Abstract
Sweet potato cultivars obtained from different nursery farmers were cultivated in an experimental field from seedling-stage to harvest, and the acetylene reduction activity (ARA) of different parts of the plant as well as the nifH genes associated with the sweet potatoes were examined. The relationship between these parameters and the plant weights, nitrogen contents, and natural abundance of 15N was also considered. The highest ARA was detected in the tubers and in September. Fragments of a single type of nitrogenase reductase gene (nifH) were amplified, and most of them had similarities with those of Enterobacteriaceae in γ-Proteobacteria. In sweet potatoes from one nursery farm, Dickeya nifH was predominantly detected in all of the cultivars throughout cultivation. In sweet potatoes from another farm, on the other hand, a transition to Klebsiella and Phytobacter nifH was observed after the seedling stage. The N2-fixing ability contributed to plant growth, and competition occurred between autochthonous and allochthonous bacterial communities in sweet potatoes.
1. Introduction
The sweet potato (Ipomoea batatas L.) is a dicotyledonous plant that belongs to the family Convolvulaceae and is a subsistence crop with huge economic importance, especially in developing countries. It is known that the sweet potato grows well in nitrogen-poor infertile soils, and it is believed that this growth ability is partially due to the functions of diazotrophic growth-promoting bacteria that contain endo- and epiphytic microorganisms of this plant [1,2,3].
A diverse range of endophytic bacteria have been isolated from sweet potatoes—namely, Azospirillum sp. [4], Gluconacetobacter sp. (formerly Acetobacter sp.) [5], Klebsiella sp. [6], Pantoea sp. and Enterobacter sp. [7], Bradyrhizobium sp., Paenibacillus sp., and Pseudomonas sp. [8]. Khan and Doty [9] identified Enterobacter sp., Rahnella sp., Rhodanobacter sp., Pseudomonas sp., Stenotrophomonas sp., Xanthomonas sp., and Phyllobacterium sp. from sweet potato stems. In a study by Marques et al. [10], 17 different genera belonging to α- and γ-Proteobacteria, Actinobacteria, and Firmicutes were isolated from three sweet potato genotypes, with Bacillus strains predominating in two of these genotypes and strains belonging to γ-Proteobacteria predominating in the third.
The plant growth-promoting properties of the above-mentioned isolated endophytes include nitrogen fixation, phytohormone production, siderophore production, antimicrobial activity against phytopathogens, and phosphate solubilization [7,9,10]. In addition to these properties, which have been examined in vitro using artificial media, plant inoculation assays for some of the isolated endophytes indicated that they had the potential to promote plant growth in vivo [8,9].
Culture-independent analysis of endophytic DNA from sweet potatoes, which was performed using polymerase chain reaction denaturing gradient gel electrophoresis (PCR-DGGE) for 16S rRNA genes [10] and using a PCR clone library for nitrogenase reductase genes (nifH) [11], also suggested the presence of a diverse range of endophytic bacteria including diazotrophs. These studies suggested that the diverse population of endophytic bacteria interact among each other as well as with their host plant and that their community structures and potentials could be influenced by their biotic and abiotic environments.
Although there have been many examples of isolation of the diazotrophic endophytes in sweet potato, few reports are available dealing with their ecological nature through the cultivation of the plant in the field, such as changes in their communities and nitrogen-fixing activities and differences among nursery farms and cultivars. Considering the potential roles of endophytes, understanding their ecological perspectives and their utilization would contribute to developing sustainable agriculture.
In this study, we cultivated different sweet potato cultivars obtained from different nursery farmers in an experimental field from seedling-stage to harvest and examined the acetylene reduction activity (ARA) of different parts of the plant as well as the nifH genes associated with the sweet potatoes. The relationship between these parameters and the plant weights, nitrogen contents, and the natural abundance of 15N is also discussed.
2. Materials and Methods
2.1. Cultivation and Sampling of Sweet Potatoes
Scions of three sweet potato cultivars, Beniharuka (B), Purple Sweet Lord (P), and Quick Sweet (Q), were obtained from the fields of four different nursery farmers located in Saitama (Sa), Shimane (Sh), Miyazaki (Mi), and Kagoshima (Ka) prefectures in Japan. The precise location, climatic and soil parameters of the farmers are presented in Table 1. They were planted at random on ridges (1 m inter-row, 0.15 m height) at a density of 2.5 plants m−2 in a rooftop experimental field (6.7 m × 8.3 m) [12] at Shimane University in Shimane, Japan. The field was filled with artificial soil (Viva soil; Toho Leo Co., Osaka, Japan) that had high porosity (45%) and contained very little nutrition, and was applied with a chemical fertilizer (N:P2O5:K2O = 3:8:15 g m−2) at planting. The plants were cultivated from June to October in 2014 with drip irrigation (Super Typhoon NETAFIM Co., Tel Aviv, Israel). During the cultivation, five plants were taken at each of four different time points: June (before planting), July, September, and October (harvesting).

