Improving Plant Water Use Efficiency through Molecular Genetics
Abstract
:1. Introduction
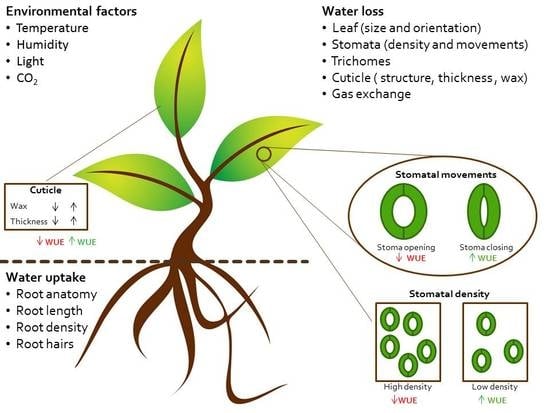
2. Physiology of Water Use and Loss
2.1. Controlling Water Uptake through Roots Architecture
2.2. Controlling Water Loss through Stomatal Density
2.3. Controlling Water Loss through the Cuticle
2.4. Controlling Water Loss through the Guard Cells Movement and Signaling Transduction
2.5. Plant Architecture and Branching Geometry
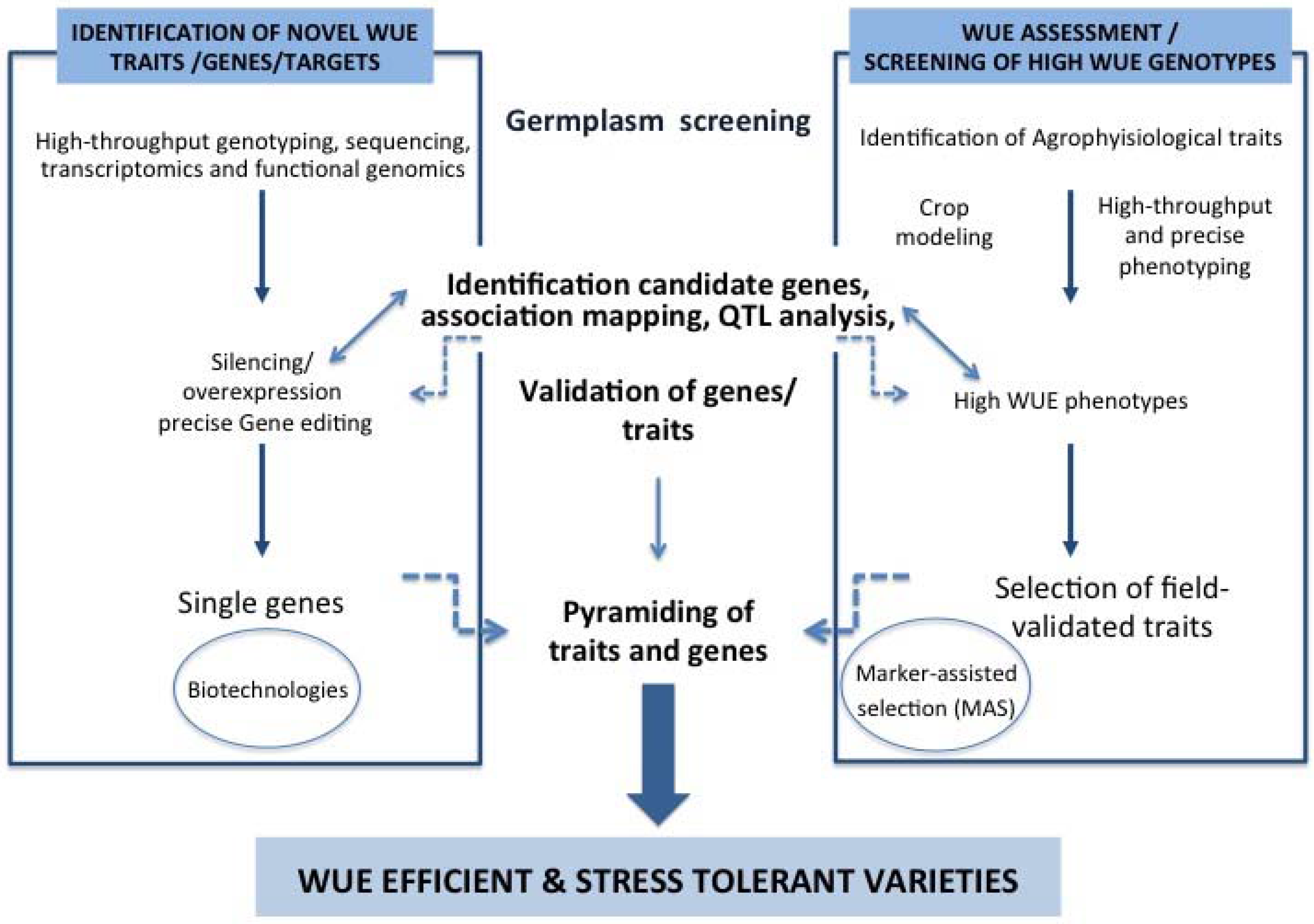
3. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- World Health Organization and UNICEF. Joint Monitoring Programme (JMP). Progress on Sanitation and Drinking Water: 2015 Update and MDG Assessment; UNICEF: Hong Kong, Chaina, 2015. [Google Scholar]
- Mir, R.R.; Zaman-Allah, M.; Sreenivasulu, N.; Trethowan, R.; Varshney, R. Integrated genomics, physiology and breeding approached for improving drought tolerance in crops. Theor. Appl. Genet. 2012, 125, 625–645. [Google Scholar] [CrossRef] [PubMed]
- Cobb, J.N.; Declerck, G.; Greenberg, A.; Clark, R.; McCouch, S. Next-generation phenotyping: Requirements and strategies for enhancing our understanding of genotype-phenotype relationships and its relevance to crop improvement. Theor. Appl. Genet. 2013, 126, 867–887. [Google Scholar] [CrossRef] [PubMed]
- Krannich, C.; Malesky, L.; Kurowsky, C.; Horn, R. Network candidate genes in breeding for drought tolerant crops. Inter. J. Mol. Sci. 2015, 16, 16378–16400. [Google Scholar] [CrossRef] [PubMed]
- Food and Agriculture Organisation (FAO) of the United Nations. Declaration of the World Summit on Food Security. In Proceedings of the World Summit on Food Security, Rome, Italy, 16–18 November 2009. [Google Scholar]
- Evans, R.G.; Sadler, E.J. Methods and technologies to improve efficiency of water use. Water Res. Res. 2008, 44, W00E04. [Google Scholar] [CrossRef]
- Eckardt, N.A.; Cominelli, E.; Galbiati, M.; Tonelli, C. The future of science: Food and water for life. Plant Cell 2009, 21, 368–372. [Google Scholar] [CrossRef] [PubMed]
- The Intergovernmental Panel on Climate Change (IPCC) fourth assessment report. Climate Change: Impacts, Adaptation and Vulnerability; The Intergovernmental Panel on Climate Change: Geneva, Switzerland, 2007. [Google Scholar]
- Tester, M.; Langridge, P. Breeding technologies to increase crop production in a changing world. Science 2010, 12, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Shanker, A.K.; Maheswari, M.; Yadav, S.K.; Desai, S.; Bhanu, D.; Bajaj Attal, N.; Venkateswarlu, B. Drought stress responses in crops. Funct. Integr. Genom. 2014, 14, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Ann. Rev. Plant. Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef] [PubMed]
- Venuprasad, R.; Lafitte, H.R.; Atlin, G.N. Response to direct selection for grain yield under drought stress in rice. Crop. Sci. 2007, 47, 285–293. [Google Scholar] [CrossRef]
- Varshney, R.K.; Thudi, M.; Nayak, S.N.; Gaur, P.M.; Kashiwagi, J.; Krishnamurthy, L.; Jaganathan, D.; Koppolu, J.; Bohra, A.; Tripathi, S.; et al. Genetic dissection of drought tolerance in chickpea (Cicer. arietinum L.). Theor. Appl. Genet. 2014, 127, 445–462. [Google Scholar] [CrossRef] [PubMed]
- Hijmans, R.J. The effect of climate change on global potato production. Am. J. Potato Res. 2003, 80, 271–279. [Google Scholar] [CrossRef]
- Morrison, J.I.L.; Baker, N.R.; Mullineaux, P.M.; Davies, W.J. Improving water use in crop production. Phil. Trans. R. Soc. 2008, B363, 639–658. [Google Scholar] [CrossRef] [PubMed]
- Lobell, D.B.; Schlenker, W.; Costa-Roberts, J. Climate trends and global crop production since 1980. Science 2011, 333, 616–620. [Google Scholar] [CrossRef] [PubMed]
- Boyer, J.S. Drought decision-making. J. Exp. Bot. 2010, 61, 3493–3497. [Google Scholar] [CrossRef] [PubMed]
- Fleury, D.; Jefferies, S.; Kuchel, H.; Langridge, P. Genetic and genomic tools to improve drought tolerance in wheat. J. Exp. Bot. 2010, 61, 3211–3222. [Google Scholar] [CrossRef] [PubMed]
- De Wit, C.T. Transpiration and Crop Yields, Versl. Landbouwk. Onderz; Institute of Biological and Chemical Research on Field Crops and Herbage: Wageningen, The Netherlands, 1958; Volume 64.6, p. 24. [Google Scholar]
- Passioura, J.B. Drought and drought tolerance. Plant. Growth Regul. 1996, 20, 79–83. [Google Scholar] [CrossRef]
- Blum, A. Effective use of water (EUW) and not-water use efficiency (WUE) is the target of the crop yield improvement under drought stress. Field Crops Res. 2009, 112, 119–123. [Google Scholar] [CrossRef]
- Keenan, T.F.; Hollinger, D.Y.; Bohrer, G.; Dragoni, D.; Munger, J.W.; Schmid, H.P.; Richardson, A.D. Increase in forest water-use efficiency as atmospheric carbon dioxide concentrations rise. Nature 2013, 499, 324–327. [Google Scholar] [CrossRef] [PubMed]
