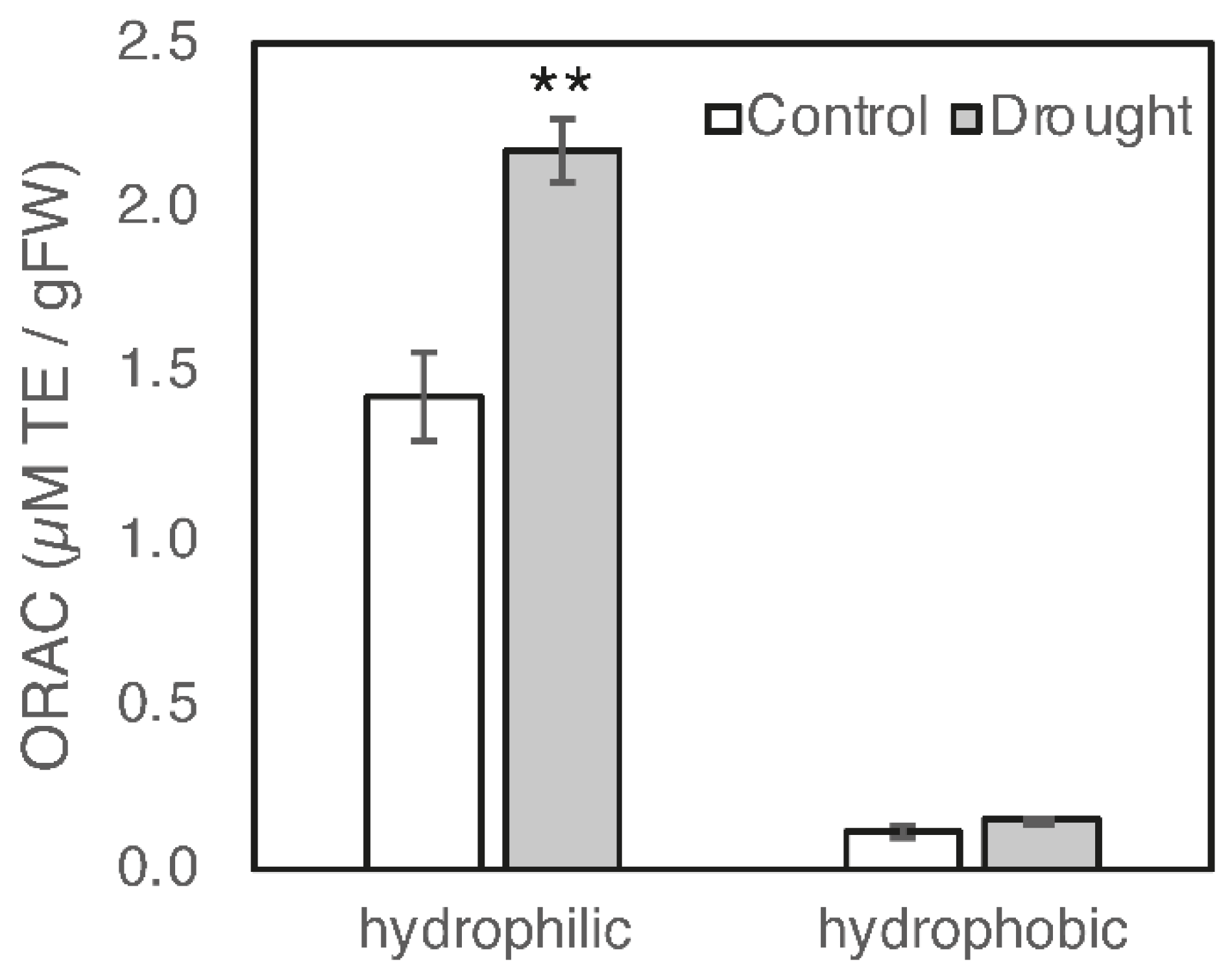

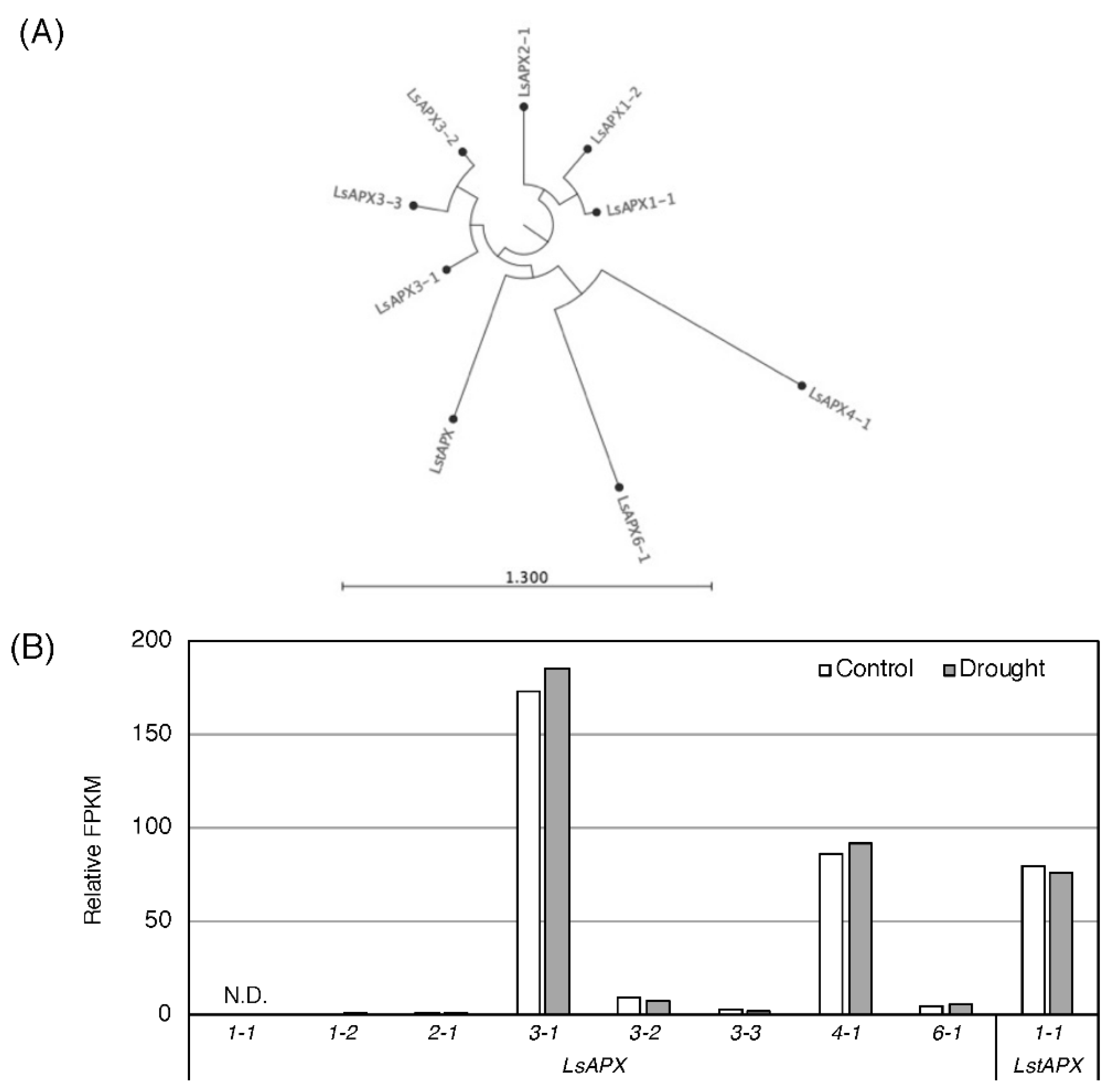
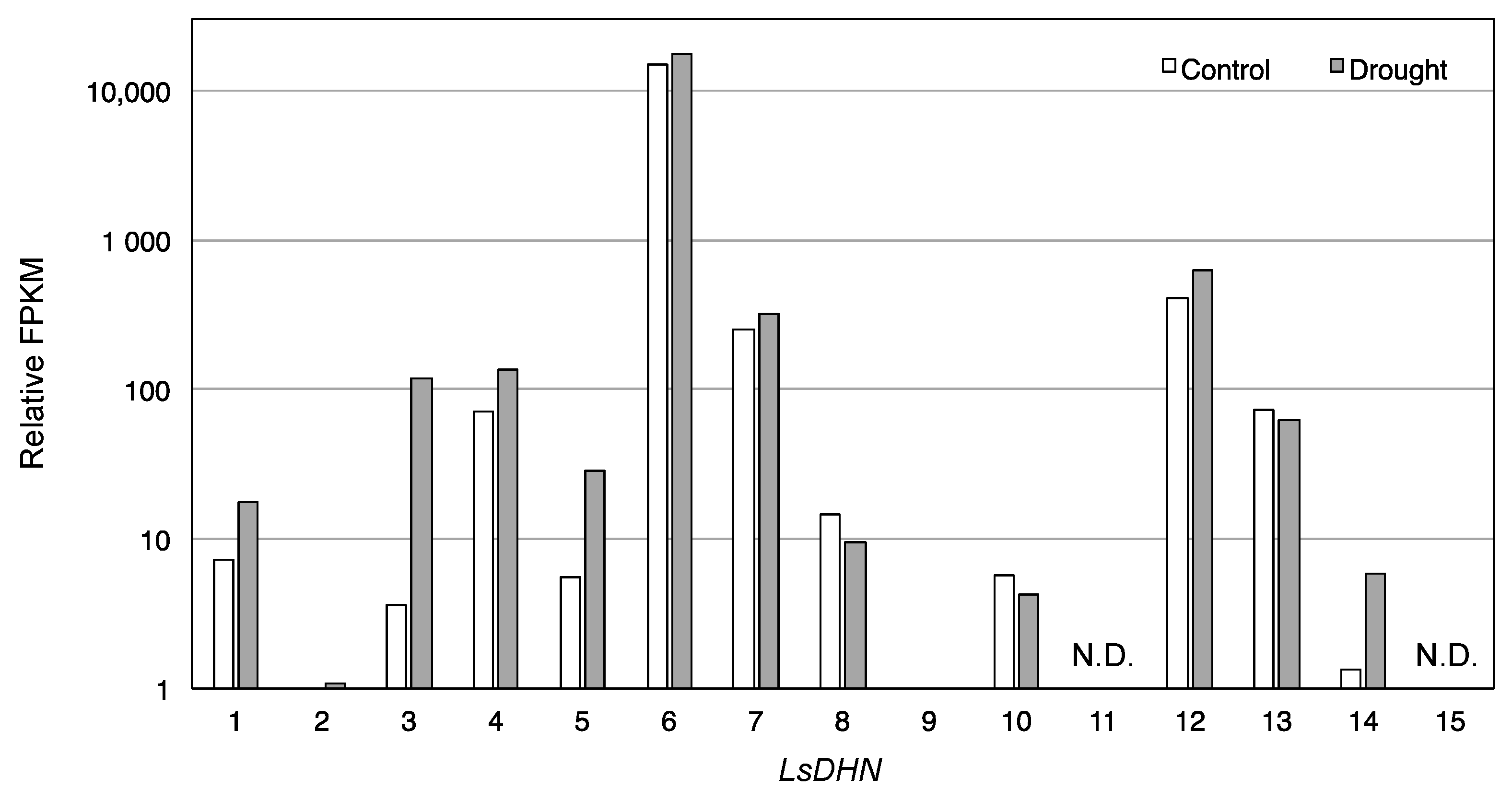

| Component | SOD (Units/mg/Protein) | APX (µmol/mg Protein/min) |
|---|---|---|
| Control | 148.82 ± 3.51 1 | 0.014 ± 0.00072 |
| Drought | 167.10 ± 3.87 ** 2 | 0.019 ± 0.00087 ** |
| Gene ID | log2 Fold Change | q-Value | Description |
|---|---|---|---|
| Lsat_2_78000 | 5.01 | 5.6 × 10−15 | Dehydrin Rab18-like 1 |
| Lsat_1_129340 | 4.85 | 2.8 × 10−4 | Stigma-specific STIG1-like protein 1 |
| Lsat_9_34320 | 3.72 | 2.0 × 10−4 | Laccase-7-like |
| Lsat_7_49380 | 2.58 | 1.7 × 10−10 | Berberine bridge enzyme-like 8 |
| Lsat_4_56281 | 2.55 | 1.2 × 10−5 | Uncharacterized |
| Lsat_2_79621 | −3.80 | 2.5 × 10−5 | Arylacetamide deacetylase |