Table 1.
Climate conditions and soil properties at the sweet potato-nursery farms.
2.2. ARA
Each plant sample was dissected into leaves, stems, roots, and tubers. After each sample was washed with tap water and its total weight was measured, a few leaves at the uppermost position (2–3 g), the upper and lower parts of the stem (8 cm, 1–4 g), the underground part of the stem (1–6 g), and a part of the root (2–3 g) were taken for the ARA assay. For the tubers, the largest tuber was cut longitudinally in quarters, and middle and both end parts of the one-quarter sample were assayed together (2–3 g). In addition to the plant samples, ca. 2 g of soil samples were taken from the rhizosphere, and ARA was measured at the same time. The remaining samples were oven-dried at 80 °C for more than 48 h to measure their nitrogen contents and natural 15N abundance.
The plant and soil samples were placed in a 100-mL glass bottle with 1 mL of sterilized distilled water. The bottle was sealed with a butyl-rubber cap and an aluminum stopper, and ca. 10% of the headspace gas was exchanged with acetylene. The bottles were incubated at 28 °C for 3 days, and 0.5 mL of the headspace gas was injected into a gas chromatograph (Shimadzu GC-14B; Shimadzu, Kyoto, Japan) equipped with a flame ionization detector and Porapak N (50/80 mesh; GL Sciences, Tokyo, Japan) for ethylene measurement. After the incubation, the samples were stored at −20 °C for the extraction of DNA. Control samples without acetylene were set under the same conditions, and no detectable ethylene production was observed.
The ARA was expressed as specific activity defined as the production of ethylene (nmol)/sample fresh weight (g)/incubation period (h), and as total activity defined as the production of ethylene (nmol)/incubation period (h)/plant by multiplying the sample fresh weight by its specific activity.
2.3. Nitrogen Content and Natural 15N Abundance
The oven-dried above-ground samples were machinery-pulverized to fine powder with a vibrating sample mill (model T-100; Cosmic Mechanical Technology. Co., Ltd., Fukushima, Japan), and then subjected to analysis of their nitrogen and ∆15N contents using a Rapid NIII element analyzed (Elementar Analysensysteme GmpH, Hanau, Germany) and an isotopic mass-spectrometer, the Flash EA1112-PELTAV ADAVANTAGE Con FLoIV system (Thermo Fisher Scientific, Waltham, MA, USA), respectively.
2.4. Analysis of nifH Genes Prepared from Plant Samples
The samples were taken after the ARA assay and further frozen in liquid nitrogen and crushed with a mortar and pestle. DNA was extracted from the samples with ISOPLANT II (Nippon Gene, Tokyo, Japan) according to the procedures described in the manual and used as a template for the PCR amplification of nifH gene fragments using a set of PolF and PolR primers designed based on the conserved sequences of the gene [13]. The PCR conditions were as follows: pre-run at 95 °C for 2 min, 40 cycles of denaturation at 95 °C for 2 min, annealing at 53 °C for 1 min and extension at 72 °C for 0.5 min, and then a post-run at 72 °C for 10 min. The components of the PCR mixture, the procedures for processing the PCR products, and the DNA sequencing and analysis were described in our previous paper [14].
2.5. Statistical Analysis
Statistical analysis was conducted using Tukey’s test after one-way ANOVA and Pearson correlation coefficient using IMB SPSS Statistics ver. 25 (IBM Co., Armonk, NY, USA).
3. Results
3.1. Plant Growth and Nitrogen Content
The total fresh weights of the sweet potato samples from September and October (harvesting) are shown in Figure 1. Sa-Q was smaller than the other cultivars due to Fusarium wilt and could not be harvested in October. The cultivar Sa-P was also smaller than the cultivar Sa-B. Among the cultivar B samples from the four different farmers, the weights were not different in September, but they differed in October in the following order: Sa > Mi > Sh = Ka. Both the aboveground and underground parts of the Sa-B plants increased after September, but only the underground parts of Mi-B increased. For Ka-B and Sh-B, growth was not observed during the period.
