Genome-Wide Analysis of Purple Acid Phosphatase Genes in Brassica rapa and Their Association with Pollen Development and Phosphorus Deprivation Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth and Pi Deprivation Treatment
2.2. Leaf Area and Phosphorus Content
2.3. Identification of BrPAP Genes in Brassica rapa
2.4. Phylogenetic Tree and Bioinformatics Analysis of BrPAPs
2.5. Expression and Co-Expression Analysis of BrPAPs
2.6. RNA Extraction, Semi-RT-PCR, and qRT-PCR
3. Results
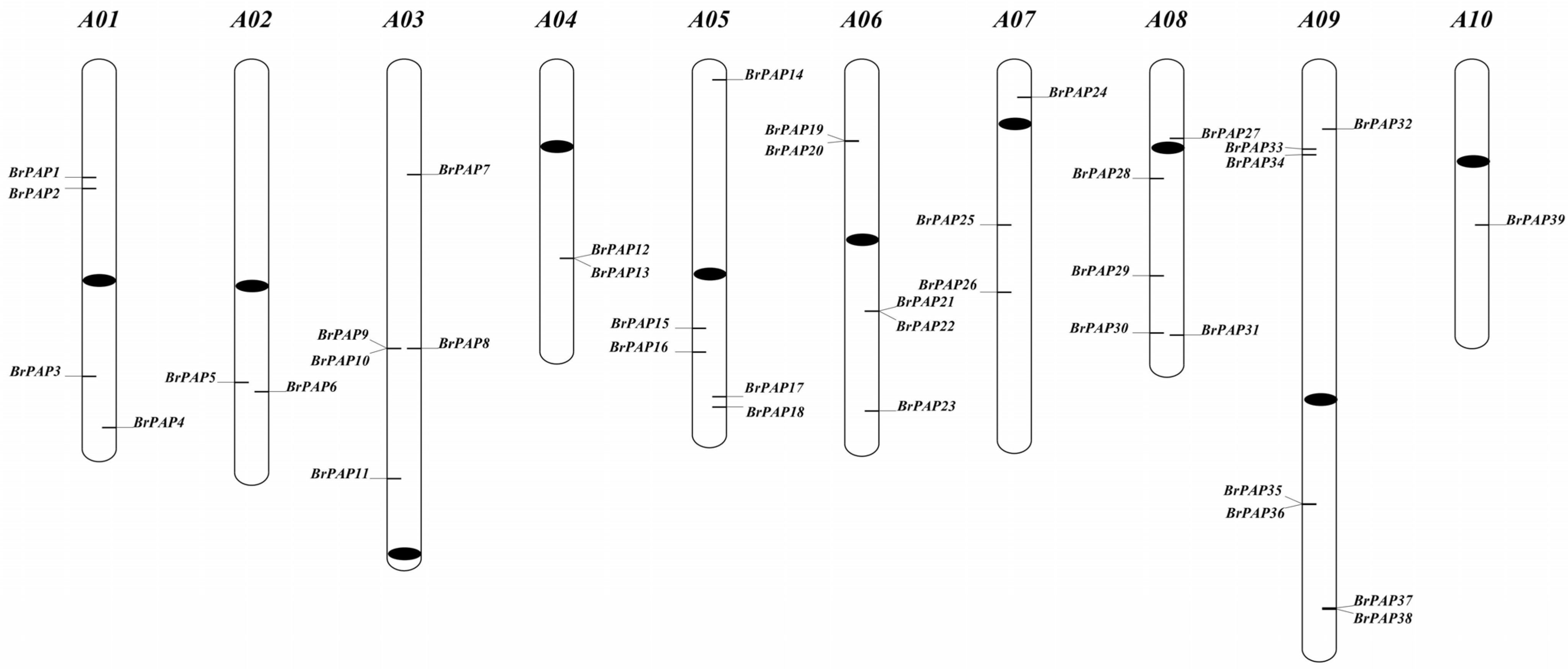
3.1. Brassica rapa PAP Genes
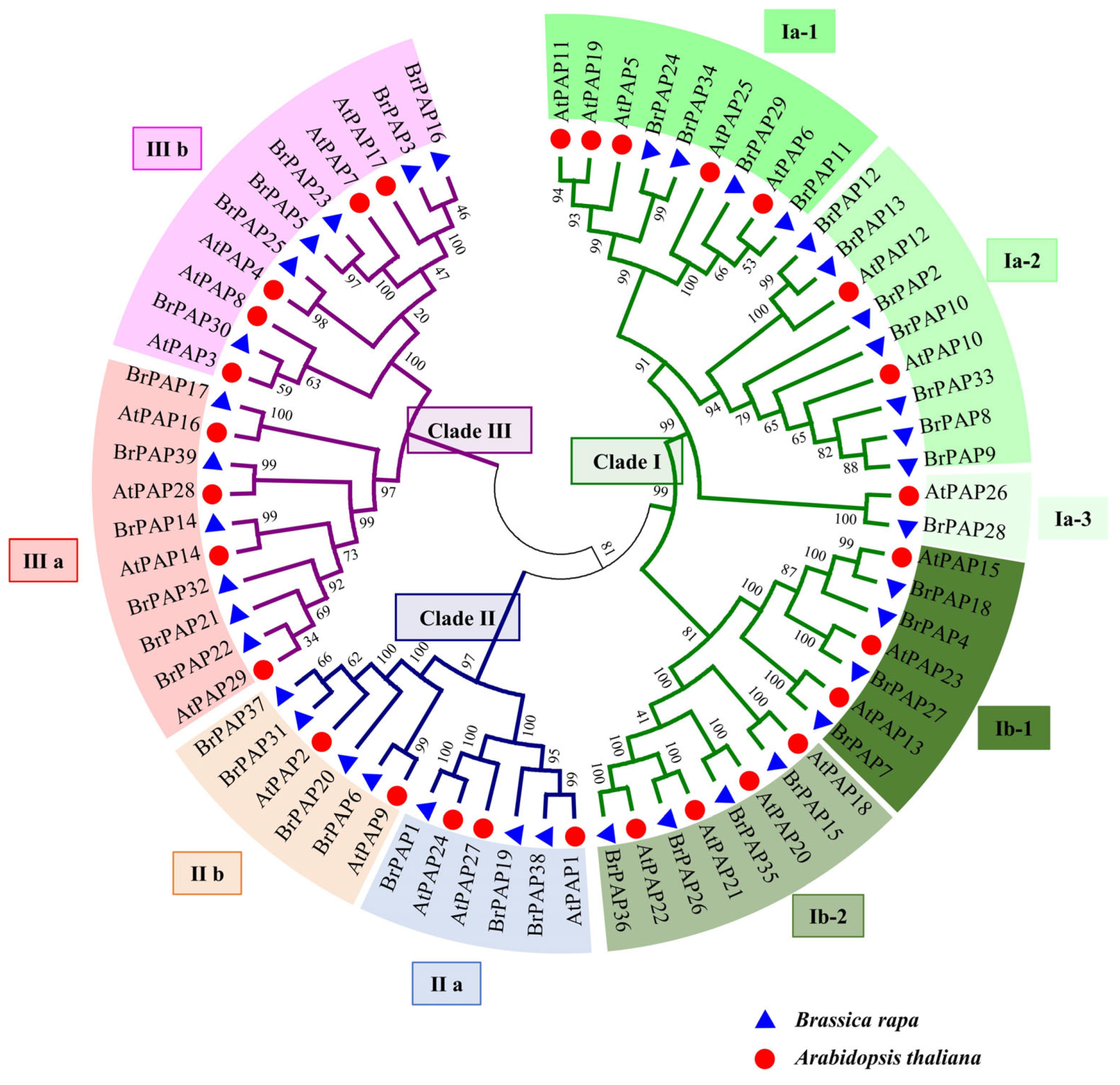
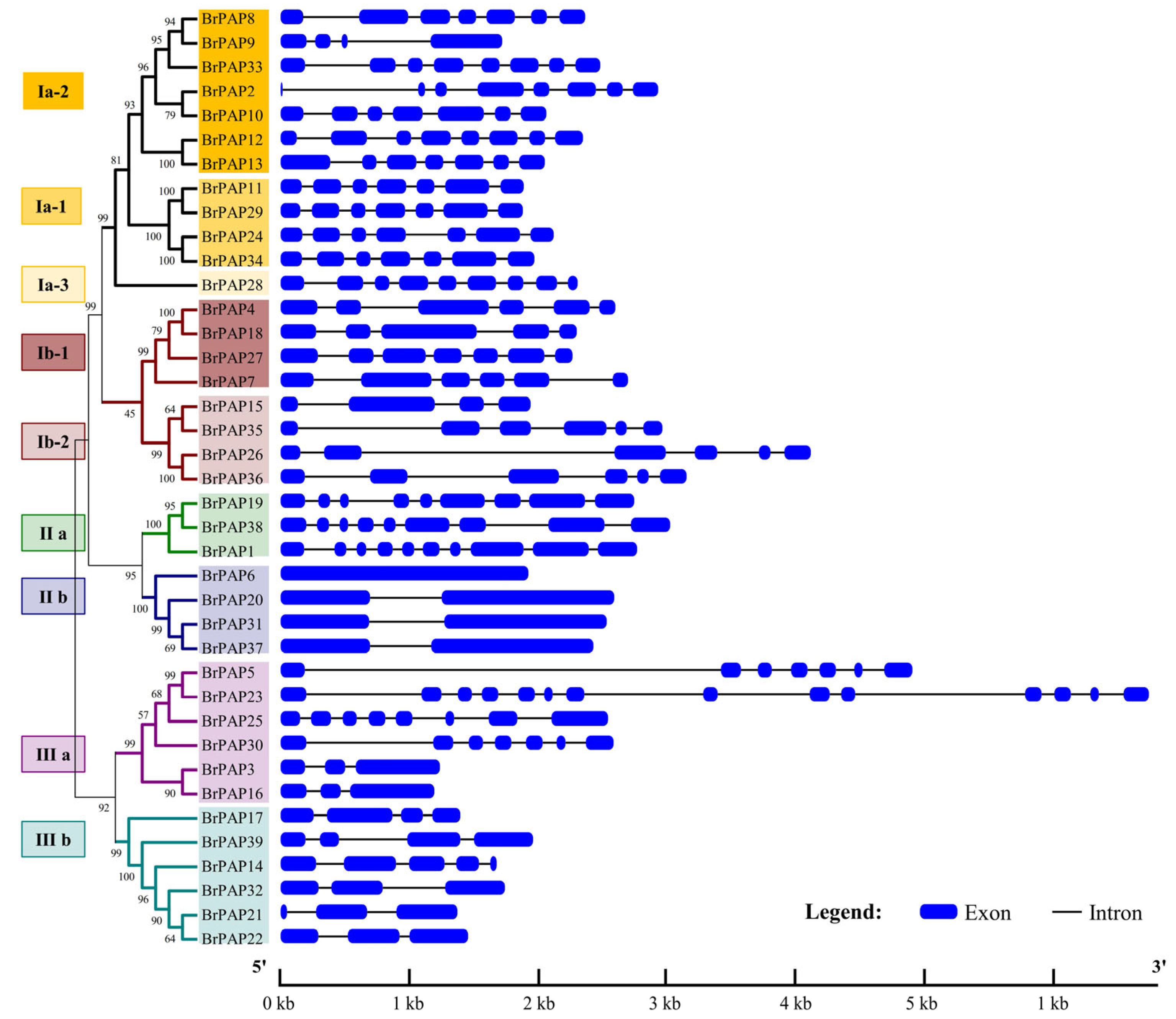
3.2. Phylogenetic and Gene Structure Analyses of BrPAPs
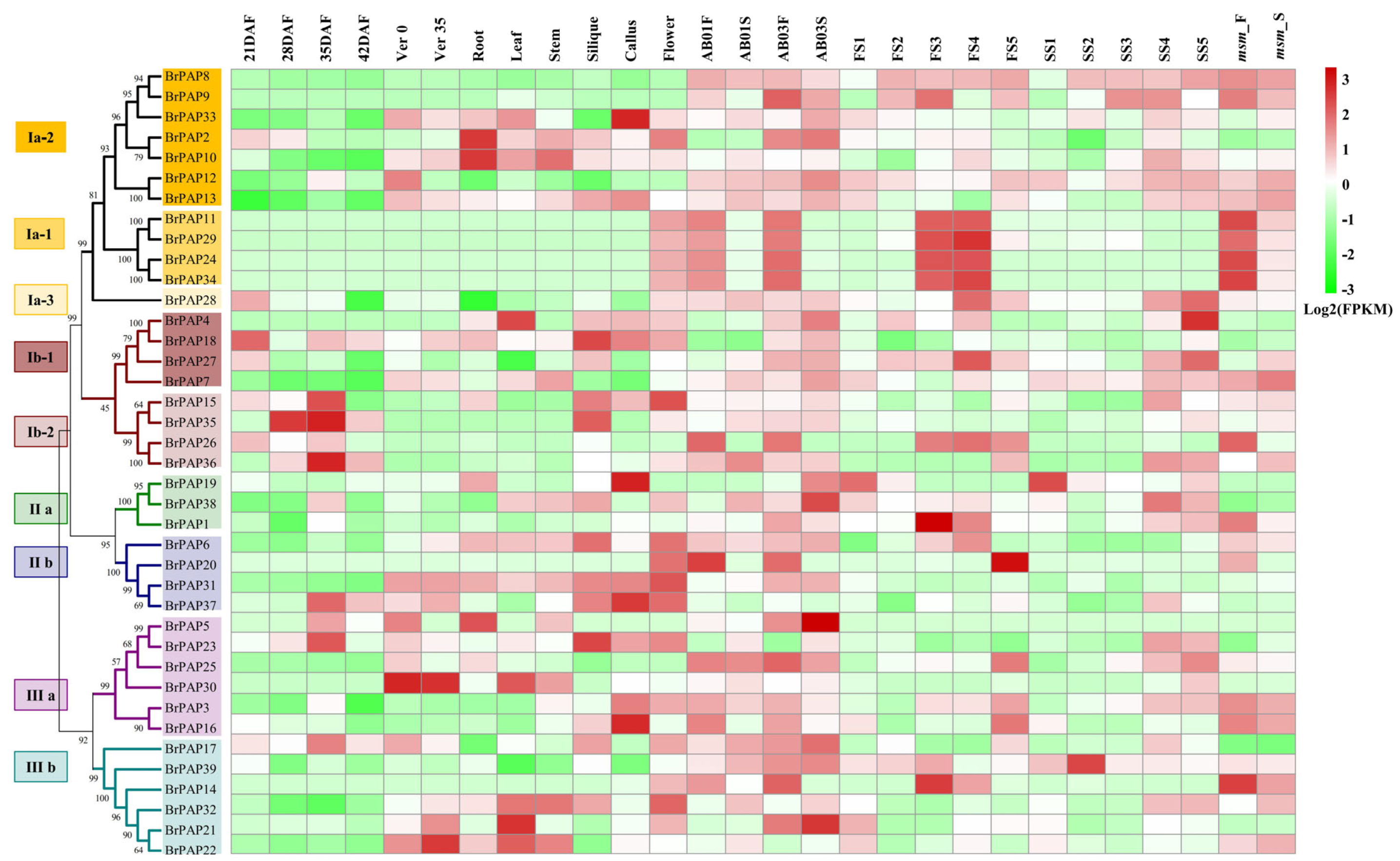
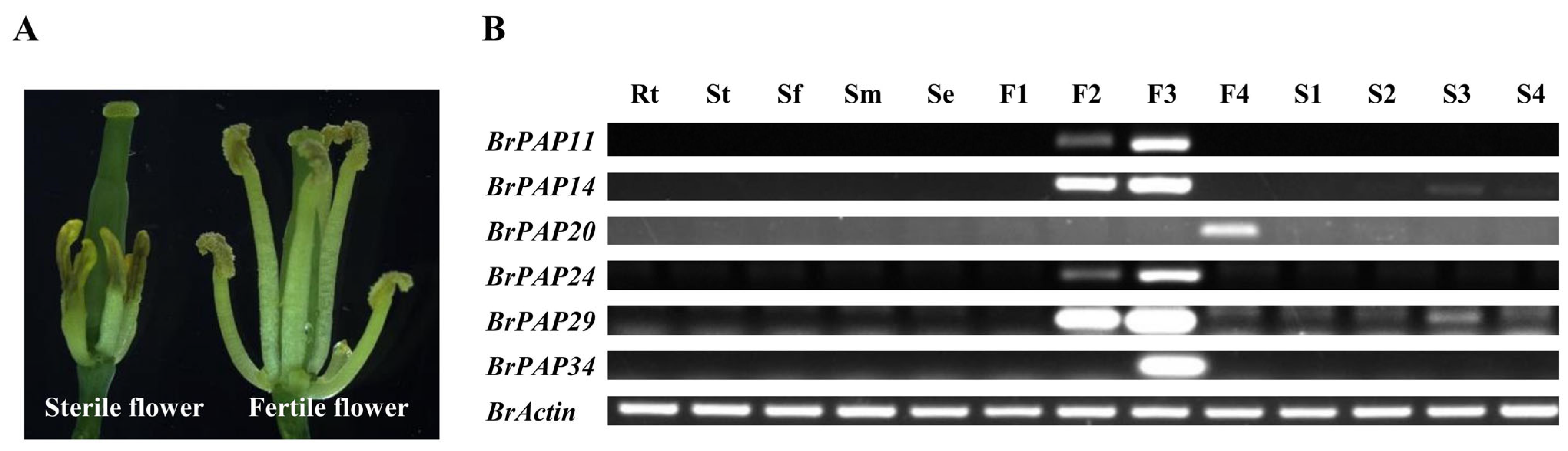
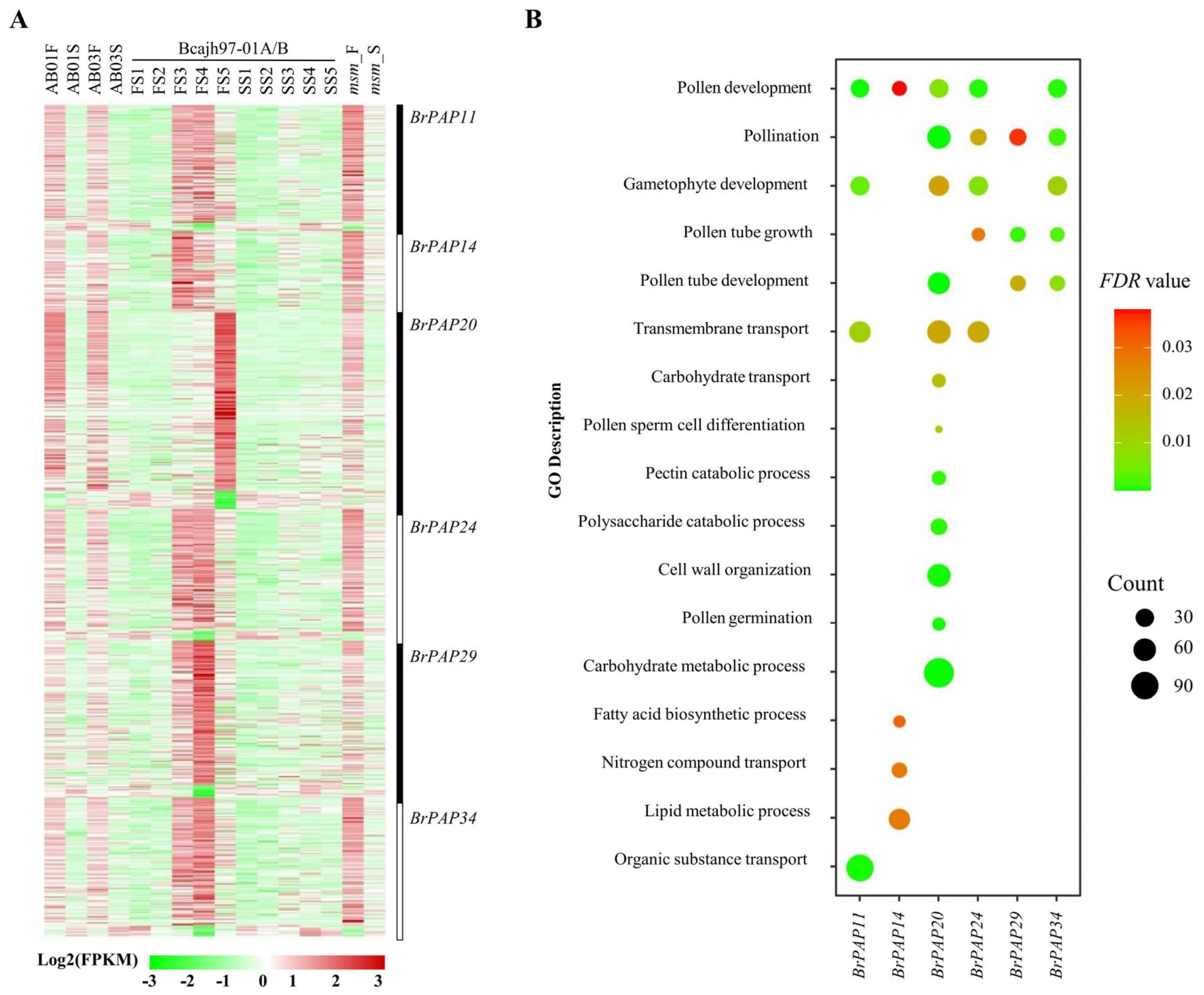
3.3. Potential Functions of BrPAPs during Pollen Development
3.4. Expression of BrPAP Genes in Response to Pi Deprivation in Brassica rapa
4. Discussion
4.1. Identification and Analysis of BrPAPs
4.2. BrPAPs and Pollen Development
4.3. Responses of Brassica rapa and BrPAPs under Pi Deprivation Conditions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Barford, D.; Das, A.K.; Egloff, M.-P. The structure and mechanism of protein phosphatases: Insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998, 27, 133–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Zhu, H.; Liu, K.; Liu, X.; Leggewie, G.; Udvardi, M.; Wang, D. Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J. Biol. Chem. 2002, 277, 27772–27781. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schenk, G.; Korsinczky, M.L.; Hume, D.; Hamilton, S.; DeJersey, J. Purple acid phosphatases from bacteria: Similarities to mammalian and plant enzymes. Gene 2000, 255, 419–424. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Qian, W.; Lu, X.; Li, D.; Liu, X.; Liu, K.; Wang, D. Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 2005, 59, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Hurley, B.A.; Plaxton, W.C. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci. 2010, 179, 14–27. [Google Scholar] [CrossRef]
- Xiao, K.; Katagi, H.; Harrison, M.; Wang, Z.-Y. Improved phosphorus acquisition and biomass production in Arabidopsis by transgenic expression of a purple acid phosphatase gene from M. truncatula. Plant Sci. 2006, 170, 191–202. [Google Scholar] [CrossRef]