- Flexas, J. Genetic improvement of leaf photosynthesis and intrinsic water use efficiency in C3 plants: Why so much little success? Plant Sci. 2016, 251, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Lawson, T.; Blatt, M.R. Stomatal size, speed and responsiveness impact on photosynthesis and water use efficiency. Plant Physiol. 2014, 64, 1556–1570. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, M.; Tuberosa, R. Translational research impacting on crop productivity in drought-prone environments. Curr. Opin. Plant Biol. 2008, 11, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Steele, K.A.; Price, A.H.; Shashidhar, H.E.; Witcombe, J.R. Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor. Appl. Genet. 2006, 112, 208–221. [Google Scholar] [CrossRef] [PubMed]
- Van Oosten, M.; Costa, A.; Punzo, P.; Landi, S.; Ruggiero, A.; Batelli, G.; Grillo, S. Genetics of Drought Stress Tolerance in Crop Plants. In Drought stress tolerance in Plants Vol. 2; Hossain, M.A., Wani, S.H., Bhattachajee, S., Burrit, D., Tran, L., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 39–70. [Google Scholar]
- Shriram, V.; Kumar, V.; Devarumath, R.M.; Khare, T.S.; Wani, S.H. MicroRNAs as potential targets for abiotic stress tolerance in plants. Front. Plant Sci. 2016, 7, 817. [Google Scholar] [CrossRef] [PubMed]
- Paez-Garcia, A.; Motes, C.M.; Scheible, W.R.; Chen, R.; Blancaflor, E.B.; Monteros, M.J. Root traits and phenotyping strategies for plant improvement. Plants 2015, 4, 334–355. [Google Scholar] [CrossRef] [PubMed]
- Bao, Y.; Aggarwal, P.; Robbins, N.E.; Sturrock, C.J.; Thompson, M.C.; Tan, H.Q.; Tham, C.; Duan, L.; Rodriguez, P.L.; Vernoux, T. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc. Natl. Acad. Sci. USA 2014, 111, 9319–9324. [Google Scholar] [CrossRef] [PubMed]
- Robbins, N.E.; Dinneny, J.R. The divining root: Moisture-driven responses of roots at the micro-and macro-scale. J. Exp. Bot. 2015. [Google Scholar] [CrossRef] [PubMed]
- Kell, D. Breeding crop plants with deep roots: Their role in sustainable carbon, nutrient and water sequestration. Ann. Bot. 2011, 108, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Palta, J.; Chen, X.; Milroy, S.; Rebetzke, G.; Dreccer, M.; Watt, M. Large root systems: Are they useful in adapting wheat to dry environments? Funct. Plant Biol. 2011, 38, 347–354. [Google Scholar] [CrossRef]
- Singh, V.; van Oosterom, E.J.; Jordan, D.R.; Messina, C.D.; Cooper, M.; Hammer, G.L. Morphological and architectural development of root systems in sorghum and maize. Plant. Soil 2010, 333, 287–299. [Google Scholar] [CrossRef]
- Jung, J.K.H.M.; McCouch, S.R.M. Getting to the roots of it: Genetic and hormonal control of root architecture. Front. Plant. Sci. 2013, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Wasson, A.; Richards, R.; Chatrath, R.; Misra, S.; Prasad, S.S.; Rebetzke, G.; Kirkegaard, J.; Christopher, J.; Watt, M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 2012, 63, 3485–3498. [Google Scholar] [CrossRef] [PubMed]
- Lynch, J.P.; Wojciechowski, T. Opportunities and challenges in the subsoil: Pathways to deeper rooted crops. J. Exp. Bot. 2015. [Google Scholar] [CrossRef] [PubMed]
- Zhan, A.; Schneider, H.; Lynch, J. Reduced lateral root branching density improves drought tolerance in maize. Plant Physiol. 2015, 168, 1603–1615. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, N.; Kato, M.; Tomioka, R.; Kurata, R.; Fukao, Y.; Aoyama, T.; Maeshima, M. Characteristics of a root hair-less line of Arabidopsis thaliana under physiological stresses. J. Exp. Bot. 2014, 65, 1497–1512. [Google Scholar] [CrossRef] [PubMed]
- Haling, R.E.; Brown, L.K.; Bengough, A.G.; Young, I.M.; Hallett, P.D.; White, P.J.; George, T.S. Root hairs improve root penetration, root-soil contact, and phosphorus acquisition in soils of different strength. J. Exp. Bot. 2013, 64, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Miguel, M.A.; Postma, J.A.; Lynch, J. Phene synergism between root hair length and basal root growth angle for phosphorus acquisition. Plant. Physiol. 2015, 167, 1430–1439. [Google Scholar] [CrossRef] [PubMed]
- Mendrinna, A.; Persson, S. Root hair growth: It’s a one way street. F1000prime Rep. 2015, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Velasquez, S.M.; Ricardi, M.M.; Dorosz, J.G.; Fernandez, P.V.; Nadra, A.D.; Pol-Fachin, L.; Egelund, J.; Gille, S.; Harholt, J.; Ciancia, M. O-glycosylated cell wall proteins are essential in root hair growth. Science 2011, 332, 1401–1403. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.M.; Quan, L.; Cannon, A.E.; Wen, J.; Blancaflor, E.B. AGD1, a class 1 ARF-GAP, acts in common signaling pathways with phosphoinositide metabolism and the actin cytoskeleton in controlling Arabidopsis root hair polarity. Plant J. 2012, 69, 1064–1076. [Google Scholar] [CrossRef] [PubMed]