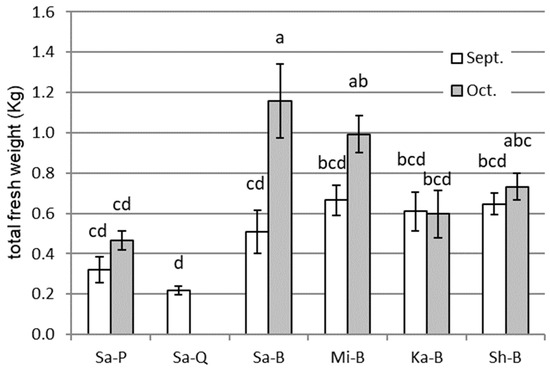
Figure 1.
Total fresh weight of different sweet potato cultivars of different origins at two measurement time points, September and October, in the course of cultivation in an experimental field. Sa, Mi, Ka, and Sh represent the nursery farms in Saitama, Miyazaki, Kagoshima, and Shimane prefectures in Japan, respectively. P, Q, and B represent the following sweet potato cultivars, respectively: Purple Sweet Lord, Quick Sweet, and Beniharuka. The error bars indicate standard errors (n = 5, and n = 4 for Sa-Q), and different letters indicate significant differences at P < 0.05 by Tukey’s test. Sa-Q could not be harvested due to plant disease.
The nitrogen contents of the aboveground parts of the sweet potato samples in September and October are shown in Figure 2. Sa-Q showed higher contents (1.85%) than the other samples (1.32–1.55%). The values were not significantly different among the other samples.
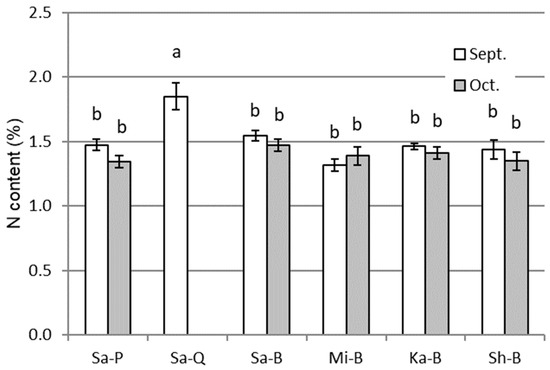
Figure 2.
Nitrogen contents of the aboveground parts of different sweet potato cultivars of different origins at two measurement time points, September and October, in the course of cultivation in an experimental field. All other details are the same as in Figure 1.
3.2. ARA
The specific ARAs of each part of the sweet potato Sa-B and the rhizosphere soil samples throughout the cultivation are shown in Figure 3. The ARA showed rather large deviation among the replicates for all of the parts and the cultivation periods. As an example, the activities of each replicate of the samples in September are presented in Figure 4. Although there were large variations among the plants, the activity was highest in September followed by July, October, and June. Regarding the plant parts, the tuber showed the highest activity in September. Considering the weight of each plant part, almost all activity existed in the tuber, as shown in Figure 5.
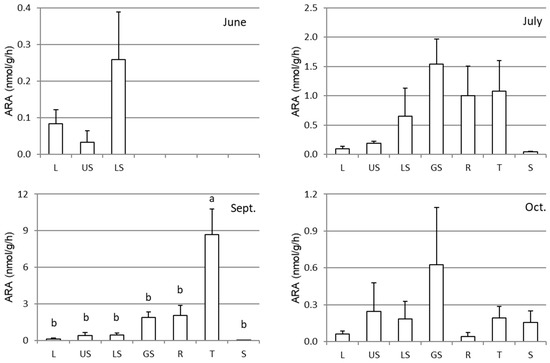
Figure 3.
Acetylene reduction activities (ARA) in different parts of sweet potatoes (Sa-B) cultivated in an experimental field from seedling-stage (June) to harvest (October). L, US, LS, GS, R, T, and S represent the leaf, upper stem, lower stem, underground stem, root, tuber, and rhizosphere soil, respectively. The error bars indicate standard errors (n = 5), and different letters indicate significant differences at P < 0.05 by Tukey’s test.