- Kong, Y.; Li, X.; Wang, B.; Li, W.; Du, H.; Zhang, C. The soybean purple acid phosphatase GmPAP14 predominantly enhances external phytate utilization in plants. Front. Plant Sci. 2018, 9, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, S.; Chen, M.; Liang, C.; Xue, Y.; Lin, S.; Tian, J. Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in soybean. Front. Plant Sci. 2020, 11, 661. [Google Scholar] [CrossRef]
- Xie, L.; Shang, Q. Genome-wide analysis of purple acid phosphatase structure and expression in ten vegetable species. BMC Genom. 2018, 19, 646. [Google Scholar] [CrossRef]
- Lu, K.; Chai, Y.R.; Zhang, K.; Wang, R.; Chen, L.; Lei, B.; Lu, J.; Xu, X.F.; Li, J.N. Cloning and characterization of phosphorus starvation inducible Brassica napus PURPLE ACID PHOSPHATASE 12 gene family, and imprinting of a recently evolved MITE-minisatellite twin structure. Theor. Appl. Genet. 2008, 117, 963–975. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, C.; Tian, J.; Li, K.; Shou, H. Identification of rice purple acid phosphatases related to posphate starvation signalling. Plant Biol. 2010, 13, 7–15. [Google Scholar] [CrossRef]
- Gonzalez-Munoz, E.; Avendano-Vazquez, A.O.; Montes, R.A.; De Folter, S.; Andres-Hernandez, L.; Abreu-Goodger, C.; Sawers, R.J. The maize (Zea mays ssp. mays var. B73) genome encodes 33 members of the purple acid phosphatase family. Front. Plant Sci. 2015, 6, 341. [Google Scholar] [CrossRef] [Green Version]
- Bhadouria, J.; Singh, A.P.; Mehra, P.; Verma, L.; Srivastawa, R.; Parida, S.K.; Giri, J. Identification of purple acid phosphatases in chickpea and potential roles of CaPAP7 in seed phytate accumulation. Sci. Rep. 2017, 7, 11012. [Google Scholar] [CrossRef] [Green Version]
- Venkidasamy, B.; Selvaraj, D.; Ramalingam, S. Genome-wide analysis of purple acid phosphatase (PAP) family proteins in Jatropha curcas L. Int. J. Biol. Macromol. 2019, 123, 648–656. [Google Scholar] [CrossRef]
- Yin, C.; Wang, F.; Fan, H.; Fang, Y.; Li, W. Identification of tea plant purple acid phosphatase genes and their expression responses to excess iron. Int. J. Mol. Sci. 2019, 20, 1954. [Google Scholar] [CrossRef] [Green Version]
- Schenk, G.; Mitić, N.; Hanson, G.R.; Comba, P. Purple acid phosphatase: A journey into the function and mechanism of a colorful enzyme. Coord. Chem. Rev. 2013, 257, 473–482. [Google Scholar] [CrossRef]