- Tian, H.; de Smet, I.; Ding, Z. Shaping a root system: Regulating lateral vs. primary root growth. Trends Plant Sci. 2014, 19, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.S.; Yao, S.Q. Transcription co-activator Arabidopsis ANGUSTIFOLIA3 (AN3) regulates water-use efficiency and drought tolerance by modulating stomatal density and improving root architecture by the transrepression of YODA (YDA). Plant Biotechnol. J. 2015, 13, 893–902. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Chen, B.; Lu, G.; Han, B. Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 2009, 379, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Redillas, M.C.; Jeong, J.S.; Kim, Y.S.; Jung, H.; Bang, S.W.; Choi, Y.D.; Ha, S.H.; Reuzeau, C.; Kim, J.K. The overexpression of OsNAC9 alters the root architecture of rice plants enhancing drought resistance and grain yield under field conditions. Plant. Biotechnol. J. 2012, 10, 792–805. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.M. Auxin homeostasis during lateral root development under drought condition. Plant Signal. Behav. 2009, 4, 1002–1004. [Google Scholar] [CrossRef] [PubMed]
- Hetherington, A.M.; Woodward, F.I. The role of stomata in sensing and driving environmental change. Nature 2003, 424, 901–908. [Google Scholar] [CrossRef] [PubMed]
- Shimazaki, K.I.; Doi, M.; Assmann, S.M.; Kinoshita, T. Light regulation of stomatal movement. Annu. Rev. Plant Biol. 2007, 58, 219–247. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Pence, H.E.; Jin, J.B.; Miura, K.; Gosney, M.J.; Hasegawa, P.M.; Mickelbart, M.V. The Arabidopsis GTL1 transcription factor regulates water use efficiency and drought tolerance by modulating stomatal density via transrepression of SDD1. Plant Cell 2010, 22, 4128–4141. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, D.C.; Sack, F.D. Stomatal development. Annu. Rev. Plant Biol. 2007, 58, 163–181. [Google Scholar] [CrossRef] [PubMed]
- Yoo, C.Y.; Pence, H.E.; Hasegawa, P.M.; Mickelbart, M.V. Regulation of transpiration to improve crop water use. Crit. Rev. Plant Sci. 2009, 28, 410–431. [Google Scholar] [CrossRef]
- Berger, D.; Altmann, T.A. Subtilisin-like serine protease involved in the regulation of stomatal density and distribution in Arabidopsis thaliana. Genes Dev. 2000, 14, 1119–1131. [Google Scholar] [PubMed]
- Büssis, D.; von Groll, U.; Fisahn, J.; Altmann, T. Stomatal aperture can compensate altered stomatal density in Arabidopsis thaliana at growth light conditions. Funct. Plant Biol. 2006, 33, 1037–1043. [Google Scholar] [CrossRef]
- Kaplan-Levy, R.N.; Brewer, P.B.; Quon, T.; Smyth, D.R. The trihelix family of transcription factors-light, stress and development. Trends Plant Sci. 2012, 17, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Kajita, R.; Torii, K.U.; Bergmann, D.C.; Kakimoto, T. The secretory peptide gene EPF1 enforces the stomatal one-cell-spacing rule. Genes Dev. 2007, 21, 1720–1725. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yokoo, T.; Kajita, R.; Onishi, T.; Yahata, S.; Peterson, K.M.; Torii, K.U.; Kakimoto, T. Epidermal cell density is autoregulated via a secretory peptide, EPIDERMAL PATTERNING FACTOR 2 in Arabidopsis leaves. Plant Cell Physiol. 2009, 50, 1019–1031. [Google Scholar] [CrossRef] [PubMed]
- Franks, P.J.; W Doheny-Adams, T.; Britton-Harper, Z.J.; Gray, J.E. Increasing water-use efficiency directly through genetic manipulation of stomatal density. New Phytol. 2015, 207, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Torii, K.U. Mix-and-match: Ligand-receptor pairs in stomatal development and beyond. Trends Plant Sci. 2012, 17, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kuroha, T.; Hnilova, M.; Khatayevich, D.; Kanaoka, M.M.; McAbee, J.M.; Sarikaya, M.; Tamerler, C.; Torii, K.U. Direct interaction of ligand–receptor pairs specifying stomatal patterning. Genes Dev. 2012, 26, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Sack, F.D. The too many mouths and four lips mutations affect stomatal production in Arabidopsis. Plant Cell 1995, 7, 2227–2239. [Google Scholar] [CrossRef] [PubMed]
- Abrash, E.B.; Davies, K.A.; Bergmann, D.C. Generation of signaling specificity in Arabidopsis by spatially restricted buffering of ligand-receptor interactions. Plant Cell 2011, 23, 2864–2879. [Google Scholar] [CrossRef] [PubMed]
- Shpak, E.D. Diverse roles of ERECTA family genes in plant development. J. Integr. Plant Biol. 2013, 55, 1238–1250. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, D.C.; Lukowitz, W.; Somerville, C.R. Stomatal development and pattern controlled by a MAPKK kinase. Science 2004, 304, 1494–1497. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, X.; Xiang, C.B. Stomatal density and bio-water saving. J. Integr. Plant Biol. 2007, 49, 1435–1444. [Google Scholar] [CrossRef]
- Ishimaru, K.; Yano, M.; Aoki, N.; Ono, K.; Hirose, T.; Lin, S.Y.; Sasaki, T.; Ohsugi, R. Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor. Appl. Gen. 2001, 102, 793–800. [Google Scholar] [CrossRef]
- Laza, M.R.C.; Kondo, M.; Ideta, O.; Barlaan, E.; Imbe, T. Quantitative trait loci for stomatal density and size in lowland rice. Euphytica 2010, 172, 149–158. [Google Scholar] [CrossRef]
- Nobel, P.S. Environmental Biology. In Agroecology, Cultivation and Uses of Cactus Pear. Plant Production and Protection Paper; Barbera, G., Inglese, P., Pimienta-Barrios, E., Arias-Jiménez, E.J., Eds.; FAO: Rome, Italy, 1995; pp. 36–57. [Google Scholar]
