Deciphering the Molecular Mechanisms That Control Ovule Development in Pomegranate
Abstract
1. Introduction
2. Overview of Ovule Development in Plants
3. Key Genes Involved in Pomegranate Ovule Development
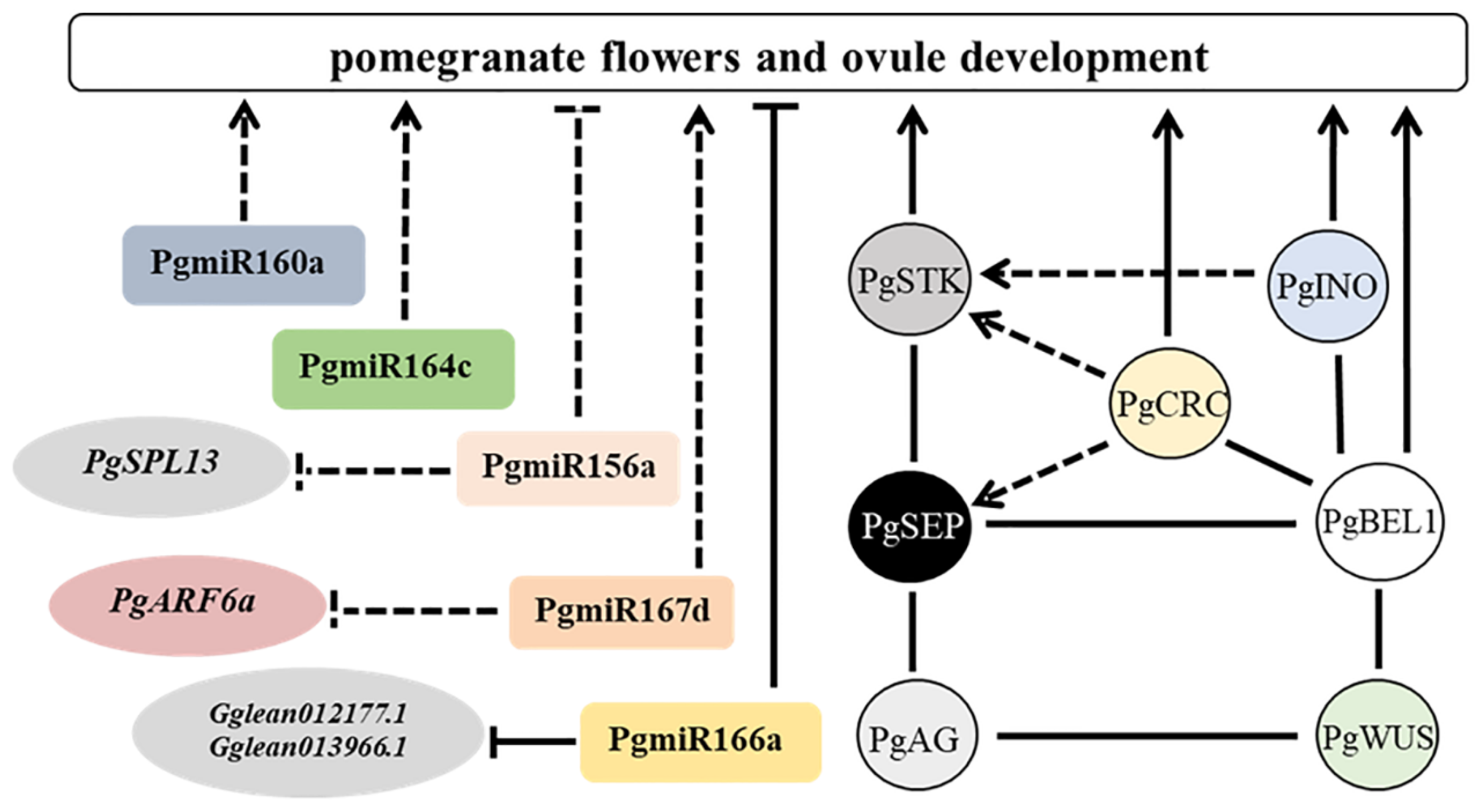
3.1. BEL1—A Regulator of Pomegranate Ovule Development
3.2. MADS-Box Genes—The Regulators of Ovule Identity and Development in Pomegranate
3.3. INO and CRC Genes—The Positive Regulator of Pomegranate Ovule Development
4. MicroRNA Regulates Ovule Development in Pomegranate
4.1. MiR166—The Negative Regulator of Pomegranate Ovule Development
4.2. MiR167—The Putative Positive Regulator of Pomegranate Ovule Development
4.3. MiR164—The Putative Positive Regulator of Pomegranate Ovule Development
4.4. MiR160—The Predicted Positive Regulator of Pomegranate Ovule Development
5. Implications for Fruit Production and Quality
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wetzstein, H.Y.; Ravid, N.; Wilkins, E.; Martinelli, A.P. A morphological and histological characterization of bisexual and male flower types in pomegranate. J. Am. Soc. Hortic. Sci. 2011, 136, 83–92. [Google Scholar] [CrossRef]
- Engin, H.; Gokbayrak, Z. Micromorphology of pollen grains from bisexual and functional male flowers of pomegranate. AGROFOR Int. J. 2017, 2, 40–46. [Google Scholar] [CrossRef]
- Chen, L.N.; Zhang, J.; Li, H.X.; Niu, J.; Xue, H.; Liu, B.B.; Wang, Q.; Luo, X.; Zhang, F.H.; Zhao, D.G.; et al. Transcriptomic analysis reveals candidate genes for female sterility in pomegranate flowers. Front. Plant Sci. 2017, 8, 1430. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Wang, Y.Y.; Yan, M.; Liu, C.Y.; Yuan, Z.H. BELL1 interacts with CRABS CLAW and INNER NO OUTER to regulate ovule and seed development in pomegranate. Plant Physiol. 2023, 191, 1066–1083. [Google Scholar] [CrossRef]
- Wetzstein, H.Y.; Yi, W.G.; Porter, J.A.; Ravid, N. Flower position and size impact ovule number per flower, fruitset, and fruit size in pomegranate. J. Am. Soc. Hortic. Sci. 2013, 138, 159–166. [Google Scholar] [CrossRef]
- Luo, X.; Li, H.X.; Wu, Z.K.; Yao, W.; Zhao, P.; Cao, D.; Yu, H.Y.; Li, K.D.; Poudel, K.; Zhao, D.G.; et al. The pomegranate (Punica granatum L.) draft genome dissects genetic divergence between soft- and hard-seeded cultivars. Plant Biotechnol. J. 2020, 18, 955–968. [Google Scholar] [CrossRef]
- Yuan, Z.H.; Fang, Y.M.; Zhang, T.K.; Fei, Z.J.; Han, F.M.; Liu, C.Y.; Liu, M.; Xiao, W.; Zhang, W.J.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2017, 16, 1363–1374. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.J.; Wang, Y.Y.; Zhao, X.Q.; Yan, M.; Ren, Y.; Yuan, Z.H. ARF6s identification and function analysis provide insights into flower development of Punica granatum L. Front. Plant Sci. 2022, 13, 833747. [Google Scholar] [CrossRef]
- Overholt, A.M.; Pierce, C.E.; Paleologos, C.S.; Shpak, E.D. ERECTA family signaling controls cell fate specification during ovule initiation in Arabidopsis. bioRxiv, 2024; preprint. [Google Scholar] [CrossRef]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arabidopsis. Plant Cell 1990, 2, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.X.; Zhou, L.W.; Hu, L.Q.; Jiang, Y.T.; Zhang, Y.J.; Feng, S.L.; Jiao, Y.L.; Xu, L.; Lin, W.H. Asynchrony of ovule primordia initiation in Arabidopsis. Development 2020, 147, 196618. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zuñiga-Mayo, V.M.; Montes, R.A.C.; Marsch-Martinez, N.; de Folter, S. Inside the gynoecium: At the carpel margin. Trends Plant Sci. 2013, 18, 644–655. [Google Scholar] [CrossRef] [PubMed]
- Barro-Trastoy, D.; Carrera, E.; Baños, J.; Palau-Rodriguez, J.; Ruiz-Rivero, O.; Tornero, P.; Alonso, J.M.; López-Díaz, I.; Gómez, M.D.; Pérez-Amador, M.A. Regulation of ovule initiation by gibberellins and brassinosteroids in tomato and Arabidopsis: Two plant species, two molecular mechanisms. Plant J. 2020, 102, 1026–1041. [Google Scholar] [CrossRef]
- Bowman, J.L.; Drews, G.N.; Meyerowitz, E.M. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 1991, 3, 749–758. [Google Scholar] [CrossRef]
- Roeder, A.H.; Yanofsky, M.F. Fruit development in Arabidopsis. Arab. Book 2006, 4, e0075. [Google Scholar] [CrossRef]
- Schneitz, K.; Baker, S.C.; Gasser, C.S.; Redweik, A. Pattern formation and growth during floral organogenesis: HUELLENLOS and AINTEGUMENTA are required for the formation of the proximal region of the ovule primordium in Arabidopsis thaliana. Development 1998, 125, 2555–2563. [Google Scholar] [CrossRef] [PubMed]
- Battaglia, R.; Colombo, M.; Kater, M.M. The ins and outs of ovule development. Fruit Development and Seed Dispersal. Annu. Plant Rev. 2009, 38, 70–106. [Google Scholar] [CrossRef]
- Cucinotta, M.; Colombo, L.; Roig-Villanova, I. Ovule development, a new model for lateral organ formation. Front. Plant Sci. 2014, 5, 117. [Google Scholar] [CrossRef]
- de Folter, S.; Shchennikova, A.V.; Franken, J.; Busscher, M.; Baskar, R.; Grossniklaus, U.; Angenent, G.C.; Immink, R.G.H. A Bsister MADS-box gene involved in ovule and seed development in petunia and Arabidopsis. Plant J. 2006, 47, 934–946. [Google Scholar] [CrossRef]
- Gupta, S.K.; Barg, R.; Arazi, T. Tomato agamous-like6 parthenocarpy is facilitated by ovule integument reprogramming involving the growth regulator KLUH. Plant Physiol. 2021, 185, 969–984. [Google Scholar] [CrossRef] [PubMed]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Dreni, L.; Jacchia, S.; Fornara, F.; Fornari, M.; Ouwerkerk, P.B.F.; An, G.; Colombo, L.; Kater, M.M. The D-lineage MADS-box gene OsMADS13 controls ovule identity in rice. Plant J. 2007, 52, 690–699. [Google Scholar] [CrossRef]
- Kawamoto, N.; Carpio, D.P.D.; Hofmann, A.; Mizuta, Y.; Kurihara, D.; Higashiyama, T.; Uchida, N.; Torii, K.U.; Colombo, L.; Groth, G.; et al. A peptide pair coordinates regular ovule initiation patterns with seed number and fruit size. Curr. Biol. 2020, 30, 4352–4361. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.Q.; Li, P.X.; Zhu, D.Y.; Ma, H.C.; Li, M.; Lai, Y.X.; Peng, Y.X.; Li, H.X.; Li, S.; Wei, J.B.; et al. SlCRCa is a key D-class gene controlling ovule fate determination in tomato. Plant Biotechnol. J. 2024, 22, 1966–1980. [Google Scholar] [CrossRef]