- Wang, L.; Li, Z.; Qian, W.; Guo, W.; Gao, X.; Huang, L.; Wang, H.; Zhu, H.; Wu, J.W.; Wang, D.; et al. The Arabidopsis purple acid phosphatase AtPAP10 is predominantly associated with the root surface and plays an important role in plant tolerance to phosphate limitation. Plant Physiol. 2011, 157, 1283–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuang, R.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, H.T.; Qian, W.; Hurley, B.A.; She, Y.M.; Wang, D.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP12 and AtPAP26: The predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ. 2010, 33, 1789–1803. [Google Scholar] [CrossRef] [PubMed]
- Ghahremani, M.; Park, J.; Anderson, E.M.; Marty-Howard, N.J.; Mullen, R.T.; Plaxton, W.C. Lectin AtGAL1 interacts with high-mannose glycoform of the purple acid phosphatase AtPAP26 secreted by phosphate-starved Arabidopsis. Plant Cell Environ. 2019, 42, 1158–1166. [Google Scholar] [CrossRef]
- Tian, J.; Wang, C.; Zhang, Q.; He, X.; Whelan, J.; Shou, H. Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. J. Integr. Plant Biol. 2012, 54, 631–639. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Qiu, W.; Gao, W.; Tyerman, S.D.; Shou, H.; Wang, C. OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ. 2016, 39, 2247–2259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mehra, P.; Pandey, B.K.; Giri, J. Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol. J. 2017, 15, 1054–1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, S.; Lu, L.; Li, J.; Du, Z.; Liu, T.; Li, W.; Xu, F.; Shi, L.; Shou, H.; Wang, C. Purple acid phosphatase 10c encodes a major acid phosphatase that regulates plant growth under phosphate-deficient conditions in rice. J. Exp. Bot. 2020, 71, 4321–4332. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Sun, F.; Fettke, J.; Schottler, M.A.; Ramsden, L.; Fernie, A.R.; Lim, B.L. Heterologous expression of AtPAP2 in transgenic potato influences carbon metabolism and tuber development. FEBS Lett. 2014, 588, 3726–3731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaida, R.; Satoh, Y.; Bulone, V.; Yamada, Y.; Kaku, T.; Hayashi, T.; Kaneko, T.S. Activation of beta-glucan synthases by wall-bound purple acid phosphatase in tobacco cells. Plant Physiol. 2009, 150, 1822–1830. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, H.; Wong, F.-L.; Phang, T.-H.; Cheung, M.-Y.; Li, W.-Y.F.; Shao, G.; Yan, X.; Lam, H.-M. GmPAP3, a novel purple acid phosphatase-like gene in soybean induced by NaCl stress but not phosphorus deficiency. Gene 2003, 318, 103–111. [Google Scholar] [CrossRef]
- Lan, P.; Li, W.; Schmidt, W. Genome-wide co-expression analysis predicts protein kinases as important regulators of phosphate deficiency-induced root hair remodeling in Arabidopsis. BMC Genom. 2013, 14, 210. [Google Scholar] [CrossRef] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circular. Calif. Agric. Exp. Stn. 1950, 347, 32. [Google Scholar]