- Borland, A.M.; Griffith, H.; Hartwell, H.; Smith, A.C. Exploiting the potential plants with crassulacean acid metabolism for bioenergy production on marginal land. J. Exp. Bot. 2009, 10, 2879–2896. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, P.G.; Johnson, D.A.; Asay, K.H. Epicuticular wax production, water status and leaf temperature in triticeae range grasses of contrasting visible glaucousness. Can. J. Plant. Sci. 1989, 69, 513–520. [Google Scholar] [CrossRef]
- Goodwin, S.M.; Jenks, M.A. Plant Cuticle Function as a Barrier to Water Loss. In Plant Abiotic Stress; Jenks, M.A., Hasegawa, P.M., Eds.; Blackwell Publishing Ltd.: Oxford, UK, 2005; pp. 14–36. [Google Scholar]
- Riederer, M.; Schreiber, L. Protecting against water loss: Analysis of the barrier properties of plant cuticles. J. Exp. Bot. 2001, 52, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Burghardt, M.; Riederer, M. Cuticular Transpiration. In Biology of the Plant Cuticle; Riederer, M., Muller, C., Eds.; Wiley-Blackwell: Hoboken, NJ, USA, 2006; pp. 292–311. [Google Scholar]
- Jenks, M.A.; Tuttle, H.A.; Eigenbrode, S.D.; Feldmann, K.A. Leaf epicuticular waxes of the eceriferum mutants in Arabidopsis. Plant. Physiol. 1995, 108, 369–377. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M. Thermodynamics of the water permeability of plant cuticles: Characterization of the polar pathway. J. Exp. Bot. 2006, 57, 2937–2942. [Google Scholar] [CrossRef] [PubMed]
- Kunst, L.; Samuels, A. Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 2003, 42, 51–80. [Google Scholar] [CrossRef]
- Bengtson, C.; Larsson, S.; Liljenberg, C. Effects of water stress on cuticular transpiration rate and amount and composition of epicuticular wax in seedlings of six oat varieties. Physiol. Plant 1978, 44, 319–324. [Google Scholar] [CrossRef]
- Jordan, W.; Shouse, P.J.; Blum, A.; Miller, F.R.; Monk, R.L. Environmental physiology of sorghum. II. Epicuticular wax load and cuticular transpiration. Crop. Sci. 1984, 24, 1168–1173. [Google Scholar]
- Islam, M.A.; Du, H.; Ning, J.; Ye, H.; Xiong, L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 2009, 70, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Komatsuda, T.; Ma, J.F.; Li, C.; Yamaji, N.; Nevo, E. A functional cutin matrix is required for plant protection against water loss. Plant Signal. Behav. 2011, 6, 1297–1299. [Google Scholar] [CrossRef] [PubMed]
- González, A.; Ayerbe, L. Effect of terminal water stress on leaf epicuticular wax load, residual transpiration and grain yield in barley. Euphytica 2009, 172, 341–349. [Google Scholar] [CrossRef]
- Leide, J.; Hildebrandt, U.; Reussing, K.; Riederer, M.; Vogg, G. The developmental pattern of tomato fruit wax accumulation and its impact on cuticular transpiration barrier properties: Effects of a deficiency in a beta-ketoacyl-coenzyme A synthase (LeCER6). Plant Physiol. 2007, 144, 1667–1679. [Google Scholar] [CrossRef] [PubMed]
- Leide, J.; Hildebrandt, U.; Vogg, G.; Riederer, M. The positional sterile (PS) mutation affects cuticular transpiration and wax biosynthesis of tomato fruits. J. Plant Physiol. 2011, 168, 871–877. [Google Scholar] [CrossRef] [PubMed]
- Kurdyukov, S.; Faust, A.; Nawrath, C.; Bär, S.; Voisin, D.; Efremova, N.; Franke, R.; Schreiber, L.; Saedler, H.; Métraux, J.P.; et al. The epidermis-specific extracellular BODYGUARD controls cuticle development and morphogenesis in Arabidopsis. Plant Cell 2006, 18, 321–339. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes LONG-CHAIN ACYL-COA SYNTHETASE 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Zhao, H.Y.; Parsons, E.P.; Xu, C.C.; Kosma, D.K.; Xu, X.; Chao, D.; Lohrey, G.; Bangarusamy, D.K.; Wang, G.; et al. The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiol. 2011, 157, 1079–1092. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Jetter, R.; Kunst, L. Plant surface lipid biosynthetic pathways and their utility for metabolic engineering of waxes and hydrocarbon biofuels. Plant J. 2008, 54, 670–683. [Google Scholar] [CrossRef] [PubMed]
- Samuels, L.; De Bono, A.; Lam, P.; Wen, M.; Jetter, R.; Kunst, L. Use of Arabidopsis eceriferum mutants to explore plant cuticle biosynthesis. J. Vis. Exp. 2008, 31, 709. [Google Scholar] [CrossRef] [PubMed]
- Aharoni, A.; Dixit, S.; Jetter, R.; Thoenes, E.; van Arkel, G.; Pereira, A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell 2004, 16, 2463–2480. [Google Scholar] [CrossRef] [PubMed]
- Broun, P.; Poindexter, P.; Osborne, E.; Jiang, C.Z.; Riechmann, J.L. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc. Natl. Acad. Sci. USA 2004, 101, 4706–4711. [Google Scholar] [CrossRef] [PubMed]