- Losa, A.; Colombo, M.; Brambilla, V.; Colombo, L. Genetic interaction between AINTEGUMENTA (ANT) and the ovule identity genes SEEDSTICK (STK), SHATTERPROOF1 (SHP1) and SHATTERPROOF2 (SHP2). Sex. Plant Reprod. 2010, 23, 115–121. [Google Scholar] [CrossRef]
- D’Apice, G.; Moschin, S.; Nigris, S.; Ciarle, R.; Muto, A.; Bruno, L.; Baldan, B. Identification of key regulatory genes involved in the sporophyte and gametophyte development in Ginkgo biloba ovules revealed by in situ expression analyses. Am. J. Bot. 2022, 109, 887–898. [Google Scholar] [CrossRef]
- Gonçalves, B.; Hasson, A.; Belcram, K.; Cortizo, M.; Morin, H.; Nikovics, K.; Vialette-Guiraud, A.; Takeda, S.; Aida, M.; Laufs, P.; et al. A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J. 2015, 83, 732–742. [Google Scholar] [CrossRef]
- Xu, M.; Tang, D.; Cheng, X.J.; Zhang, J.X.; Tang, Y.J.; Tao, Q.D.; Shi, W.Q.; You, A.Q.; Gu, M.H.; Cheng, Z.K.; et al. OsPINOID regulates stigma and ovule initiation through maintenance of the floral meristem by auxin signaling. Plant Physiol. 2019, 180, 952–965. [Google Scholar] [CrossRef]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-box protein complexes control carpel and ovule development in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [PubMed]
- Angenent, G.C.; Franken, J.; Busscher, M.; Van Dijken, A.; Van Went, J.L.; Dons, H.J.; Van Tunen, A.J. A novel class of MADS-box genes is involved in ovule development in petunia. Plant Cell 1995, 7, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.; Franken, J.; Van der Krol, A.R.; Wittichy, P.E.; Donsy, H.J.M.; Angenenta, G.C. Downregulation of ovule-specific MADS-box genes from petunia results in maternally controlled defects in seed development. Plant Cell 1997, 9, 703–715. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.L.; Zhong, H.X.; Zhang, F.C.; Zheng, J.L.; Zhang, C.; Vivek, Y.; Zhou, X.M.; Nocker, S.V.; Wu, X.Y.; Wang, X.P. Identification of grapevine BASIC PENTACYSTEINE transcription factors and functional characterization of VvBPC1 in ovule development. Plant Sci. 2025, 356, 112491. [Google Scholar] [CrossRef]
- Brambilla, V.; Battaglia, R.; Colombo, M.; Masiero, S.; Bencivenga, S.; Kater, M.M.; Colombo, L. Genetic and molecular interactions between BELL1 and MADS-box factors support ovule development in Arabidopsis. Plant Cell 2007, 19, 2544–2556. [Google Scholar] [CrossRef] [PubMed]
- Gross-Hardt, R.; Lenhard, M.; Laux, T. WUSCHEL signaling functions in interregional communication during Arabidopsis ovule development. Genes Dev. 2002, 16, 1129–1138. [Google Scholar] [CrossRef]
- Sieber, P.; Gheyselinck, J.; Gross-Hardt, R.; Laux, T.; Grossniklaus, U.; Schneitz, K. Pattern formation during early ovule development in Arabidopsis thaliana. Dev. Biol. 2004, 273, 321–334. [Google Scholar] [CrossRef]
- Kelley, D.R.; Skinner, D.J.; Gasser, C.S. Roles of polarity determinants in ovule development. Plant J. 2009, 57, 1054–1064. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Hsiao, Y.Y.; Li, C.I.; Yeh, C.M.; Mitsuda, N.; Yang, H.X.; Chiu, C.C.; Chang, S.B.; Liu, Z.J.; Tsai, W.C. The ancestral duplicated DL/CRC orthologs, PeDL1 and PeDL2, function in orchid reproductive organ innovation. J. Exp. Bot. 2021, 72, 5442–5461. [Google Scholar] [CrossRef] [PubMed]
- Gong, P.C.; Song, C.J.; Liu, H.Y.; Li, P.G.; Zhang, M.S.; Zhang, J.S.; Zhang, S.H.; He, C.Y. Physalis floridana CRABS CLAW mediates neofunctionalization of GLOBOSA genes in carpel development. J. Exp. Bot. 2021, 72, 6882–6903. [Google Scholar] [CrossRef]
- Lan, J.Q.; Wang, N.; Wang, Y.T.; Jiang, Y.D.; Yu, H.; Cao, X.F.; Qin, G.J. Arabidopsis TCP4 transcription factor inhibits high temperature-induced homeotic conversion of ovules. Nat. Commun. 2023, 14, 5673. [Google Scholar] [CrossRef]
- Western, T.L.; Haughn, G.W. BELL1 and AGAMOUS genes promote ovule identity in Arabidopsis thaliana. Plant J. 1999, 18, 329–336. [Google Scholar] [CrossRef]
- Lu, H.W.; Klocko, A.L.; Brunner, A.M.; Ma, C.; Magnuson, A.C.; Howe, G.T.; An, X.M.; Strauss, S.H. RNA interference suppression of AGAMOUS and SEEDSTICK alters floral organ identity and impairs floral organ determinacy, ovule differentiation, and seed-hair development in Populus. New Phytol. 2019, 222, 923–937. [Google Scholar] [CrossRef] [PubMed]
- Osnato, M.; Lacchini, E.; Pilatone, A.; Dreni, L.; Grioni, A.; Chiara, M.; Horner, D.; Pelaz, S.; Kater, M.M. Transcriptome analysis reveals rice MADS13 as an important repressor of the carpel development pathway in ovules. J. Exp. Bot. 2021, 72, 398–414. [Google Scholar] [CrossRef] [PubMed]
- Colombo, L.; Franken, J.; Koetje, E.; Van Went, J.; Dons, H.J.; Angenent, G.C.; Van Tunen, A.J. The petunia MADS-box gene FBP11 determines ovule identity. Plant Cell 1995, 7, 1859–1868. [Google Scholar] [CrossRef]
- Wang, X.; Liu, J.X.; Shang, E.L.; Hawar, A.; Ito, T.; Sun, B. Brassinosteroid signaling represses ZINC FINGER PROTEIN11 to regulate ovule development in Arabidopsis. Plant Cell 2024, 36, 5004–5022. [Google Scholar] [CrossRef] [PubMed]
- di Rienzo, V.; Imanifard, Z.; Mascio, I.; Gasser, C.S.; Skinner, D.J.; Pierri, C.L.; Marini, M.; Fanelli, V.; Sabetta, W.; Montemurro, C.; et al. Functional conservation of the grapevine candidate gene INNER NO OUTER for ovule development and seed formation. Hortic. Res. 2021, 8, 29. [Google Scholar] [CrossRef]
- Skinner, D.J.; Brown, R.H.; Kuzoff, R.K.; Gasser, C.S. Conservation of the role of INNER NO OUTER in development of unitegmic ovules of the Solanaceae despite a divergence in protein function. BMC Plant Biol. 2016, 16, 143. [Google Scholar] [CrossRef]
- Lora, J.; Hormaza, J.I.; Herrero, M. Transition from two to one integument in Prunus species: Expression pattern of INNER NO OUTER (INO), ABERRANT TESTA SHAPE (ATS) and ETTIN (ETT). New Phytol. 2015, 208, 584–595. [Google Scholar] [CrossRef]