- Dong, X.; Feng, H.; Xu, M.; Lee, J.; Kim, Y.K.; Lim, Y.P.; Piao, Z.; Park, Y.D.; Ma, H.; Hur, Y. Comprehensive analysis of genic male sterility-related genes in Brassica rapa using a newly developed Br300K oligomeric chip. PLoS ONE 2013, 8, e72178. [Google Scholar] [CrossRef] [Green Version]
- Miao, X.; Miao, Y.; Gong, H.; Tao, S.; Chen, Y.; Chen, Z.; Liao, W. Digestion methods for determining phosphorus content in plants. Chin. Agric. Sci. Bull. 2019, 35, 132–137. [Google Scholar]
- Zhang, L.; Cai, X.; Wu, J.; Liu, M.; Grob, S.; Cheng, F.; Liang, J.; Cai, C.; Liu, Z.; Liu, B.; et al. Improved Brassica rapa reference genome by single-molecule sequencing and chromosome conformation capture technologies. Hortic. Res. 2018, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein identification and analysis tools in the ExPASy server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Armenteros, J.J.A.; Tsirigos, K.D.; Sonderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; Von Heijne, G.; Nielsen, H.B. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, C.; Wang, X.; Yu, J.; Wu, J.; Li, W.; Huang, J.; Dong, C.; Hua, W.; Liu, S. Comprehensive analysis of RNA-seq data reveals the complexity of the transcriptome in Brassica rapa. BMC Genom. 2013, 14, 689. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Liu, Z.; Li, C.; Zhang, Y.; Feng, H. Comparative transcriptome analysis of fertile and sterile buds from a genetically male sterile line of Chinese cabbage. Vitr. Cell. Dev. Biol. Plant 2016, 52, 130–139. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, Z.; Ji, R.; Feng, H. Comparative transcript profiling of fertile and sterile flower buds from multiple-allele-inherited male sterility in Chinese cabbage (Brassica campestris L. ssp. pekinensis). Mol. Genet. Genom. 2017, 292, 967–990. [Google Scholar] [CrossRef]
- Lee, Y.H.; Kim, K.S.; Lee, J.E.; Cha, Y.L.; Moon, Y.H.; Song, Y.S.; Jeong, E.G.; Ahn, S.J.; Park, W. Comprehensive transcriptome profiling in relation to seed storage compounds in tetralocular Brassica rapa. J. Plant Growth Regul. 2018, 37, 867–882. [Google Scholar] [CrossRef]