- Cominelli, E.; Sala, T.; Calvi, D.; Gusmaroli, G.; Tonelli, C. Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 2008, 53, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Javelle, M.; Vernoud, V.; Depege-Fargeix, N.; Arnould, C.; Oursel, D.; Domergue, F.; Sarda, X.; Rogowsky, P.M. Overexpression of the epidermis-specific homeodomain-leucine zipper IV transcription factor outer cell layer1 in maize identifies target genes involved in lipid metabolism and cuticle biosynthesis. Plant Physiol. 2010, 154, 273–286. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Y.; Broeckling, C.D.; Blancaflor, E.B.; Sledge, M.K.; Sumner, L.W.; Wang, Z.Y. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 2005, 42, 689–707. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhang, Y.; Xu, C.; Ren, J.; Liu, X.; Black, K.; Gai, X.; Wang, Q.; Ren, H. Cucumber ECERIFERUM1 (CsCER1), which influences the cuticle properties and drought tolerance of cucumber, plays a key role in VLC alkanes biosynthesis. Plant Mol. Biol. 2015, 87, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Al-Abdallat, A.M.; Al-Debei, H.S.; Ayad, J.Y.; Hasan, S. Over-Expression of SlSHN1 gene improves drought tolerance by increasing cuticular wax accumulation in Tomato. Int. J. Mol. Sci. 2014, 15, 19499–19515. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ni, E.; Yang, J.; Zhou, H.; Liang, H.; Li, J.; Jiang, D.; Wang, Z.; Liu, Z.; Zhuang, C. Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS ONE 2013, 8, e65139. [Google Scholar] [CrossRef] [PubMed]
- Lü, S.; Zhao, H.; Des Marais, D.L.; Parsons, E.P.; Wen, X.; Xu, X.; Bangarusamy, D.K.; Wang, G.; Rowland, O.; Juenger, T.; et al. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Stalfelt, M.G. Die Abhaengigkeit der Spaltoeffnungsreaktionen von der Wasserbalanz. Planta 1929, 8, 287–340. [Google Scholar] [CrossRef]
- Sharpe, J.H.; Wu, H. Stomatal mechanics: Volume changes during opening. Plant Cell Environ. 1978, 1, 259–268. [Google Scholar] [CrossRef]
- Kolb, H.A.; Marten, I.; Hedrich, R. Hodgkin-Huxley analysis of aGCAC1anion channel in the plasma-membrane of guard cells. J. Membr. Biol. 1995, 146, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Linder, B.; Raschke, K. A slow anion channel in guard-cells, activating at larg hyperpolarization, may be principal for stomatal closing. FEBS Lett. 1992, 313, 27–30. [Google Scholar] [CrossRef]
- Vahisalu, T.; Kollist, H.; Wang, Y.F.; Nishimura, N.; Chan, W.Y.; Valerio, G.; Lamminmäki, A.; Brosché, M.; Moldau, H.; Desikan, R.; et al. SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 2008, 452, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Geiger, D.; Scherzer, S.; Mumm, P.; Stange, A.; Marten, I.; Bauer, H.; Ache, P.; Matschi, S.; Liese, A.; Al-Rasheid, K.A.S.; et al. Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. USA 2009, 106, 21425–21430. [Google Scholar] [CrossRef] [PubMed]
- Merilo, E.; Laanemets, K.; Hu, H.; Xue, S.; Jakobsen, L.; Tulva, I.; Gonzales-Guzman, M.; Rodriguez, P.L.; Schroeder, J.I.; Brosche, M.; et al. PYR/RCAR receptors contribute to ozone-, reduced air humidity-, darkness- and CO2-induced stomatal regulation. Plant Physiol. 2013, 162, 1652–1668. [Google Scholar] [CrossRef] [PubMed]
- Geiger, D.; Maierhofer, T.; Al-Rasheid, K.A.; Scherzer, S.; Mumm, P.; Liese, A.; Ache, P.; Wellmann, C.; Marten, I.; Grill, E.; et al. Stomatal closure by fast abscisic acid signaling is mediated by the guard cell anion channel SLAH3 and the receptor. Sci. Signal. 2011, 4, ra32. [Google Scholar] [CrossRef] [PubMed]
- Dreyer, I.; Gomez-Porras, J.L.; Riano-Pachon, D.M.; Hedrich, R.; Geiger, D. Molecular evolution of slow and quick anion channels (SLACs and QUACs/ALMTs). Front. Plant Sci. 2012, 3, 263. [Google Scholar] [CrossRef] [PubMed]
- Meyer, S.; Mumm, P.; Imes, D.; Endler, A.; Weder, B.; Al-Rasheid, K.A.; Geiger, D.; Marten, I.; Martinoia, E.; Hedrich, R. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010, 63, 1054–1062. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, T.; Mori, I.C.; Furuichi, T.; Munemasa, S.; Toyooka, K.; Matsuoka, K.; Murata, Y.; Yamamoto, Y. Closing plant stomata requires a homolog of an aluminum activated malate transporter. Plant Cell Physiol. 2010, 51, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Choi, Y.; Burla, B.; Kim, Y.Y.; Jeon, B.; Maeshima, M.; Yoo, J.Y.; Martinoia, E.; Lee, Y. The ABC transporter AtABCB14 is a malate importer and modulates stomatal response to CO2. Nat. Cell Biol. 2008, 10, 1217–1223. [Google Scholar] [CrossRef] [PubMed]
- Eisenach, C.; Papanatsiou, M.; Hillert, E.K.; Blatt, M.R. Clustering of the K+ channel GORK of Arabidopsis parallels its gating by extracellular K+. Plant J. 2014, 78, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Hosy, E.; Vavasseur, A.; Mouline, K.; Dreyer, I.; Gaymard, F.; Pore, F.; Boucherez, J.; Lebaudy, A.; Bouchez, D.; Very, A.; et al. The Arabidopsis outward K+ channel GORK is involved in regulation of stomatal movements and plant transpiration. Proc. Natl. Acad. Sci. USA 2003, 100, 5549–5554. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, Y.; Arinaga, N.; Umezawa, T.; Katsura, S.; Nagamachi, K.; Tanaka, H.; Ohiraki, H.; Yamada, K.; Seo, S.U.; Abo, M.; et al. Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 2013, 25, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Navarro, A.; Rubio, F. High-affinity potassium and sodium transport systems in plants. J. Exp. Bot. 2006, 57, 1149–1160. [Google Scholar] [CrossRef] [PubMed]
- Hedrich, R. Ion channels in plants. Physiol. Rev. 2012, 92, 1777–1811. [Google Scholar] [CrossRef] [PubMed]