- Uemura, A.; Yamaguchi, N.; Xu, Y.F.; Wee, W.Y.; Ichihashi, Y.; Suzuki, T.; Shibata, A.; Shirasu, K.; Ito, T. Regulation of floral meristem activity through the interaction of AGAMOUS, SUPERMAN, and CLAVATA3 in Arabidopsis. Plant Reprod. 2018, 31, 89–105. [Google Scholar] [CrossRef]
- Liu, Z.; Franks, R.G.; Klink, V.P. Regulation of gynoecium marginal tissue formation by LEUNIG and AINTEGUMENTA. Plant Cell 2000, 12, 1879–1891. [Google Scholar] [CrossRef]
- Simon, M.K.; Skinner, D.J.; Gallagher, T.L.; Gasser, C.S. Integument development in Arabidopsis depends on interaction of YABBY protein INNER NO OUTER with coactivators and corepressors. Genetics 2017, 207, 1489–1500. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Kalve, S.; Wolabu, T.W.; Nakashima, J.; Golz, J.F.; Tadege, M. Control of leaf blade outgrowth and floral organ development by LEUNIG, ANGUSTIFOLIA3 and WOX transcriptional regulators. New Phytol. 2019, 223, 2024–2038. [Google Scholar] [CrossRef]
- Wynn, A.N.; Seaman, A.A.; Jones, A.L.; Franks, R.G. Novel functional roles for PERIANTHIA and SEUSS during floral organ identity specification, floral meristem termination, and gynoecial development. Front. Plant Sci. 2014, 5, 130. [Google Scholar] [CrossRef] [PubMed]
- Azhakanandam, S.; Nole-Wilson, S.; Bao, F.; Franks, R.G. SEUSS and AINTEGUMENTA mediate patterning and ovule initiation during gynoecium medial domain development. Plant Physiol. 2008, 146, 1165–1181. [Google Scholar] [CrossRef] [PubMed]
- Das, P.; Ito, T.; Wellmer, F.; Vernoux, T.; Dedieu, A.; Traas, J.; Meyerowitz, E.M. Floral stem cell termination involves the direct regulation of AGAMOUS by PERIANTHIA. Development 2009, 136, 1605–1611. [Google Scholar] [CrossRef]
- Maier, A.T.; Stehling-Sun, S.; Wollmann, H.; Demar, M.; Hong, R.L.; Haubeiß, S.; Weigel, D.; Lohmann, J.U. Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 2009, 136, 1613–1620. [Google Scholar] [CrossRef]
- Skinner, D.J.; Baker, S.C.; Meister, R.J.; Broadhvest, J.; Schneitz, K.; Gasser, C.S. The Arabidopsis HUELLENLOS gene, which is essential for normal ovule development, encodes a mitochondrial ribosomal protein. Plant Cell 2001, 13, 2719–2730. [Google Scholar] [CrossRef]
- Huang, H.Y.; Jiang, W.B.; Hu, Y.W.; Wu, P.; Zhu, J.Y.; Liang, W.Q.; Wang, Z.Y.; Lin, W.H. BR signal influences Arabidopsis ovule and seed number through regulating related genes expression by BZR1. Mol. Plant 2013, 6, 456–469. [Google Scholar] [CrossRef]
- Broadhvest, J.; Baker, S.C.; Gasser, C.S. SHORT INTEGUMENTS 2 promotes growth during Arabidopsis reproductive development. Genetics 2000, 155, 899–907. [Google Scholar] [CrossRef]
- Hill, T.A.; Broadhvest, J.; Kuzoff, R.K.; Gasser, C.S. Arabidopsis SHORT INTEGUMENTS 2 is a mitochondrial DAR GTPase. Genetics 2006, 174, 707–718. [Google Scholar] [CrossRef]
- Hugouvieux, V.; Blanc-Mathieu, R.; Janeau, A.; Paul, M.; Lucas, J.; Xu, X.C.; Ye, H.L.; Lai, X.L.; Hir, S.L.; Guillotin, A.; et al. SEPALLATA-driven MADS transcription factor tetramerization is required for inner whorl floral organ development. Plant Cell 2024, 36, 3435–3450. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Hu, X.W.; Wei, H.; Cheng, Y.Q.; Liu, J.F. Identification of ChSEP genes in the whole genome of hazelnut, preparation of polyclonal antibody and expression in ovules. J. Beijing For. Univ. 2023, 45, 123–131. [Google Scholar] [CrossRef]
- Hoong, L.; Ireland, H.S.; Tomes, S.; Gunaseelan, K.; Ruslan, M.; McKenzie, C.; Hallett, I.; David, K.M.; Schaffer, R.J. Overexpression of the apple SEP1/2-like gene MdMADS8 promotes floral determinacy and enhances fruit flesh tissue and ripening. Planta 2025, 261, 53. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, Z.H.; Wu, J.; Hong, L.; Liang, J.J.; Ren, Y.M.; Guan, P.Y.; Hu, J.F. MADS-box protein complex VvAG2, VvSEP3 and VvAGL11 regulates the formation of ovules in Vitis vinifera L. cv. ‘Xiangfei’. Genes 2021, 12, 647. [Google Scholar] [CrossRef]
- Ditta, G.; Pinyopich, A.; Robles, P.; Pelaz, S.; Yanofsky, M.F. The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 2004, 14, 1935–1940. [Google Scholar] [CrossRef]
- Ehlers, K.; Bhide, A.S.; Tekleyohans, D.G.; Wittkop, B.; Snowdon, R.J.; Becker, A. The MADS Box Genes ABS, SHP1, and SHP2 are essential for the coordination of cell divisions in ovule and seed coat development and for endosperm formation in Arabidopsis thaliana. PLoS ONE 2016, 11, e0165075. [Google Scholar] [CrossRef]
- Lee, J.Y.; Baum, S.F.; Alvarez, J.; Patel, A.; Chitwood, D.H.; Bowman, J.L. Activation of CRABS CLAW in the nectaries and carpels of Arabidopsis. Plant Cell 2005, 17, 25–36. [Google Scholar] [CrossRef]
- Zhang, S.Q.; Tan, F.Q.; Chung, C.H.; Slavkovic, F.; Devani, R.S.; Troadec, C.; Marcel, F.; Morin, H.; Camps, C.; Roldan, M.V.G.; et al. The control of carpel determinacy pathway leads to sex determination in cucurbits. Science 2022, 378, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Sieber, P.; Petrascheck, M.; Barberis, A.; Schneitz, K. Organ polarity in Arabidopsis. NOZZLE Physically Interacts with Members of the YABBY Family. Plant Physiol. 2004, 135, 2172–2185. [Google Scholar] [CrossRef] [PubMed]
- Kamiuchi, Y.; Yamamoto, K.; Furutani, M.; Tasaka, M.; Aida, M. The CUC1 and CUC2 genes promote carpel margin meristem formation during Arabidopsis gynoecium development. Front. Plant Sci. 2014, 5, 165. [Google Scholar] [CrossRef] [PubMed]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164a and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Cucinotta, M.; Manrique, S.; Cuesta, C.; Benkova, E.; Novak, O.; Colombo, L. CUP-SHAPED COTYLEDON1 (CUC1) and CUC2 regulate cytokinin homeostasis to determine ovule number in Arabidopsis. J. Exp. Bot. 2018, 69, 5169–5176. [Google Scholar] [CrossRef]
- Chen, J.J.; Wang, W.; Qin, W.Q.; Men, S.Z.; Li, H.L.; Mitsuda, N.; Ohme-Takagi, M.; Wu, A.M. Transcription factors KNAT3 and KNAT4 are essential for integument and ovule formation in Arabidopsis. Plant Physiol. 2023, 191, 463–478. [Google Scholar] [CrossRef]
- Wu, M.F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef]