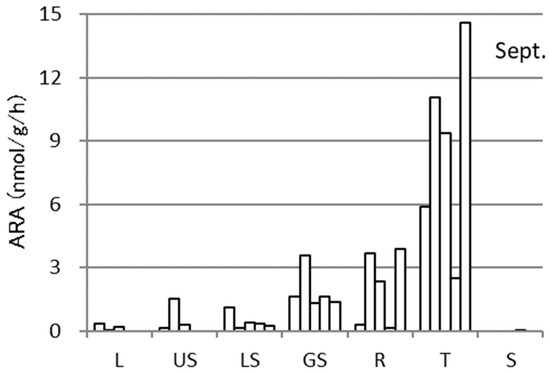
Figure 4.
Deviation of acetylene reduction activities (ARA) among five replicates in different parts of sweet potatoes (Sa-B) at one measurement time point, September, over the course of cultivation in an experimental field. All other details are the same as in Figure 3.
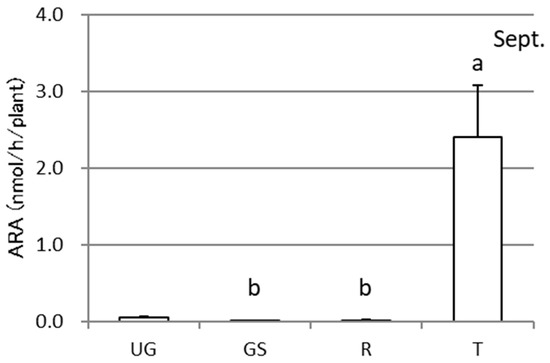
Figure 5.
Total acetylene reduction activities (ARA) in different plant parts of sweet potatoes (Sa-B) at one measurement time point, September, over the course of cultivation in an experimental field. UG, GS, R, and T represent the aboveground, underground stem, root, and tuber, respectively. The error bars indicate standard errors (n = 5), and different letters indicate significant differences at P < 0.05 by Tukey’s test.
The same trend of higher specific ARA activity in tubers and in September was observed in the other samples with the exceptions being tubers of Ka-B and Sh-B, and the activity showed a tendency to increase in the Sa farm compared to others, but no significant difference were observed due to the large deviations (Figure 6).
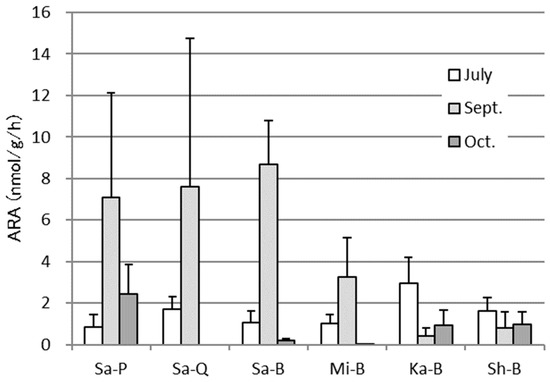
Figure 6.
Acetylene reduction activities (ARA) in tubers from sweet potato cultivars of different origins at three measurement time points, July, September, and October, during the course of cultivation in an experimental field. The same footnote as Figure 1. The error bars indicate standard errors (n = 5), and no significant difference was detected at P < 0.05 by Tukey’s test.
3.3. Relationship of ARA with Plant Growth and ∆15N Content
When the relationship between the ARA and the sample fresh weight was examined over the specific cultivation periods for the same cultivar from different farmers, no correlation was observed (Figure 7).
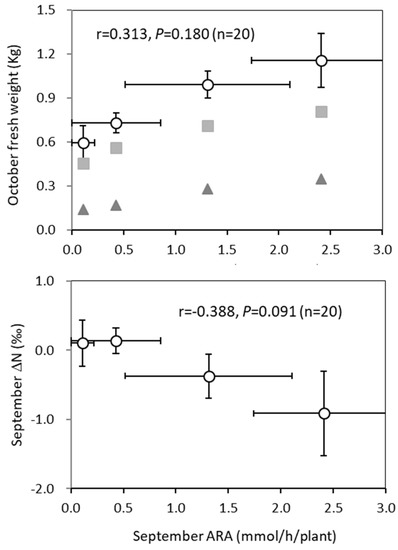
Figure 7.
Relationship of ARA in September with fresh weight in October and ∆15N content in September in sweet potato tubers (Beniharuka) with different origins during the course of cultivation in an experimental field. Closed squares and triangles represent the fresh weights of the aboveground and underground parts of the sweet potatoes, respectively. The error bars indicate standard errors (n = 5), and Pearson correlation coefficient was examined (n = 20).
In the case of ∆15N content, there was no correlation between the ARA in September and the ∆15N content in September (Figure 7).