- Merlot, S.; Leonhardt, N.; Fenzi, F.; Valon, C.; Costa, M.; Piette, L.; Vavasseur, A.; Genty, B.; Boivin, K.; Muller, A.; et al. Constitutive activation of a plasma membrane H+-ATPase prevents abscisic acid-mediated stomatal closure. Embo J. 2007, 26, 3216–3226. [Google Scholar] [CrossRef] [PubMed]
- Imes, D.; Mumm, P.; Bohm, J.; Al-Rasheid, K.A.; Marten, I.; Geiger, D.; Hedrich, R. Open stomata 1 (OST1) kinase controls R-type anion channel QUAC1 in Arabidopsis guard cells. Plant J. 2013, 74, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Inoue, S.; Takahashi, K.; Kinoshita, T. Immunohistochemical detection of blue light-induced phosphorylation of the plasma membrane H+-ATPase in stomatal guard cells. Plant Cell Physiol. 2011, 52, 1238–1248. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, A.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Fung, P.; Nishimura, N. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.P.; Chen, P.; Ren, J.; Ji, K.; Li, Q.; Li, P.; Dai, S.; Leng, P. Transcriptional regulation of SlPYL, SlPP2C, and SlSnRK2 gene families encoding ABA signal core components during tomato fruit development and drought stress. J. Exp. Bot. 2011, 62, 5659–5669. [Google Scholar] [CrossRef] [PubMed]
- González-Guzmán, M.; Rodríguez, L.; Lorenzo-Orts, L.; Pons, C.; Sarrión-Perdigones, A.; Fernández, M.A.; Peirats-Llobet, M.; Forment, J.; Moreno-Alvero, M.; Cutler, S.R.; et al. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, X.; Modrego, A.; Rodríguez, D.; Gonzales-Garcia, M.P.; Sanz, L.; Nicolas, G.; Lorenzo, O. The nuclear interactor PYL8/RCAR3 of Fagus sylvatica FsPP2C1 is a positive regulator of abscisic acid signaling in seeds and stress. Plant Physiol. 2010, 152, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Chai, Y.M.; Jia, H.F.; Li, C.L.; Dong, Q.H.; Shen, Y.Y. FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 2011, 62, 5079–5089. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Hwang, H.; Hong, J.W.; Lee, Y.N.; Ahn, I.P.; Yoon, I.S.; Yoo, S.D.; Lee, S.; Lee, S.C.; Kim, B.G. A rice orthologue of the ABA receptor, OsPYL/RCAR5, is a positive regulator of the ABA signal transduction pathway in seed germination and early seedling growth. J. Exp. Bot. 2012, 63, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.; Lafuente, M.T.; Rodrigo, M.J. The citrus ABA signalosome: Identification and transcriptional regulation during sweet orange fruit ripening and leaf dehydration. J. Exp. Bot. 2012, 63, 4931–4945. [Google Scholar] [CrossRef] [PubMed]
- Bai, G.; Yang, D.H.; Zhao, Y.; Ha, S.; Yang, F.; Ma, J.; Gao, X.S.; Wang, Z.M.; Zhu, J.K. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 2013, 83, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Chinnusamy, V.; Rodrigues, A.; Rubio, S.; Antoni, R.; Park, S.; Cutler, S.R.; Sheen, J.; Rodriguez, P.L.; Zhu, J.K. In vitro reconstitution of an ABA signaling pathway. Nature 2009, 462, 660–664. [Google Scholar] [CrossRef] [PubMed]
- Fujii, H.; Zhu, J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction and stress. Proc. Natl. Acad. Sci. USA 2009, 106, 8380–8385. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Nakashima, K.; Yoshida, T.; Katagiri, T.; Kidokoro, S.; Kanamori, N.; Umezawa, T.; Fujita, M.; Maruyama, K.; Ishiyama, K.; et al. Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 2009, 50, 2123–2132. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Sato, Y.; Fukao, Y.; Fujiwara, M.; Umezawa, T.; Shinozaki, K.; Hibi, T.; Taniguchi, M.; Miyake, H.; Goto, D.B.; Uozumi, N. Threonine at position 306 of the KAT1 potassium channel is essential for channel activity and is a target site for ABA-activated SnRK2/OST1/SnRK2.6 protein kinase. Biochem. J. 2009, 424, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Roelfsema, M.R.G.; Konrad, K.R.; Marten, H.; Psaras, G.K.; Hartung, W.; Hedrich, R. Guard cells in albino leaf patches do not respond to photosynthetically active radiation, but are sensitive to blue light, CO2 and abscisic acid. Plant Cell Environ. 2006, 29, 1595–1605. [Google Scholar] [CrossRef] [PubMed]
- Christie, J.M. Phototropin blue-light receptors. Ann. Rev. Plant Biol. 2007, 58, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Takemiya, A.; Kinoshita, T.; Asanuma, M.; Shimazaki, K.I. Protein phosphatase 1 positively regulates stomatal opening in response to blue light in Vicia faba. Proc. Natl. Acad. Sci. USA 2006, 103, 13549–13554. [Google Scholar] [CrossRef] [PubMed]
- Marten, H.; Hedrich, R.; Roelfsema, M.R.G. Blue light inhibits guard cell plasma membrane anion channels in a phototropin-dependent manner. Plant J. 2007, 50, 29–39. [Google Scholar] [CrossRef] [PubMed]
- Roelfsema, M.R.G.; Hanstein, S.; Felle, H.H.; Hedrich, R. CO2 provides an intermediate link in the red light response of guard cells. Plant J. 2002, 32, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Matrosova, A.; Bogireddi, H.; Mateo-Peñas, A.; Hashimoto-Sugimoto, M.; Iba, K.; Schroeder, J.I.; Israelsson-Nordström, M. The HT1 protein kinase is essential for red light-induced stomatal opening and genetically interacts with OST1 in red light and CO2-induced stomatal movement responses. New Phytol. 2015, 208, 1126–1137. [Google Scholar] [CrossRef] [PubMed]