- Liu, N.; Wu, S.; Van Houten, J.; Wang, Y.; Ding, B.; Fei, Z.J.; Clarke, T.H.; Reed, J.W.; van der Knaap, E. Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 2014, 65, 2507–2520. [Google Scholar] [CrossRef]
- Goetz, M.; Hooper, L.C.; Johnson, S.D.; Rodrigues, J.C.M.; Vivian-Smith, A.; Koltunow, A.M. Expression of aberrant forms of AUXIN RESPONSE FACTOR8 stimulates parthenocarpy in Arabidopsis and Tomato. Plant Physiol. 2007, 145, 351–366. [Google Scholar] [CrossRef]
- Hu, L.Q.; Chang, J.H.; Yu, S.X.; Jiang, Y.T.; Li, R.H.; Zheng, J.X.; Zhang, Y.J.; Xue, H.W.; Lin, W.H. PIN3 positively regulates the late initiation of ovule primordia in Arabidopsis thaliana. PLoS Genet. 2022, 18, e1010077. [Google Scholar] [CrossRef] [PubMed]
- Petrella, R.; Gabrieli, F.; Cavalleri, A.; Schneitz, K.; Colombo, L.; Cucinotta, M. Pivotal role of STIP in ovule pattern formation and female germline development in Arabidopsis thaliana. Development 2022, 149, dev201184. [Google Scholar] [CrossRef] [PubMed]
- Gomez, M.D.; Ventimilla, D.; Sacristan, R.; Perez-Amador, M.A. Gibberellins regulate ovule integument development by interfering with the Transcription Factor ATS. Plant Physiol. 2016, 172, 2403–2415. [Google Scholar] [CrossRef]
- Wang, H.B.; Zhang, H.Q.; Liang, F.F.; Cong, L.; Song, L.Y.; Li, X.Y.; Zhai, R.; Yang, C.Q.; Wang, Z.G.; Ma, F.W.; et al. PbEIL1 acts upstream of PbCysp1 to regulate ovule senescence in seedless pear. Hortic. Res. 2021, 8, 59. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Lin, J.Y.; Hsu, W.H.; Kung, C.T.; Dai, S.Y.; Yang, J.Y.; Tan, C.M.; Yang, C.H. OAF is a DAF-like gene that controls ovule development in plants. Commun. Biol. 2023, 6, 498. [Google Scholar] [CrossRef]
- Wu, J.Q.; Li, P.X.; Li, M.; Zhu, D.Y.; Ma, H.C.; Xu, H.M.; Li, S.; Wei, J.B.; Bian, X.X.; Wang, M.Y.; et al. Heat stress impairs floral meristem termination and fruit development by affecting the BR-SlCRCa cascade in tomato. Plant Commun. 2024, 5, 100790. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Deka, M.; Sebastian, D.; Rosanna, P.; Veronica, G.; Li, J.R.; Wu, L.; Sonia, G. Spatiotemporal restriction of FUSCA3 expression by Class I BPCs promotes ovule development and coordinates embryo and endosperm growth. Plant Cell 2020, 32, 1886–1904. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.N.; Zhang, J.; Niu, J.; Li, H.X.; Xue, H.; Liu, B.B.; Xia, X.C.; Zhang, F.H.; Zhao, D.G.; Cao, S.Y. Cloning and functional verification of gene PgAGL11 associated with the development of flower organs in pomegranate plant. Acta Hortic. Sin. 2017, 44, 2089–2098. [Google Scholar] [CrossRef]
- Chen, L.N.; Wang, H.; Xu, T.T.; Liu, R.T.; Zhu, J.L.; Li, H.X.; Zhang, H.W.; Tang, L.Y.; Jing, D.; Yang, X.W.; et al. A telomere-to-telomere gap-free assembly integrating multi-omics uncovers the genetic mechanism of fruit quality and important agronomic trait associations in pomegranate. Plant Biotechnol. J. 2025, 23, 2852–2870. [Google Scholar] [CrossRef]
- Balasubramanian, S.; Schneitz, K. NOZZLE links proximal distal and adaxial abaxial pattern formation during ovule development in Arabidopsis thaliana. Development 2002, 129, 4291–4300. [Google Scholar] [CrossRef]
- Modrusan, Z.; Reiser, L.; Feldmann, K.A.; Fischer, R.L.; Haughn, G.W. Homeotic transformation of ovules into carpel-like structures in Arabidopsis. Plant Cell 1994, 6, 333–349. [Google Scholar] [CrossRef]
- Ray, A.; Robinson-Beers, K.; Ray, S.; Baker, S.C.; Lang, J.D.; Preuss, D.; Milligan, S.B.; Gasser, C.S. Arabidopsis floral homeotic gene BELL1 (BEL1) controls ovule development through negative regulation of AGAMOUS gene (AG). Proc. Natl. Acad. Sci. USA 1994, 91, 5761–5765. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Zhao, Y.J.; Yan, M.; Zhao, H.L.; Zhang, X.H.; Yuan, Z.H. Genome-wide identification and expression analysis of TALE gene family in pomegranate (Punica granatum L.). Agronomy 2020, 10, 829. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Liu, C.Y.; Zhao, X.Q.; Wang, Y.Y.; Yan, M.; Yuan, Z.H. Cloning and spatiotemporal expression analysis of PgWUS and PgBEL1 in Punica granatum. Acta Hortic. Sin. 2021, 48, 355–366. [Google Scholar] [CrossRef]
- Zhao, Y.J. Study on the Mechanism of Pomegranate Ovule Development and the Function of Key Transcription Factors. Ph.D. Thesis, Nanjing Forestry University, Nanjing, China, 2023. [Google Scholar]
- Zhao, Y.J.; Zhao, H.L.; Wang, Y.Y.; Zhang, X.H.; Zhao, X.Q.; Yuan, Z.H. Genome-wide identification and expression analysis of MIKC-Type MADS-box gene family in Punica granatum L. Agronomy 2020, 10, 1197. [Google Scholar] [CrossRef]
- Vandenbussche, M.; Zethof, J.; Souer, E.; Koes, R.; Tornielli, G.B.; Pezzotti, M.; Ferrario, S.; Angenent, G.C.; Gerats, T. Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D floral organ identity functions require SEPALLATA-like MADS-box genes in petunia. Plant Cell 2003, 15, 2680–2693. [Google Scholar] [CrossRef]
- Immink, R.G.H.; Tonaco, I.A.N.; de Folter, S.; Shchennikova, A.; van Dijk, A.D.J.; Busscher-Lange, J.; Borst, J.W.; Angenent, G.C. SEPALLATA3: The ‘glue’ for MADS-box transcription factor complex formation. Genome Biol. 2009, 10, R24. [Google Scholar] [CrossRef] [PubMed]
- Schoof, H.; Lenhard, M.; Haecker, A.; Mayer, K.F.X.; Jürgens, G.; Laux, T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 2000, 100, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Mathews, S.; Kramer, E.M. The evolution of reproductive structures in seed plants: A re-examination based on insights from developmental genetics. New Phytol. 2012, 194, 910–923. [Google Scholar] [CrossRef]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef]
- Liu, X.G.; Kim, Y.J.; Müller, R.; Yumul, R.E.; Liu, C.Y.; Pan, Y.Y.; Cao, X.F.; Goodrich, J.; Chen, X.M. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group Proteins. Plant Cell 2011, 23, 3654–3670. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.C.; Robinson-Beers, K.; Villanueva, J.M.; Gaiser, J.C.; Gasser, C.S. Interactions among genes regulating ovule development in Arabidopsis thaliana. Genetics 1997, 145, 1109–1124. [Google Scholar] [CrossRef]