3.4. nifH Genes in Sweet Potato
The PCR primer set used in this study amplified nifH fragments from 61% of the sweet potato samples (Table 2). The percentage of amplification differed among nursery farms, plant cultivars, and sampling times. That is, the value was lower in Sa-Q (27%) than Sa-P (75%) and Sa-B (80%) from the same Sa farm, and the values in Ka-B (50%) and Mi-B (50%) were lower than those in Sa-B (80%) and Sh-B (75%) for the same cultivar from different farms. The average values were lower for all of the samples collected in July (53%) and October (48%) than for those collected in the nursery (in June) (77%) and in September (63%). Almost all the amplified bands (84% on average) were successfully sequenced as they were directly used as templates, and the appearance of the nucleotide signals in the charts and the results of the BLAST (Basic Local Alignment Search Tool) search indicated that a specific type of nifH fragment was dominant in 86% of the amplified bands. In the other samples, the signals were mixed with multiple nucleotides in several positions, indicating that the amplified bands consisted of various nifH fragments. The presence of a mixed community was obvious in the sample Mi-B, in which the dominant nifH fragment comprised only 43%.

Table 2.
Percentages of sweet potato tubers exhibiting PCR amplification for nifH, sequenced PCR fragments, PCR fragments with dominant nifH, and nifH genes similar to Enterobacteriaceae, Dickeya, Klebsiella, and Phytobacter. The same abbreviations as in Figure 1.
The results of a BLAST search for the amplified nifH fragments are summarized in Table 3. Among them, 29 samples (57%) showed sequences most similar to those of nifH of Dickeya dadantii 3937 belonging to Enterobacteriaceae in γ-Proteobacteria. The sequences of the second-most abundant fragments (12%) were closest to those of Klebsiella sp. and Phytobacter ursingii CAV1151 in Enterobacteriaceae. Including the other samples that were closest to nifH of Enterobactor sp. and Pantoea sp., nifH of Enterobacteriaceae strains was detected as a major fragment in 86% of the samples. The other minor nifH sequences detected included sequences similar to those of Paenibacillus, Azotobacter, Bradyrhizobium, Azohydromonas, and Variovorax. Phylogenetic trees of the representative nifH genes are presented in Supplementary Figure S1.

Table 3.
Similarities of sweet potato nifH gene sequences to the most similar sequences from known diazotrophic bacteria.
The dominance of the Enterobacteriaceae nifH was different among sweet potato samples as well as at a genus level (Table 2). The Mi-B samples showed less dominance (33%) than the other samples (70%–100%), among which Dickeya nifH was more dominant in all of the cultivars from the Sa farm (71%–100%) than in the cultivars from the other farms (29%–43%). In Ka-B and Sh-B, Klebsiella and Phytobacter nifH (14%–43%) were present at levels comparable to those of Dickeya nifH.
The presence of nifH of the dominant genera was associated with the cropping stage. Dickeya nifH was constantly detected from the seedling to harvest stages (63%–71%); on the other hand, Klebsiella nifH was not detected at the seedling stage, and Phytobacter nifH was mostly detected at the seedling stage (Table 2).
4. Discussion
Differences in the growth of the sweet potato plants were observed among different cultivars obtained from the same nursery farm and among identical cultivars obtained from different nursery farms. The differences were clear after September, when carbohydrates were accumulated in the tubers. Several studies have reported that endophytic N2-fixation significantly contributed to the nitrogen supply of the sweet potato [1,2,3]. The ARA in September, when most samples presented the highest activity, showed no correlation with either the weight in September or the 15N content in October, and the ARA in October showed no correlation with any parameters. We conjectured that sweet potatoes showed higher ARA fixed atmospheric nitrogen mostly in September (the high temperature season, Supplementary Table S1), which caused lower plant 15N content in September and produced the heavier tubers at harvest in October as a result. However, the relationship between ARA and the fresh weight increase was low and insignificant due to the large deviation among the samples. In addition to N2-fixation, other beneficial effects like phytohormone production could have contributed to the fresh weight increase.
The ARA was mainly distributed in the tuber parts of the sweet potatoes grown in the field. In this study, the samples were washed with tap water to remove almost all soil and then underwent the assay. Surface sterilization was not conducted in the process; therefore, microorganisms residing inside as well as on the surface of the samples were targeted for the survey because both N2-fixing bacteria would contribute to the nitrogen supply. Previous reports suggested that the microbial communities were different inside the sweet potato tubers than in the rhizosphere of the tubers [10,15], but separation was not considered in this study.