- Shen, X.; Xu, L.; Liu, Y.; Dong, H.; Zhou, D.; Zhang, Y.; Lin, S.; Cao, J.; Huang, L. Comparative transcriptome analysis and ChIP-sequencing reveals stage-specific gene expression and regulation profiles associated with pollen wall formation in Brassica rapa. BMC Genom. 2019, 20, 264. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Peng, S.; Liu, Z.; Li, C.; Tan, C.; Yao, R.; Li, D.; Li, X.; Hou, L.; Feng, H. Investigation of the genes associated with a male sterility mutant (msm) in Chinese cabbage (Brassica campestris ssp. pekinensis) using RNA-Seq. Mol. Genet. Genom. 2020, 295, 233–249. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Galaxy, T. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, A.I.; Bhagabati, N.K.; Braisted, J.C.; Liang, W.; Sharov, V.; Howe, E.A.; Li, J.; Thiagarajan, M.; White, J.A.; Quackenbush, J. TM4 microarray software suite. Methods Enzymol. 2006, 411, 134–193. [Google Scholar] [CrossRef]
- Tian, T.; Liu, Y.; Yan, H.; You, Q.; Yi, X.; Du, Z.; Xu, W.; Su, Z. agriGO v2.0: A GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 2017, 45, W122–W129. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Li, C.; Gui, S.; Yang, T.; Walk, T.; Wang, X.; Liao, H. Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann. Bot. 2011, 109, 275–285. [Google Scholar] [CrossRef] [PubMed]
- George, T.; Richardson, A. Potential and limitations to improving crops for enhanced phosphorus utilization. In The Ecophysiology of Plant-Phosphorus Interactions; Springer: Dordrecht, The Netherlands, 2008; pp. 247–270. [Google Scholar]
- Sun, F.; Liang, C.; Whelan, J.; Yang, J.; Zhang, P.; Lim, B.L. Global transcriptome analysis of AtPAP2-overexpressing Arabidopsis thaliana with elevated ATP. BMC Genom. 2013, 14, 752. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurley, B.A.; Tran, H.T.; Marty, N.J.; Park, J.; Snedden, W.A.; Mullen, R.T.; Plaxton, W.C. The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol. 2010, 153, 1112–1122. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, X.; Lu, S.; Liu, D. A major root-associated acid phosphatase in Arabidopsis, AtPAP10, is regulated by both local and systemic signals under phosphate starvation. J. Exp. Bot. 2014, 65, 6577–6588. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Gene ID | Gene Name | PAP Conserved Motifs | ||||
|---|---|---|---|---|---|---|
| GDXG | GDXXY | GNH(D/E) | VXXH | GHXH | ||
| BraA01g015600.3C | BrPAP1 | GDMG | GDITY | GNHE | FIAH | GHVH |
| BraA01g017190.3C | BrPAP2 | GDLG | GDLSY | GNHE | VLNH | GHVH |
| BraA01g034870.3C | BrPAP3 | GDWG | GDNFY | GNHD | VVGH | GHDH |
| BraA01g041630.3C | BrPAP4 | GDTG | GDVSY | GNHE | VTWH | GHVH |
| BraA02g035210.3C | BrPAP5 | GDWG | GDNFY | GNHD | VVGH | GHDH |