- Villanueva, J.M.; Broadhvest, J.; Hauser, B.A.; Meister, R.J.; Schneitz, K.; Gasser, C.S. INNER NO OUTER regulates abaxial-adaxial patterning in Arabidopsis ovules. Genes Dev. 1999, 13, 3160–3169. [Google Scholar] [CrossRef]
- Meister, R.J.; Kotow, L.M.; Gasser, C.S. SUPERMAN attenuates positive INNER NO OUTER autoregulation to maintain polar development of Arabidopsis ovule outer integument. Development 2002, 129, 4281–4289. [Google Scholar] [CrossRef]
- Gallagher, T.L.; Gasser, C.S. Independence and interaction of regions of the INNER NO OUTER protein in growth control during ovule development. Plant Physiol. 2008, 147, 306–315. [Google Scholar] [CrossRef]
- Jia, D.D.; Chen, L.G.; Yin, G.M.; Yang, X.R.; Gao, Z.H.; Guo, Y.; Sun, Y.; Tang, W.Q. Brassinosteroids regulate outer ovule integument growth in part via the control of INNER NO OUTER by BRASSINOZOLE-RESISTANT family transcription factors. J. Integr. Plant Biol. 2020, 62, 1093–1111. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Liu, C.Y.; Ge, D.P.; Yan, M.; Ren, Y.; Huang, X.B.; Yuan, Z.H. Genome-wide identification and expression of YABBY genes family during flower development in Punica granatum L. Gene 2020, 752, 144784. [Google Scholar] [CrossRef]
- Gross, T.; Broholm, S.; Becker, A. CRABS CLAW acts as a bifunctional transcription factor in flower development. Front. Plant Sci. 2018, 9, 835. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA genes regulate growth and pattern formation during gynoecium development in Arabidopsis thaliana. Int. J. Plant Sci. 2002, 163, 17–41. [Google Scholar] [CrossRef]
- Castañeda, L.; Giménez, E.; Pineda, B.; García-Sogo, B.; Ortiz-Atienza, A.; Micol-Ponce, R.; Angosto, T.; Capel, J.; Moreno, V.; Yuste-Lisbona, F.J.; et al. Tomato CRABS CLAW paralogues interact with chromatin remodelling factors to mediate carpel development and floral determinacy. New Phytol. 2022, 234, 1059–1074. [Google Scholar] [CrossRef]
- Sablowski, R. Control of patterning, growth, and differentiation by floral organ identity genes. J. Exp. Bot. 2015, 66, 1065–1073. [Google Scholar] [CrossRef] [PubMed]
- Axtell, M.J. Evolution of microRNAs and their targets: Are all microRNAs biologically relevant? Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2008, 1779, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Cao, D.; Zhang, J.F.; Chen, L.; Xia, X.C.; Li, H.X.; Zhao, D.G.; Zhang, F.H.; Xue, H.; Chen, L.N.; et al. Integrated microRNA and mRNA expression profiling reveals a complex network regulating pomegranate (Punica granatum L.) seed hardness. Sci. Rep. 2018, 8, 9292. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Li, Z.W.; An, X.L.; Zhu, T.T.; Yan, T.W.; Wu, S.W.; Tian, Y.H.; Li, J.P.; Wan, X.Y. Discovering and constructing ceRNA-miRNA-target gene regulatory networks during anther development in Maize. Int. J. Mol. Sci. 2019, 20, 3480. [Google Scholar] [CrossRef]
- Quesada, V.; Dean, C.; Simpson, G.G. Regulated RNA processing in the control of Arabidopsis flowering. Int. J. Dev. Biol. 2005, 49, 773–780. [Google Scholar] [CrossRef] [PubMed]
- Abdurakhmonov, I.Y.; Devor, E.J.; Buriev, Z.T.; Huang, L.Y.; Makamov, A.; Shermatov, S.E.; Bozorov, T.; Kushanov, F.N.; Mavlonov, G.T.; Abdukarimov, A. Small RNA regulation of ovule development in the cotton plant, G. hirsutum L. BMC Plant Biol. 2008, 8, 93. [Google Scholar] [CrossRef]
- Wang, Z.M.; Xue, W.; Dong, C.J.; Jin, L.G.; Bian, S.M.; Wang, C.; Wu, X.Y.; Liu, J.Y. A comparative miRNAome analysis reveals seven fiber initiation-related and 36 novel miRNAs in developing cotton ovules. Mol. Plant 2012, 4, 889–900. [Google Scholar] [CrossRef]
- Petrella, R.; Cucinotta, M.; Mendes, M.A.; Underwood, C.J.; Colombo, L. The emerging role of small RNAs in ovule development, a kind of magic. Plant Reprod. 2021, 34, 335–351. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Miyashima, S.; Sato-Nara, K.; Yamada, T.; Nakajima, K. Functionally diversified members of the MIR165/6 gene family regulate ovule morphogenesis in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1017–1026. [Google Scholar] [CrossRef]
- Zhao, Y.J.; Huang, J.Y.; Li, M.; Ren, H.F.; Jiao, J.; Wan, R.; Liu, Y.; Wang, M.M.; Shi, J.L.; Zhang, K.X.; et al. Exploring microRNAs associated with pomegranate pistil development: An identification and analysis study. Horticulturae 2024, 10, 85. [Google Scholar] [CrossRef]
- Saminathan, T.; Bodunrin, A.; Singh, N.V.; Devarajan, R.; Nimmakayala, P.; Jeff, M.; Aradhya, M.; Reddy, U.K. Genome-wide identification of microRNAs in pomegranate (Punica granatum L.) by high-throughput sequencing. BMC Plant Biol. 2016, 16, 122. [Google Scholar] [CrossRef] [PubMed]
- Li, B.B.; Zhao, Y.J.; Wang, S.; Zhang, X.H.; Wang, Y.W.; Shen, Y.; Yuan, Z.H. Genome-wide identification, gene cloning, subcellular location and expression analysis of SPL gene family in P. granatum L. BMC Plant Biol. 2021, 21, 400. [Google Scholar] [CrossRef]
- Chen, L.N.; Luo, X.; Yang, X.W.; Jing, D.; Xia, X.C.; Li, H.X.; Poudel, K.; Cao, S.Y. Small RNA and mRNA sequencing reveal the roles of microRNAs involved in pomegranate female sterility. Int. J. Mol. Sci. 2020, 21, 558. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Rieater, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Nodine, M.D.; Bartel, D.P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis. Genes Dev. 2010, 24, 2678–2692. [Google Scholar] [CrossRef] [PubMed]
- Palatnik, J.F.; Allen, E.; Wu, X.; Schommer, C.; Schwab, R.; Carrington, J.C.; Weigel, D. Control of leaf morphogenesis by microRNAs. Nature 2003, 425, 257–263. [Google Scholar] [CrossRef]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef]
- Liu, J.F.; Luo, Q.Z.; Zhang, X.Z.; Zhang, Q.; Cheng, Y.Q. Identification of vital candidate microRNA/mRNA pairs regulating ovule development using high-throughput sequencing in hazel. BMC Dev. Biol. 2020, 20, 13. [Google Scholar] [CrossRef]
- Mallory, A.C.; Bartel, D.P.; Bartel, B. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 2005, 17, 1360–1375. [Google Scholar] [CrossRef]