- Messinger, S.M.; Buckley, T.N.; Mott, K.A. Evidence for involvement of photosynthetic processes in the stomatal response to CO2. Plant Physiol. 2006, 140, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Roelfsema, M.R.G.; Hedrich, R. In the light of stomatal opening: New insights into ‘the Watergate’. New Phytol. 2005, 167, 665–691. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Hou, C.; Ren, Z.; Pan, Y.; Jia, J.; Zhang, H.; Bai, F.; Zhang, P.; Zhu, H.; He, Y.; et al. A molecular pathway for CO2 response in Arabidopsis guard cells. Nat. Commun. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, M.; Negi, J.; Young, J.; Israelsson, M.; Schroeder, J.I.; Iba, K. Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 2006, 8, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.M.; Mackintosh, C.A.; Shin, S.; Gilding, E.; Kravchenko, S.; Baldridge, G.; Zeyen, R.; Muehlbauer, G.J. Overexpression of the maize Teosinte Branched1 gene in wheat suppresses tiller development. Plant Cell Rep. 2008, 27, 1217–1225. [Google Scholar] [CrossRef] [PubMed]
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant. J. 2011, 67, 701–714. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Lin, Z.; Li, H.; Li, X.; Li, J.; Wang, Y.; Zhang, X.; Zhu, Z.; Zhai, W.; Wang, X.; et al. TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 2007, 52, 891–898. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Huang, W.; Gao, J.P.; Yang, J.; Shi, M.; Zhu, M.Z.; Luo, D.; Lin, H.X. Genetic control of rice plant architecture under domestication. Nat. Genet. 2008, 40, 1365–1369. [Google Scholar] [CrossRef] [PubMed]
- Juenger, T.E.; Mckay, J.K.; Hausmann, N.; Keurentjes, J.J.B.; Sen, S.; Stowe, K.A.; Dawson, T.E.; Simms, E.L.; Richards, J.H. Identification and Characterization of QTL Underlying Whole-Plant Physiology in Arabidopsis Thaliana: δ13C, Stomatal Conductance and Transpiration Efficiency. Plant Cell Environ. 2005, 28, 697–708. [Google Scholar] [CrossRef]
- Vasseur, F.; Bontpart, T.; Dauzat, M.; Granier, C.; Vile, D. Multivariate Genetic Analysis of Plant Responses to Water Deficit and High Temperature Revealed Contrasting Adaptive Strategies. J. Exp. Bot. 2014, 65, 6457–6469. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.E.; Ewers, B.E.; McClung, C.R.; Lou, P.; Weinig, C. Quantitative Variation in Water-Use Efficiency across Water Regimes and Its Relationship with Circadian, Vegetative, Reproductive, and Leaf Gas-Exchange Traits. Mol. Plant 2012, 5, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Edwards, C.E.; Ewers, B.E.; Weinig, C. Genotypic Variation in Biomass Allocation in Response to Field Drought Has a Greater Affect on Yield than Gas Exchange or Phenology. BMC Plant Biol. 2016, 16, 185. [Google Scholar] [CrossRef] [PubMed]
- Campitelli, B.E.; Des Marais, D.L.; Juenger, T.E. Ecological Interactions and the Fitness Effect of Water-Use Efficiency: Competition and Drought Alter the Impact of Natural MPK12 Alleles in Arabidopsis. Ecol. Lett. 2016, 19, 424–434. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.; Bailey, K.J.; Gray, J.E. The signalling peptide EPFL9 is a positive regulator of stomatal development. New Phytol. 2010, 186, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Masle, J.; Gilmore, S.R.; Farquhar, G.D. The ERECTA gene regulates plant transpiration efficiency in Arabidopsis. Nature 2005, 436, 866–870. [Google Scholar] [CrossRef] [PubMed]
- Negi, J.; Matsuda, O.; Nagasawa, T.; Oba, Y.; Takahashi, H.; Kawai-Yamada, M.; Uchimiya, H.; Hashimoto, M.; Iba, K. CO2 regulator SLAC1 and its homologues are essential for anion homeostasis in plant cells. Nature 2008, 452, 483–486. [Google Scholar] [CrossRef] [PubMed]
- Kwak, J.M.; Murata, Y.; Baizabal-Aguirre, V.M.; Merrill, J.; Wang, M.; Kemper, A.; Hawke, S.D.; Tallman, G.; Schroeder, J.I. Dominant negative guard cell K+ channel mutants reduce inward-rectifying K+ currents and light-induced stomatal opening in Arabidopsis. Plant Physiol. 2001, 127, 473–485. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Guzman, M.; Pizzio, G.A.; Antoni, R.; Vera-Sirera, F.; Merilo, E.; Bassel, G.W.; Fernández, M.A.; Holdsworth, M.J.; Perez-Amador, M.A.; Kollist, H.; et al. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell 2012, 24, 2483–2496. [Google Scholar] [CrossRef] [PubMed]
- Saez, A.; Robert, N.; Maktabi, M.H.; Schroeder, J.I.; Serrano, R.; Rodriguez, P.L. Enhancement of abscisic acid sensitivity and reduction of water consumption in Arabidopsis by combined inactivation of the protein phosphatases type 2C ABI1 and HAB1. Plant Physiol. 2006, 141, 1389–1399. [Google Scholar] [CrossRef] [PubMed]
- Thompson, A.J.; Mulholland, B.J.; Jackson, A.C.; McKee, J.M.; Hilton, H.W.; Symonds, R.C.; Sonneveld, T.; Burbidge, A.; Stevenson, P.; Taylor, I.B. Regulation and manipulation of ABA biosynthesis in roots. Plant Cell Environ. 2007, 30, 67–78. [Google Scholar] [CrossRef] [PubMed]