The highest specific and total ARAs were observed in the tubers, where favorable environmental conditions would be supposed for N2-fixing bacteria because abundant carbohydrates were available and nitrogen sources were limited. In addition, microaerobic conditions in the tubers were expected because Terakado-Tonooka et al. [8] reported that the ARA of the endophytic isolate from the sweet potato was detected only under oxygen levels of less than 5%. The heavier weight of the tubers in the whole plant would also contribute to the high total ARA in this part of the sweet potato.
For the measurement of ARA in large samples such as tubers, the samples were cut into small pieces, and a few pieces were combined in a bottle for the assay. It should be considered that some changes in conditions may have been caused by the sample processing and that the diffusion efficiency of gases, including acetylene and ethylene, and the uneven distribution of bacteria in and on the sample may have affected the data and contributed to the frequent deviation in data among the replicates in this study.
Transitions of sweet potato nifH in different cultivars and origins during the course of cultivation are summarized in Table 4. nifH was detected in 67% of the nursery sweet potato samples (Table 2); these included the samples from all nursery farmers and cultivars examined. The extraction of DNA was conducted before these plants were transplanted to the field. Therefore, we considered that nifH-containing bacteria resided in the nurseries at the start of the experiment, and that they had originated from their mother plants and/or soils in their nursery farms. In support of this idea, nifH similar to Dickeya’s was predominantly detected in all of the nurseries of the different cultivars from the same Sa farm. The results suggested that nursery environments should be taken into account when examining the endophytic bacterial communities of sweet potatoes. Phylogenetically similar nifH genes were also predominant in the nurseries from the Ka farmer, while different nifH genes similar to Phytobacter’s were predominant in the samples from the Sh and Mi farmers, which might reflect the different environments in the different nursery farms.

Table 4.
Transitions of sweet potato nifH in different cultivars of different origins during the course of cultivation in an experimental field. The same abbreviations as in Figure 1.
More than 85% of the detected nifH genes were identified as members of the Enterobacteriaceae family due to the dominant presence of Dickeya’s nifH in the Sa samples. The nifH genes were detected throughout the cultivation stage for all of the cultivars. Different compositions of nifH genes were previously detected in Japanese and African sweet potatoes. In two cultivars of field-grown sweet potatoes in Japan, nifH genes of 14 genera were detected; these genera included four genera in Cyanobacteria and β-Proteobacteria and five genera in α-Proteobacteria, of which the α-Proteobacteria genera dominated [16]. Seven cultivars of African sweet potatoes collected from three regions showed the presence of nifH in eight genera belonging to five classes (α-, β-, and γ-Proteobacteria, Firmicutes, and Clostridia), among which α-Proteobacteria also dominated [11]. Rhizobium, Bradyrhizobium, and Shinorhizobium were common genera in α-Proteobacteria in both of the studies, but only Bradyrhizobium was detected as a minor genus in this study. The reason for the difference in the nifH diversity remains unclear. Regarding the specificity of the universal primer set for nifH [13] used in this study, the results in the other reports using this primer set for analyzing diversity of nifH in sweet potatoes [16], an epiphyte orchid, Dendrobium catenatum Lindl. [17], and Himalayan soils [18] suggested that there seemed to be no apparently biased PCR amplification by the primer set.
In addition to the direct DNA sequencing of the amplified nifH fragments used in this study, the construction of a clone library for the gene fragments would make more precise analyses possible for the purpose of examining minor components. The direct sequencing indicated the presence of a dominant nifH in the samples in most cases. No further method was therefore applied in this study. Due to the nature of the nucleic acid sequence data obtained in this study, the sequences are not deposited into the database but supplied as a supplemental material with the results of their BLAST search (Supplementary Table S2).
To the best of our knowledge, this is the first study to examine the transition of sweet-potato-associated nifH-containing bacteria from the seedling stage to harvest. In all of the cultivars from the Sa farm, Dickeya’s nifH was predominantly detected throughout the cultivation. On the other hand, the predominant Dickeya’s nifH in the nurseries from the Ka farm was not detected thereafter, and the nifH genes of Klebsiella and Phytobacter, both belonging to γ Proteobacteria, appeared instead in September and October, respectively. It was considered that the transition occurred as a result of competition between autochthonous and allochthonous bacterial communities. The transitions were also observed in Sh-B and Mi-B, and nifH genes of Klebsiella and Paenibacillus were detected only after planting, suggesting that they originated from the soil of the experimental field.