| BraA02g036110.3C | BrPAP6 | GDMG | GDISY | GNHE | VQGH | GHVH |
| BraA03g017330.3C | BrPAP7 | GDLG | CDSCY | GEHE | ATWS | |
| BraA03g043110.3C | BrPAP8 | GDLG | GDFSY | GNHE | VLMH | GHVH |
| BraA03g043120.3C | BrPAP9 | GDLE | YDTKY | GNCD | ||
| BraA03g043130.3C | BrPAP10 | GDLG | GDLSY | GNHE | VLNH | GHVH |
| BraA03g059950.3C | BrPAP11 | GDLG | GDLSY | GNHE | VIVH | GHVH |
| BraA04g019300.3C | BrPAP12 | GDLG | GDLSY | GNHE | VLVH | GHVH |
| BraA04g019310.3C | BrPAP13 | GDLG | GDLSY | GNHE | VLVH | GHVH |
| BraA05g001000.3C | BrPAP14 | SDMHY | GNHD | VYVH | GHDH | |
| BraA05g026730.3C | BrPAP15 | GDLG | GDLSY | GNHE | ALFH | GHVH |
| BraA05g029790.3C | BrPAP16 | GDWG | GDNFY | GNHD | VVGH | GHDH |
| BraA05g036550.3C | BrPAP17 | GDVVT | GNHD | IFWH | GHNH | |
| BraA05g038510.3C | BrPAP18 | GDLG | GDVSY | GNHE | VSWH | GHVH |
| BraA06g009980.3C | BrPAP19 | GDMG | GDICY | GNHE | FLAH | GHAH |
| BraA06g010040.3C | BrPAP20 | GDMG | GDISY | GNHE | VQGH | GHVH |
| BraA06g026750.3C | BrPAP21 | GDNIF | GNHD | AYFH | GHDH | |
| BraA06g026760.3C | BrPAP22 | ADMH | GDNIF | GNHD | AYFH | GHDH |
| BraA06g040030.3C | BrPAP23 | GDWG | GDNFY | GNHD | VVGH | GHDH |
| BraA07g002610.3C | BrPAP24 | GDLG | GDLSY | GNHE | VLVH | GHVH |
| BraA07g012620.3C | BrPAP25 | GDWG | GDNIY | GNHD | VVGH | GHDH |
| BraA07g020920.3C | BrPAP26 | GDLG | GDLSY | GNHE | AVMH | GHIH |
| BraA08g006360.3C | BrPAP27 | GDLG | GDFTY | GNHE | ATMH | GHVH |
| BraA08g009450.3C | BrPAP28 | GDLG | GDLSY | GNHE | VLMH | GHVH |
| BraA08g021060.3C | BrPAP29 | GDLG | GDLAY | GNHE | VMVH | GHVH |
| BraA08g030080.3C | BrPAP30 | GDWG | GDNFY | GNHD | VVGH | GHDH |
| BraA08g030430.3C | BrPAP31 | GDMG | GDISY | GNHE | VQGH | GHVH |
| BraA09g007730.3C | BrPAP32 | ADMH | GDNIF | GNHD | AYFH | GHDH |
| BraA09g010460.3C | BrPAP33 | GDLG | GDFSY | GNHE | VLMH | GHVH |
| BraA09g011240.3C | BrPAP34 | GDLG | GDLSY | GNHE | VLVH | GHVH |
| BraA09g043870.3C | BrPAP35 | GDLG | GDLSY | GNHE | AVIH | GHVH |
| BraA09g043900.3C | BrPAP36 | GDLG | GDLSY | GNHE | VLLH | GHVH |
| BraA09g059830.3C | BrPAP37 | GDMG | GDISY | GNHE | VQGH | GHVH |
| BraA09g059970.3C | BrPAP38 | GDMG | GDICY | GNHE | FLAH | GHAH |
| BraA10g015040.3C | BrPAP39 | ADMH | GDNIF | GNHD | AFFH | GHDH |
| Gene Name | Chromosome Location | No. of Exons | Gene Length(bp) | M.W. (kDa) | Protein Length (a.a.) | PI | Signal Peptide (Length, Cleave Site) | Number of N-Glycosylation Sites | Best Hit to Arabidopsis | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| At_ID | At_Name | E-Value | |||||||||