| Lsat_9_13940 | −3.60 | 3.4 × 10−9 | Expansin |
| Lsat_7_10721 | −3.40 | 4.8 × 10−19 | Jasmonate O-methyltransferase |
| Lsat_7_94901 | −3.26 | 2.7 × 10−4 | Nitrate reductase |
| Lsat_1_16900 | −3.15 | 1.5 × 10−6 | Jasmonate O-methyltransferase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koyama, R.; Yoshimoto, A.; Ishibashi, M.; Itoh, H.; Uno, Y. Enzymatic Activities and Gene Transcript Levels Associated with the Augmentation of Antioxidant Constituents during Drought Stress in Lettuce. Horticulturae 2021, 7, 444. https://doi.org/10.3390/horticulturae7110444
Koyama R, Yoshimoto A, Ishibashi M, Itoh H, Uno Y. Enzymatic Activities and Gene Transcript Levels Associated with the Augmentation of Antioxidant Constituents during Drought Stress in Lettuce. Horticulturae. 2021; 7(11):444. https://doi.org/10.3390/horticulturae7110444
Chicago/Turabian StyleKoyama, Ryohei, Aika Yoshimoto, Misaki Ishibashi, Hiromichi Itoh, and Yuichi Uno. 2021. "Enzymatic Activities and Gene Transcript Levels Associated with the Augmentation of Antioxidant Constituents during Drought Stress in Lettuce" Horticulturae 7, no. 11: 444. https://doi.org/10.3390/horticulturae7110444
APA StyleKoyama, R., Yoshimoto, A., Ishibashi, M., Itoh, H., & Uno, Y. (2021). Enzymatic Activities and Gene Transcript Levels Associated with the Augmentation of Antioxidant Constituents during Drought Stress in Lettuce. Horticulturae, 7(11), 444. https://doi.org/10.3390/horticulturae7110444