Although the endophytic nifH of Enterobacteriaceae is not often found in Japanese [16] and African [11] sweet potatoes, several endophytic strains belonging to Pantoea, Klebsiella, Enterobacter, and Phytobacter, all of which are Enterobacteriaceae, have been isolated from the sweet potatoes as major constituents [6,9,10,12]. The last two studies were conducted using sweet potato samples cultivated in the same experimental field in Japan, but the field was different from ours. The soil microbial community at the sites of nursery preparation and/or cultivation might determine the endophytic microbial community of sweet potatoes. When three sweet potato genotypes were planted on a research farm in Brazil, Bacillus predominated (72–79%) in two of them, while Enterobacteriaceae predominated (44%) in the other [10], suggesting an influence of the sweet potato genotype on its endophytes.
Dickeya, previously named Erwinia chrysanthemi and then Pectobacterium chrysanthemi, affects a wide range of plant hosts worldwide [19]. Among the six species of this genus [20], nifH similar to that of Dickeya dadantii was predominantly detected in the sweet potato tubers, especially those that originated from the Sa farm. The sweet potato has been listed in the literature as one of its pathogenic host plants [19], but none of the disease symptoms were observed, although its presence was suggested throughout the cultivation stage. Positive effects, instead, such as higher ARA, higher nitrogen contents, and better growth, were observed in the Sa samples. Hardoim et al. [21] suggested in their review that the nature of plant-endophytic interactions ranges from mutualism to pathogenicity, depending on a set of several abiotic and biotic conditions. The boundaries between mutualists and pathogens might not always be clear because they both have the potential to enter and reside in plants. Recently, Gaby and Buckley [22] comprehensively evaluated previously reported nifH PCR primers in silico, and the results showed unfortunately that the PolF-PolR primer set used in this study only encompassed 25% of the genes in the database. In this study, we at first tried more universal degenerated primers sets as used in Teraoka-Tonooka et al. [16] under the recommended conditions, but several unspecific PCR products were observed. Then, considering that the universal primer set PolF/PolR was designed using test strains consisting of α-, β-, and γ-Proteobacteria, Firmicutes, and Actinobacteria [13], which have been generally reported as endophytes, and that the poor coverage of the primer set is based on the in silico analysis, giving possibly uneven distribution of the nifH sequence in the database used, we used the PolF-PolR primer set under the rather lower annealing temperature to improve the low coverage caused by a few base pairs mismatch.
In addition to biological nitrogen fixation, the production of phytohormones and siderophores, antimicrobial activity against phytopathogens, and phosphate solubilization have been reported as a variety of mechanisms by which endophytes promote plant growth [9,10,23], and the economic importance of their usage has also been suggested [24]. Because the endophytic presence of diverse microbes seems to be common [10], understanding their ecology in plants, such as the interactions among them as well as their interactions with abiotic and biotic environmental conditions, is essential for their effective usage.
Supplementary Materials
The following are available online at https://www.mdpi.com/2311-7524/5/3/53/s1, Figure S1: Phylogenetic tree of nifH genes of the representative sweet potato-associated microbes and similar sequences in the database with their accession numbers, Table S1: Average monthly temperatures and total rainfalls during the sweet potato cultivation at the study site, Table S2: Partial nifH sequences in the sweet potato samples and their results of BLAST search.