| BrPAP1 | A01 | 10 | 2778 | 72.67 | 642 | 6.09 | 24, GHA-SN | 1 | AT4G24890.1 | AtPAP24 | 0 |
| BrPAP2 | A01 | 8 | 2942 | 45.42 | 393 | 5.81 | ― | 4 | AT2G16430.1 | AtPAP10 | 4.05 × 10−171 |
| BrPAP3 | A01 | 3 | 1240 | 37.80 | 334 | 5.28 | 26, TNG-EL | 1 | AT3G17790.1 | AtPAP17 | 0 |
| BrPAP4 | A01 | 6 | 2607 | 60.35 | 541 | 5.23 | 32, SDA-IP | 6 | AT3G07130.1 | AtPAP15 | 0 |
| BrPAP5 | A02 | 7 | 4923 | 37.60 | 331 | 6.3 | 24, SFS-KL | 2 | AT2G01880.1 | AtPAP7 | 0 |
| BrPAP6 | A02 | 1 | 1929 | 72.89 | 643 | 6.07 | 18, VHS-TP | 4 | AT2G03450.1 | AtPAP9 | 0 |
| BrPAP7 | A03 | 6 | 2705 | 60.27 | 536 | 4.97 | 21, VDA-FP | 6 | AT2G32770.3 | AtPAP13 | 0 |
| BrPAP8 | A03 | 7 | 2372 | 56.76 | 489 | 7.16 | 24, CHG-GT | 1 | AT2G16430.2 | AtPAP10 | 0 |
| BrPAP9 | A03 | 4 | 1726 | 34.34 | 309 | 9.37 | ― | 1 | AT2G16430.2 | AtPAP10 | 2.48 × 10−51 |
| BrPAP10 | A03 | 7 | 2070 | 53.73 | 467 | 7.76 | 25, CNG-GI | 5 | AT2G16430.2 | AtPAP10 | 0 |
| BrPAP11 | A03 | 7 | 1894 | 52.19 | 461 | 6.22 | 22, ING-GM | 3 | AT1G56360.1 | AtPAP6 | 0 |
| BrPAP12 | A04 | 8 | 2354 | 55.49 | 482 | 6.16 | 28, CDG-GI | 4 | AT2G27190.1 | AtPAP12 | 0 |
| BrPAP13 | A04 | 7 | 2057 | 54.08 | 470 | 6.25 | 28, CDG-GI | 3 | AT2G27190.1 | AtPAP12 | 0 |
| BrPAP14 | A05 | 5 | 1682 | 44.56 | 394 | 8.47 | 24, VDA-YG | 1 | AT2G46880.1 | AtPAP14 | 0 |
| BrPAP15 | A05 | 4 | 1947 | 47.48 | 414 | 5.68 | 19, VAA-DD | 0 | AT3G20500.1 | AtPAP18 | 0 |
| BrPAP16 | A05 | 3 | 1196 | 38.33 | 338 | 5.57 | ― | 1 | AT3G17790.1 | AtPAP17 | 0 |
| BrPAP17 | A05 | 4 | 1399 | 42.91 | 385 | 5.86 | 23, AVG-WE | 1 | AT3G10150.2 | AtPAP16 | 0 |
| BrPAP18 | A05 | 5 | 2307 | 61.43 | 541 | 5.64 | 26, SSA-DY | 4 | AT3G07130.1 | AtPAP15 | 0 |
| BrPAP19 | A06 | 9 | 2755 | 69.52 | 621 | 7.9 | 22, GGA-IQ | 2 | AT1G13750.1 | AtPAP1 | 0 |
| BrPAP20 | A06 | 2 | 2599 | 75.79 | 680 | 5.6 | 22, ANA-EA | 3 | AT1G13900.1 | AtPAP2 | 0 |
| BrPAP21 | A06 | 3 | 1378 | 34.08 | 308 | 6.24 | ― | 0 | AT5G63140.1 | AtPAP29 | 0 |
| BrPAP22 | A06 | 3 | 1460 | 42.19 | 383 | 9.01 | 31, ASA-QG | 1 | AT5G63140.1 | AtPAP29 | 0 |
| BrPAP23 | A06 | 14 | 6766 | 69.10 | 607 | 5.51 | 29, SRA-EL | 3 | AT2G01890.1 | AtPAP8 | 0 |
| BrPAP24 | A07 | 7 | 2127 | 52.10 | 460 | 5.86 | 24, SHA-GV | 4 | AT4G36350.1 | AtPAP25 | 0 |
| BrPAP25 | A07 | 8 | 2551 | 52.34 | 468 | 7.17 | ― | 2 | AT1G25230.3 | AtPAP4 | 0 |
| BrPAP26 | A07 | 6 | 4130 | 50.56 | 437 | 8.25 | 22, SQA-YN | 1 | AT3G52810.1 | AtPAP21 | 0 |
| BrPAP27 | A08 | 7 | 2273 | 61.20 | 548 | 5.02 | 31, AGG-ES | 4 | AT4G13700.1 | AtPAP23 | 0 |