- Wang, J.W.; Wang, L.J.; Mao, Y.B.; Cai, W.J.; Xue, H.W.; Chen, X.Y. Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 2005, 17, 2204–2216. [Google Scholar] [CrossRef]
- Laufs, P. MicroRNA regulation of the CUC genes is required for boundary size control in Arabidopsis meristems. Development 2004, 131, 4311–4322. [Google Scholar] [CrossRef] [PubMed]
- Baker, C.C.; Sieber, P.; Wellmer, F.; Meyerowitz, E.M. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005, 15, 303–315. [Google Scholar] [CrossRef]
- Guo, H.S.; Xie, Q.; Fei, J.F.; Chua, N.H. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development. Plant Cell 2005, 17, 1376–1386. [Google Scholar] [CrossRef]
- Williams, L.; Grigg, S.P.; Xie, M.; Christensen, S.; Fletcher, J.C. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development 2005, 132, 3657–3668. [Google Scholar] [CrossRef]
- Zhou, G.K.; Kubo, M.; Zhong, R.; Demura, T.; Ye, Z.H. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007, 48, 391–404. [Google Scholar] [CrossRef] [PubMed]
- Carlsbecker, A.; Lee, J.Y.; Roberts, C.J.; Dettmer, J.; Lehesranta, S.; Zhou, J.; Lindgren, O.; Moreno-Risueno, M.A.; Vaten, A.; Thitamadee, S.; et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature 2010, 465, 316–321. [Google Scholar] [CrossRef] [PubMed]
- Christian, C.; Ravi, S.D.; Rachid, B.; Hao, Y.W.; Halima, M.; Fabien, M.; Marion, V.; Brahim, M.; Gwilherm, B.; Sylvie, C.; et al. The miR166-SlHB15A regulatory module controls ovule development and parthenocarpic fruit set under adverse temperatures in tomato. Mol. Plant 2021, 14, 1185–1198. [Google Scholar] [CrossRef]
- Yao, X.Z.; Chen, J.L.; Zhou, J.; Yu, H.; Ge, C.N.; Zhang, M.; Gao, X.H.; Dai, X.H.; Yang, Z.N.; Zhao, Y.D. An essential role for miRNA167 in maternal control of embryonic and seed development. Plant Physiol. 2019, 180, 453–464. [Google Scholar] [CrossRef]
- Combier, J.P.; Frugier, F.; De Billy, F.; Boualem, A.; El-Yahyaoui, F.; Moreau, S.; Vernie, T.; Ott, T.; Gamas, P.; Crespi, M.; et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 2006, 20, 3084–3088. [Google Scholar] [CrossRef]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Lauter, N.; Kampani, A.; Carlson, S.; Goebel, M.; Moose, S.P. MicroRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc. Natl. Acad. Sci. USA 2005, 102, 9412–9417. [Google Scholar] [CrossRef] [PubMed]
- Wollmann, H.; Mica, E.; Todesco, M.; Long, J.A.; Weigel, D. On reconciling the interactions between APETALA2, miR172 and AGAMOUS with the ABC model of flower development. Development 2010, 137, 3633–3642. [Google Scholar] [CrossRef]
- Koyama, T.; Furutani, M.; Tasaka, M.; Ohme-Takagi, M. TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 2007, 19, 473–484. [Google Scholar] [CrossRef]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chetelat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.H.; Zhang, B.B.; Ma, R.J.; Yu, M.L.; Guo, S.L.; Guo, L. Isolation and expression analysis of four HD-ZIP III family genes targeted by microRNA166 in peach. Genet. Mol. Res. 2015, 14, 14151–14161. [Google Scholar] [CrossRef] [PubMed]
- Prigge, M.J.; Otsuga, D.; Alonso, J.M.; Ecker, J.R.; Drews, G.N.; Clark, S.E. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic and distinct roles in Arabidopsis development. Plant Cell 2005, 17, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.F.; Ning, K.; Che, G.; Yan, S.S.; Han, L.J.; Gu, R.; Li, Z.; Weng, Y.Q.; Zhang, X.L. CsSPL functions as an adaptor between HD-ZIP III and CsWUS transcription factors regulating anther and ovule development in cucumber. Plant J. 2018, 94, 535–547. [Google Scholar] [CrossRef]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef]
- Zheng, L.J.; Nagpal, P.; Villarino, G.; Trinidad, B.; Bird, L.; Huang, Y.B.; Reed, J.W. MiR167 limits anther growth to potentiate anther dehiscence. Development 2019, 146, 174375. [Google Scholar] [CrossRef]
- Schubert, R.; Dobritzsch, S.; Gruber, C.; Hause, G.; Athmer, B.; Schreiber, T.; Marillonnet, S.; Okabe, Y.; Ezura, H.; Acosta, I.F.; et al. Tomato MYB21 acts in ovules to mediate Jasmonate-regulated fertility. Plant Cell 2019, 31, 1043–1062. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Y.; McCaig, B.C.; Wingerd, B.A.; Wang, J.H.; Whalon, M.E.; Pichersky, E.; Howe, G.A. The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 2004, 16, 126–143. [Google Scholar] [CrossRef]
- Ishiguro, S.; Kawai-Oda, A.; Ueda, J.; Nishida, I.; Okada, K. The DEFECTIVE IN ANTHER DEHISCIENCE gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 2001, 13, 2191–2209. [Google Scholar] [CrossRef]
- Gupta, S.K.; Vishwakarma, A.; Kenea, H.D.; Galsurker, O.; Cohen, H.; Aharoni, A.; Arazi, T. CRISPR/Cas9 mutants of tomato MICRORNA164 genes uncover their functional specialization in development. Plant Physiol. 2021, 187, 1636–1652. [Google Scholar] [CrossRef] [PubMed]
- Galbiati, F.; Roy, D.S.; Simonini, S.; Cucinotta, M.; Ceccato, L.; Cuesta, C.; Simaskova, M.; Benkova, E.; Kamiuchi, Y.; Aida, M.; et al. An integrative model of the control of ovule primordia formation. Plant J. 2013, 76, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.P.; Montgomery, T.A.; Fahlgren, N.; Kasschau, K.D.; Nonogaki, H.; Carrington, J.C. Repression of AUXIN RESPONSE FACTOR10 by microRNA160 is critical for seed germination and post-germination stages. Plant J. 2007, 52, 133–146. [Google Scholar] [CrossRef]
- Damodharan, S.; Zhao, D.Z.; Arazi, T. A common miRNA160-based mechanism regulates ovary patterning, floral organ abscission and lamina outgrowth in tomato. Plant J. 2016, 86, 458–471. [Google Scholar] [CrossRef]
- Damodharan, S.; Corem, S.; Gupta, S.K.; Arazi, T. Tuning of SlARF10A dosage by sly-miR160a is critical for auxin-mediated compound leaf and flower development. Plant J. 2018, 96, 855–868. [Google Scholar] [CrossRef]
- Yang, J.; Tian, L.; Sun, M.X.; Huang, X.Y.; Zhu, J.; Guan, Y.F.; Jia, Q.S.; Yang, Z.N. AUXIN RESPONSE FACTOR17 is essential for pollen wall pattern formation in Arabidopsis. Plant Physiol. 2013, 162, 720–731. [Google Scholar] [CrossRef]