| Gene | Gene Name | Protein Function | Species | Mutant Type | Expected Effect on WUE | Ref. |
|---|---|---|---|---|---|---|
| Uptake through roots | ||||||
| NAC9 | NAC DOMAIN CONTAINING PROTEIN 9 | NAM-ATAF-CUC | O. sativa | overexpression | increase | [49] |
| MYB96 | MYB DOMAIN PROTEIN 96 | R2R3-type MYB TF | A. thaliana | overexpression | increase | [155] |
| AN3 | ANGUSTIFOLIA3 | A. thaliana | loss of function | increase | [47] | |
| Stomatal density | ||||||
| SDD1 | STOMATAL DENSITY AND DISTRIBUTION 1 | serine protease | A. thaliana | overexpression | increase | [57] |
| GTL1 | GT2-like 1 | GT2-like TF | A. thaliana | loss of function | increase | [53] |
| EPF1 | EPIDERMAL PATTERNING FACTOR 1 | cysteine-rich peptide | A. thaliana | loss of function | decrease | [61] |
| EPF2 | EPIDERMAL PATTERNING FACTOR 2 | cysteine-rich peptide | A. thaliana | loss of function | decrease | [61] |
| EPFL9 | EPIDERMAL PATTERNING FACTOR-LIKE 9 | cysteine-rich peptide/signalling pepetide | A. thaliana | overexpression | decrease | [156] |
| ERECTA | QRP1, QUANTITATIVE RESISTANCE TO PLECTOSPHAERELLA 1 | LRR-like kinase | A. thaliana | loss of function | decrease | [157] |
| YODA | MAPKKK4, MAP KINASE KINASE KINASE 4 | MAPKK kinase | A. thaliana | loss of function | decrease | [67] |
| AN3 | ANGUSTIFOLIA3 | A. thaliana | loss of function | increase | [47] | |
| Cuticle | ||||||
| CER1 | ECERIFERUM1 | alkane biosynthesis | C. sativus | knockdown mutant | increase | [98] |
| SHN1 | WAX INDUCER1/SHINE1 | AP2/EREBP transcription factor | S. lycopersicum | overexpression | increase | [99] |
| CER9 | ECERIFERUM9 | cuticle biosynthesis | A. thaliana | loss of function | increase | [101] |
| Stomata aperture and responses | ||||||
| SLAC1 | SLOW ANION CHANNEL-ASSOCIATED 1 | S-type anion channel | A. thaliana | loss of function | decrease | [158] |
| SLAH3 | SLAC1 HOMOLOGUE 3 | S-type anion channel | A. thaliana | loss of function | decrease | [109] |
| ALMT12 or QUAC1 | ALUMINIUM-ACTIVATED ANION CHANNEL 12 OR QUICK ANION CHANNEL 1 | R-type anion channel | A. thaliana | loss of function | decrease | [112] |
| ABCB14 | ABC TRANSPORTER B FAMILY MEMBER 14 | ATP-binding cassette (ABC) transporter | A. thaliana | loss of function | increase | [113] |
| GORK | GUARD CELL OUTWARD RECTIFYING K+ CHANNEL | outward potassium channel | A. thaliana | loss of function | increase | [115] |
| KAT1 | K+ CHANNEL ARABIDOPSIS THALIANA 1 | Potassium channel protein | A. thaliana | dominant negative | increase | [159] |
| OST2 or AHA1 | OPEN STOMATA 2 OR H(+)-ATPASE 1 | proton ATPase | A. thaliana | dominant mutant | decrease | [119] |
| pyr1/pyl1/pyl2/pyl4/pyl5/pyl8 | PYRABACTIN RESISTANCE/PYR1-LIKE | ABA receptor | A. thaliana | sextuple mutant | decrease | [160] |
| ABI1 | ABA INSENSITIVE 1 | protein phosphatase 2C | A. thaliana | loss of function | increase | [161] |
| HAB1 | HOMOLOGY TO ABI1 | protein phosphatase 2C | A. thaliana | loss of function | increase | [161] |
| OST1 or SnRK2.6 | OPEN STOMATA 1 OR SNF1-RELATED PROTEIN KINASE 2.6 | Ser/Thr kinase | A. thaliana | loss of function | decrease | [132] |
| HT1 | HIGH TEMPERATURE 1 | protein kinase | A. thaliana | dominant-negative | increase | [144] |
| NCED1 | 9-CIS-EPOXYCAROTENOID DEOXYGENASE | dioxygenase | S. lycopersicum | overexpression | increase | [162] |
| Gene | WUE Related Trait | Species | WUE Measurment | Range of Variation | Tested Conditions | Ref. |
|---|---|---|---|---|---|---|
| AN3 | Stomatal density | A. Thaliana | Leaf RWC | 44% reduction in mutant | drought (19 days) | [47] |
| GTL1 | Stomatal density | A. Thaliana | Integrated WUE, fresh shoot weight/water used (g/Kg) | 2 times more in mutant | well-watered | [53] |
| EPF1 | Stomatal density | A. Thaliana | WUE, assimilation rate/transpiration rate (mmol/mol) | 3 times more in mutant | well-watered | [61] |
| ERECTA | Stomatal density | A. Thaliana | WUE, plant dry weight/water used (g/g) | 1 time less in mutant | drought (16 days) | [157] |
| CER9 | Wax accumulation | A. Thaliana | WUE, δ13C | 50% reduction in mutant | well-watered | [101] |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ruggiero, A.; Punzo, P.; Landi, S.; Costa, A.; Van Oosten, M.J.; Grillo, S. Improving Plant Water Use Efficiency through Molecular Genetics. Horticulturae 2017, 3, 31. https://doi.org/10.3390/horticulturae3020031
Ruggiero A, Punzo P, Landi S, Costa A, Van Oosten MJ, Grillo S. Improving Plant Water Use Efficiency through Molecular Genetics. Horticulturae. 2017; 3(2):31. https://doi.org/10.3390/horticulturae3020031
Chicago/Turabian StyleRuggiero, Alessandra, Paola Punzo, Simone Landi, Antonello Costa, Michael J. Van Oosten, and Stefania Grillo. 2017. "Improving Plant Water Use Efficiency through Molecular Genetics" Horticulturae 3, no. 2: 31. https://doi.org/10.3390/horticulturae3020031
APA StyleRuggiero, A., Punzo, P., Landi, S., Costa, A., Van Oosten, M. J., & Grillo, S. (2017). Improving Plant Water Use Efficiency through Molecular Genetics. Horticulturae, 3(2), 31. https://doi.org/10.3390/horticulturae3020031