| BrPAP28 | A08 | 9 | 2313 | 55.40 | 481 | 7.35 | 27, GEG-GI | 1 | AT5G34850.1 | AtPAP26 | 0 |
| BrPAP29 | A08 | 7 | 1887 | 52.42 | 460 | 6.6 | 19, ING-GI | 1 | AT1G56360.1 | AtPAP6 | 0 |
| BrPAP30 | A08 | 7 | 2595 | 38.03 | 335 | 5.81 | 29, STA-EL | 1 | AT1G14700.1 | AtPAP3 | 0 |
| BrPAP31 | A08 | 2 | 2540 | 73.13 | 652 | 5.95 | 2, ANA-KA | 6 | AT1G13900.1 | AtPAP2 | 0 |
| BrPAP32 | A09 | 3 | 1746 | 42.92 | 386 | 9.2 | 32, TSS-HR | 2 | AT5G63140.1 | AtPAP29 | 0 |
| BrPAP33 | A09 | 8 | 2490 | 54.12 | 469 | 6.77 | 29, CHG-GR | 3 | AT2G16430.2 | AtPAP10 | 0 |
| BrPAP34 | A09 | 7 | 1977 | 52.82 | 468 | 5.49 | 23, SHA-GV | 3 | AT2G18130.1 | AtPAP11 | 0 |
| BrPAP35 | A09 | 6 | 2974 | 46.99 | 415 | 6.01 | 21, VSS-YD | 1 | AT3G52780.1 | AtPAP20 | 0 |
| BrPAP36 | A09 | 6 | 3163 | 51.08 | 448 | 5.79 | 35, SQA-DV | 2 | AT3G52820.1 | AtPAP22 | 0 |
| BrPAP37 | A09 | 2 | 2436 | 73.36 | 653 | 6.18 | 21, ANA-KA | 4 | AT1G13900.1 | AtPAP2 | 0 |
| BrPAP38 | A09 | 9 | 3035 | 69.22 | 622 | 6.16 | 24, ALG-GR | 4 | AT1G13750.1 | AtPAP1 | 0 |
| BrPAP39 | A10 | 4 | 1966 | 45.34 | 404 | 6.83 | ― | 3 | AT5G57140.1 | AtPAP28 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, Y.; Qi, J.; Li, C.; Miao, K.; Jiang, B.; Yang, X.; Han, W.; Wang, Y.; Gao, J.; Dong, X. Genome-Wide Analysis of Purple Acid Phosphatase Genes in Brassica rapa and Their Association with Pollen Development and Phosphorus Deprivation Stress. Horticulturae 2021, 7, 363. https://doi.org/10.3390/horticulturae7100363
Cai Y, Qi J, Li C, Miao K, Jiang B, Yang X, Han W, Wang Y, Gao J, Dong X. Genome-Wide Analysis of Purple Acid Phosphatase Genes in Brassica rapa and Their Association with Pollen Development and Phosphorus Deprivation Stress. Horticulturae. 2021; 7(10):363. https://doi.org/10.3390/horticulturae7100363
Chicago/Turabian StyleCai, Yongfang, Jiao Qi, Chun Li, Kehui Miao, Baixue Jiang, Xiaoshuang Yang, Wenyu Han, Yang Wang, Jing Gao, and Xiangshu Dong. 2021. "Genome-Wide Analysis of Purple Acid Phosphatase Genes in Brassica rapa and Their Association with Pollen Development and Phosphorus Deprivation Stress" Horticulturae 7, no. 10: 363. https://doi.org/10.3390/horticulturae7100363
APA StyleCai, Y., Qi, J., Li, C., Miao, K., Jiang, B., Yang, X., Han, W., Wang, Y., Gao, J., & Dong, X. (2021). Genome-Wide Analysis of Purple Acid Phosphatase Genes in Brassica rapa and Their Association with Pollen Development and Phosphorus Deprivation Stress. Horticulturae, 7(10), 363. https://doi.org/10.3390/horticulturae7100363