- Hodson, R. Food security. Nature 2017, 544, S5. [Google Scholar] [CrossRef]
- Jagtap, A.B.; Yadav, I.S.; Vikal, Y.; Praba, U.P.; Kaur, N.; Gill, A.S.; Joha, G.S. Transcriptional dynamics of maize leaves, pollens and ovules to gain insights into heat stress-related responses. Front. Plant Sci. 2023, 14, 1117136. [Google Scholar] [CrossRef]
- Yang, Y.; Tilman, D.; Jin, Z.N.; Smith, P.; Barrett, C.B.; Zhu, Y.G.; Burney, J.; D’Odorico, P.; Fantke, P.; Fargione, J.; et al. Climate change exacerbates the environmental impacts of agriculture. Science 2024, 385, eadn3747. [Google Scholar] [CrossRef]
- Martre, P.; Dueri, S.; Guarin, J.R.; Ewert, F.; Webber, H.; Calderini, D.; Molero, G.; Reynolds, M.; Miralles, D.; Garcia, G.; et al. Global needs for nitrogen fertilizer to improve wheat yield under climate change. Nat. Plants 2024, 10, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.Q.; Zhang, Y.Y.; Li, G.; Shi, J.; Wang, D.X.; Zhu, W.W.; Yang, X.J.; Dreni, L.; Tucker, M.R.; Zhang, D.B. MADS8 is indispensable for female reproductive development at high ambient temperatures in cereal crops. Plant Cell 2023, 36, 65–84. [Google Scholar] [CrossRef]
- Yang, X.J.; Tucker, M.R. Establishing a regulatory blueprint for ovule number and function during plant development. Curr. Opin. Plant Biol. 2021, 63, 102095. [Google Scholar] [CrossRef]
- Rudall, P.J. Evolution and patterning of the ovule in seed plants. Biol. Rev. 2021, 96, 943–960. [Google Scholar] [CrossRef]
- Zu, S.H.; Jiang, Y.T.; Chang, J.H.; Zhang, Y.J.; Xue, H.W.; Lin, W.H. Interaction of brassinosteroid and cytokinin promotes ovule initiation and increases seed number per silique in Arabidopsis. J. Integr. Plant Biol. 2022, 64, 702–716. [Google Scholar] [CrossRef]
- Cucinotta, M.; Marzo, M.D.; Guazzotti, A.; de Folter, S.; Kater, M.M.; Colombo, L. Gynoecium size and ovule number are interconnected traits that impact seed yield. J. Exp. Bot. 2020, 71, 2479–2489. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.Q.; Xie, M.Y.; Jiang, Y.M.; Yuan, T. Abortion occurs during double fertilization and ovule development in Paeonia ludlowii. J. Plant Res. 2022, 135, 295–310. [Google Scholar] [CrossRef]
- Cui, X.Y.; Liu, Q.Y.; Luo, Y.J.; Zhu, P.P.; Guan, P.Y.; Zhang, J.X. Study on influencing factors of embryo rescue and germplasm innovation in seedless grape. Plant Cell Tissue Organ Cult. (PCTOC) 2024, 157, 24. [Google Scholar] [CrossRef]
- Chu, Y.C.; Lin, T.S.; Chang, J.C. Pollen effects on fruit set, seed weight, and shriveling of ‘73-S-20’ Litchi-with special reference to artificial induction of parthenocarpy. Hort. Sci. 2015, 50, 369–373. [Google Scholar] [CrossRef]
- Ma, Y.W.; Li, Q.L.; Hu, G.B.; Qin, Y.H. Comparative transcriptional survey between self-incompatibility and self-compatibility in Citrus reticulata Blanco. Gene 2017, 609, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Wijesinghe, S.A.E.C.; Evans, L.J.; Kirkland, L.; Rader, R. A global review of watermelon pollination biology and ecology: The increasing importance of seedless cultivars. Sci. Hortic. 2020, 271, 109493. [Google Scholar] [CrossRef]
- Hawar, A.; Chen, W.; Zhu, T.; Wang, X.; Liu, J.X.; Xiong, S.Q.; Ito, T.; Chen, D.J.; Sun, B. The histone acetyltransferase GCN5 regulates floral meristem activity and flower development in Arabidopsis. Plant Cell 2025, 37, koaf135. [Google Scholar] [CrossRef]
- Sun, S.N.; Jia, P.F.; Wang, W.; Chen, L.C.; Gong, X.R.; Lin, H.F.; Wu, R.; Yang, W.C.; Li, H.J.; Zuo, J.R.; et al. S-Sulfenylation-mediated inhibition of the GSNOR1 activity regulates ovule development in Arabidopsis. J. Genet. Genom. 2025, 52, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
| Species | Gene Name | Gene Number | Biological Function | References |
|---|---|---|---|---|
| A. thaliana, Populus alba, Oryza sativa | AG (AGAMOUS), OsMADS13 | AT4G18960, Potri.004G064300 (PtAG1), Potri.011G075800 (PtAG2) | determine ovule identity, repress carpel development | [40,41,42] |
| A. thaliana, P. alba | STK (SEEDSTICK) | AT2G01930 | determine ovule identity, floral organ determinacy, and ovule differentiation | [25,33,41] |
| A. thaliana | BEL1 (BELL1) | AT5G41410, | determine ovule identity | [33,39,40] |
| A. thaliana | SHP (SHATTERPROOF) | AT3G58780 AT2G42830 | determine ovule identity | [21,25] |
| Petunia hybrida | FBP7 (Floral Hinding Protein7) | CAA57311 | determine ovule identity | [30,31] |
| P. hybrida | FBP11 (Floral Hinding Protein 11) | CAA57445 | determine ovule identity | [30,43] |
| A. thaliana, Vitis vinifera, Solanum lycopersicum, Nicotiana tabacum, Prunus species | INO (INNER NO OUTER) | AT1G23420, VIT201s0127g00330, Solyc05g005240.1.1 | determine outer integument formation | [44,45,46,47] |
| A. thaliana | SUP (SUPERMAN) | AT3G23130 | determine integument development | [48] |
| A. thaliana, Ginkgo biloba | ANT (AINTEGUMENTA) | AT4G37750 | determine ovule primordium initiation and integument development | [16,25,26] |
| A. thaliana | LUG (LEUNIG) | AT4G32551 | regulate gynoecium and ovule development | [49,50,51] |
| A. thaliana | SEU (SEUSS) | AT1G43850 | regulate ovule primordium | [52,53] |
| A. thaliana | PAN (PERIANTHIA) | AT1G68640 | regulate inflorescence, embryo and ovule development | [52,54,55] |
| A. thaliana | HLL (HUELLENLOS) | AT1G17560 | regulate integument development | [56,57] |
| A. thaliana | SIN 2 (SHORT INTEGUMENTS 2) | AT2G41670 | regulate integument development | [58,59] |
| A. thaliana, V. vinifera, Malus domestica | SEP1 (SEPALLATA1) | AT5G15800, VIT_14s0083g01050, Cor0054610.1, MD17G1065400 | determine the identity of carpels | [29,60,61,62] |
| A. thaliana, Vitis vinifera | SEP2 (SEPALLATA2) | AT3G02310, VIT_17s0000g05000 | determine the identity of carpels | [29,60] |
| A. thaliana, V. vinifera | SEP3 (SEPALLATA3) | AT1G24260, VIT_01s0010g03900, Cor0008190.1 | determine the identity of carpels, ovules formation | [29,60,61,63] |
| A. thaliana, V. vinifera | SEP4 (SEPALLATA4) | AT2G03710, VIT_01s0011g00110, Cor0142960.1 | regulate the development of petals, stamens, carpels, and sepal | [60,61,64] |