Author Contributions
K.I. conceived and designed the experiments; K.O. and N.Y. performed the experiments and analyzed the data; F.A. and S.H. helped to conduct the field and laboratory experiments, respectively. K.I. prepared the manuscript.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Hill, W.A.; Dodo, H.; Hahn, S.K.; Mulongoy, K.; Adeyeye, S.O. Sweet potato root and biomass production with and without nitrogen fertilization. Agron. J. 1990, 82, 1120–1122. [Google Scholar] [CrossRef]
- Yonebayashi, K.; Katsumi, N.; Nishi, T.; Okazaki, M. Activation of nitrogen-fixing endophytes is associated with the tuber growth of sweet potato. Mass Spectrom. 2014, 3, A0032. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T.; Terakado, J.; Masuda, T. Natural abundance of 15N in sweet potato, pumpkin, sorghum and castor bean: Possible input of N2-derived nitrogen in sweet potato. Biol. Fertil. Soils 1998, 26, 152–154. [Google Scholar] [CrossRef]
- Hill, W.A.; Bacon-Hill, P.; Crossman, S.M.; Stevens, C. Characterization of N2-fixing bacteria associated with sweet potato roots. Can. J. Microbiol. 1983, 29, 860–862. [Google Scholar] [CrossRef]
- Paula, M.A.; Reis, V.M.; Döbereiner, J. Interactions of Glomus clarum with Acetobacter diazotrophicus in infection of sweet potato (Ipomoea batatas), sugarcane (Saccharum spp.), and sweet sorghum (Sorghum vulgare). Biol. Fertil. Soils 1991, 11, 111–115. [Google Scholar] [CrossRef]
- Adachi, K.; Nakatani, M.; Mochida, H. Isolation of an endophytic diazotroph, Klebsiella oxytoca, from sweetpotato stems in Japan. Soil Sci. Plant Nutr. 2002, 48, 889–895. [Google Scholar] [CrossRef]
- Asis, C.A., Jr.; Adachi, K. Isolation of endophytic diazotroph Pantoea agglomerans and nondiazotroph Enterobacter asburiae from sweetpotato stem in Japan. Lett. Appl. Microbiol. 2003, 38, 19–23. [Google Scholar] [CrossRef]
- Terakado-Tonooka, J.; Fujihara, S.; Ohwaki, Y. Possible contribution of Bradyrhizobium on nitrogen fixation in sweet potatoes. Plant Soil 2013, 367, 639–650. [Google Scholar] [CrossRef]
- Khan, Z.; Doty, S.L. Characterization of bacterial endophytes of sweet potato plants. Plant Soil 2009, 322, 197–207. [Google Scholar] [CrossRef]
- Marques, J.M.; da Silva, T.F.; Vollú, R.E.; de Lacerda, J.R.M.; Blank, A.F.; Smalla, K.; Seldin, L. Bacterial endophytes of sweet potato tuberous roots affected by the plant genotype and growth stage. Appl. Soil Ecol. 2015, 96, 273–281. [Google Scholar] [CrossRef]
- Reiter, B.; Bürgmann, H.; Burg, K.; Sessitsch, A. Endophytic nifH gene diversity in African sweet potato. Can. J. Microbiol. 2003, 49, 549–555. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aizaki, M.; Sumita, A. Effect of hydrophytes on the control of water temperature in model wetland type green roof garden. Environ. Sci. 2005, 18, 535–540. [Google Scholar]
- Poly, F.; Monrozier, L.J.; Bally, R. Improvement in the RFLP procedure for studying the diversity of nifH genes in communities of nitrogen fixers in soil. Res. Microbiol. 2001, 152, 95–103. [Google Scholar] [CrossRef]
- Adhikari, D.; Kaneto, M.; Itoh, K.; Suyama, K.; Pokharel, B.B.; Gaihre, Y.K. Genetic diversity of soybean-nodulation rhizobia in Nepal in relation to climate and soil properties. Plant Soil 2012, 357, 131–145. [Google Scholar] [CrossRef]
- Marques, J.M.; da Silva, T.F.; Vollu, R.E.; Blank, A.F.; Ding, G.C.; Seldin, L.; Smalla, K. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 2014, 88, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Terakado-Tonooka, J.; Ohwaki, Y.; Yamakawa, H.; Tanaka, F.; Yoneyama, T.; Fujihara, S. Expressed nifH genes of endophytic bacteria detected in field-grown sweet potatoes (Ipomoea batatas L.). Microbes Environ. 2008, 23, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Li, O.; Xiao, R.; Sun, L.; Guan, C.; Kong, D.; Hu, X. Bacterial and diazotrophic diversities of endophytes in Dendrobium catenatum determined through barcoded pyrosequencing. PLoS ONE 2017, 12, e0184717. [Google Scholar] [CrossRef] [PubMed]
- Soni, R.; Suyal, D.C.; Sai, S.; Goel, R. Exploration of nifH gene through soil metagenomes of the western Indian Himalayas. 3 Biotech 2016, 6, 25. [Google Scholar] [CrossRef]
- Samson, R.; Legendre, J.B.; Christen, R.; Fischer-Le, S.M.; Achouak, W.; Gardan, L. Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1415–1427. [Google Scholar]
- Czajkowski, R.; Pérombelon, M.; Jafra, S.; Lojkowska, E.; Potrykus, M.; van der Wolf, J.; Sledz, W. Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 2015, 166, 18–38. [Google Scholar] [CrossRef]
- Hardoim, P.R.; van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef]
- Gaby, J.C.; Buckley, D.H. A comprehensive evaluation of PCR primers to amplify the nifH gene of nitrogenase. PLoS ONE 2012, 7, e42149. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; van Overbeek, L.S.; Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).