| A. thaliana | ABS (ARABIDOPSIS BSISTER) | AT5G23260 | regulate inner seed/coat layer formation | [65] |
| A. thaliana, Cucumis melo, Phalaenopsis equestris, Physalis floridana, S. lycopersicum | CRC (CRABS CLAW) | AT1G69180, XP_020572967, MN969999, XM_004228801.4, Solyc01g01 0240, Solyc05g012050 | carpel identity, nectary development, ovule fate determination | [24,37,38,66,67] |
| A. thaliana | ZFP11 (ZINC FINGER PROTEIN11) | AT2G42410 | ensure ovule primordia and number development | [44] |
| A. thaliana | NZZ (NOZZLE) | AT4G27330 | adaxial–abaxial ovule development | [68] |
| A. thaliana | CUC (CUP-SHAPED COTYLEDON) | AT3G15170, AT5G53950, At1g76420 | promote carpel margin meristem formation, regulate ovule number and morphology | [9,69,70,71] |
| A. thaliana | KNAT3, KNAT4 (KNOTTED1-LIKE) | AT5G25220 AT5G11060 | regulate integument and ovule formation | [72] |
| A. thaliana, S. lycopersicum | ARF6 (AUXIN RESPONSE FACTOR 6) | AT1G30330, Solyc00g196060, solyc07g043610/043620 | regulate ovules, anther, and seed development | [73,74] |
| A. thaliana, S. lycopersicum | ARF8 (AUXIN RESPONSE FACTOR 8) | AT5G37020, EF667342, Solyc03g031970, Solyc02g037530 | regulate ovules, anther, and seed development | [73,74,75] |
| A. thaliana | PIN3 (PINFORMED3) | AT1G70940 | promote the late ovule initiation | [76] |
| A. thaliana | PHB (PHABULOSA) | AT2G34710 | determine inner and outer integument development | [77] |
| A. thaliana | ATS/KAN4 (ABERRANT TESTA SHAPE) | AT5G42630 | regulate integument growth | [78] |
| A. thaliana | TCP4 (TEOSINTE BRANCHED1/CYCLOIDEA/PCF) | AT3G15030 | maintain ovule identity | [39] |
| Pyrus communis | EIL1 (Ethylene-Insensitive 3-Like) | AT5G21120, LOC103928267 | regulate ovule senescence | [79] |
| A. thaliana, Phalaenopsis aphrodite | OAF (Ovule Activating Factor) | AT3G10910 | regulate ovule development | [80] |
| A. thaliana, S. lycopersicum | WUS (WUSCHEL) | Solyc02g083950 | regulate the development of ovules | [81] |
| A. thaliana | EPFL (EPIDERMAL PATTERNING FACTOR-LIKE) | At5g10310, At4g37810 | regulate ovule number and morphology, promote ovule initiation | [9] |
| A. thaliana | ERL (ERECTA-LIKE) | At5g62230, At5g07180 | promote seed density | [9] |
| A. thaliana, V. vinifera | BPC (BASIC PENTACYSTEINE) | AT2G01930 (BPC1), AT1G14685 (BPC2), AT1G68120 (BPC3), 100233138, 100255909, 100233069, 100233070 | regulate ovule development | [32,82] |
| A. thaliana Orthologue | Gene Number | Biological Function | References |
|---|---|---|---|
| STK (SEEDSTICK) | OP828677 | ovule identity, floral organ determinacy, ovule differentiation | [4,83] |
| AG (AGAMOUS) | OP828676 | regulate reproductive organ identity and meristem determinacy | [4] |
| BEL1 (BELL1) | OP828678 | ovule identity | [4] |
| INO (INNER NO OUTER) | OP828670 | ovule outer integument formation, positive regulation of ovule number | [4] |
| WUS (WUSCHEL) | OP828679 | regulate the development of ovules | [4] |
| SEP1 (SEPALLATA1) | OP828672 | the development of petals, stamens and carpels, and negative regulation of ovule development | [4] |
| SEP3 (SEPALLATA3) | OP828673, OP828674 | determine the identity of carpels, and ovule formation | [4] |
| SEP4 (SEPALLATA4) | OP828675 | regulate the development of stamens, carpels, and sepal, and negative regulation of the number of ovules | [4] |
| CRC (CRABS CLAW) | OP828671 | carpel identity, nectary development, positive regulation of ovule differentiation, and ovule fate determination | [4] |
| NST3/NAC012 | Pgr01G048970 | regulate the thickness of the inner seed coat | [84] |
| miRNA Name | Target Genes | Function | Reference |
|---|---|---|---|
| miR156/157 | SPL3/4/5/9 | vegetative-to-reproductive phase transitions and flowering | [123,124] |
| miR159 | MYB33/65/101 | flowering time; floral patterning; anther development | [125,126,127] |
| miR160 | ARF10/16/17 | carpel development; floral patterning; predicted positive regulator of ovule development | [128,129] |
| miR164 | CUC1, CUC2, NAC1 | petal number control; normal embryonic development; putative positive regulator of ovule development | [130,131,132] |
| miR165/166 | PHB, PHV, REV, HB15 | adaxial and abaxial polarity; embryo development; negative regulator of ovule development | [133,134,135,136] |
| miR167 | ARF6, ARF8 | floral induction; ovule, embryonic and anther development; putative positive regulator of ovule development | [73,127,137] |
| miR169 | NF-YA (NUCLEAR TRAN-SCRIPTION FACTOR Y SUBUNIT ALPHA) | regulate stamen and carpel development | [138] |
| miR172 | AP2, TOE1/2/3, SMZ, SNZ | flowering time; seed and floral development | [127,139,140,141] |
| miR319 | TCPs | regulation of flower development; male and female gametophyte development | [142,143] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license.
Share and Cite
Zhao, Y.; Sassa, H.; Li, M.; Miao, Y.; Zhu, X.; Hao, P.; Wan, R.; Zhang, K.; Cong, L.; Shen, Y.; et al. Deciphering the Molecular Mechanisms That Control Ovule Development in Pomegranate. Horticulturae 2026, 12, 26. https://doi.org/10.3390/horticulturae12010026
Zhao Y, Sassa H, Li M, Miao Y, Zhu X, Hao P, Wan R, Zhang K, Cong L, Shen Y, et al. Deciphering the Molecular Mechanisms That Control Ovule Development in Pomegranate. Horticulturae. 2026; 12(1):26. https://doi.org/10.3390/horticulturae12010026
Chicago/Turabian StyleZhao, Yujie, Hidenori Sassa, Ming Li, Yifei Miao, Xiaoyan Zhu, Pengbo Hao, Ran Wan, Kunxi Zhang, Liu Cong, Yawen Shen, and et al. 2026. "Deciphering the Molecular Mechanisms That Control Ovule Development in Pomegranate" Horticulturae 12, no. 1: 26. https://doi.org/10.3390/horticulturae12010026
APA StyleZhao, Y., Sassa, H., Li, M., Miao, Y., Zhu, X., Hao, P., Wan, R., Zhang, K., Cong, L., Shen, Y., Liu, Y., Wang, M., Shi, J., Song, S., Bai, T., Jiao, J., Yuan, Z., & Zheng, X. (2026). Deciphering the Molecular Mechanisms That Control Ovule Development in Pomegranate. Horticulturae, 12(1), 26. https://doi.org/10.3390/horticulturae12010026






