Ginseng Quality Identification Based on Multi-Scale Feature Extraction and Knowledge Distillation
Abstract
1. Introduction
- Replacing the original C3K2 module with the DCA module enhances the focus on key channels to capture detailed image features. It significantly enhances the ability to capture fine-grained texture and edge features, making it easier to extract small target features and thus improving detection accuracy.
- Replacing traditional standard convolutions with DWSConv for downsampling can significantly reduce the number of parameters while improving detection accuracy.
- We incorporate EMA’s attention mechanism to help the model focus on small target regions, mitigate the effects of irrelevant contextual variables, and improve detection accuracy by performing more stably, especially in complex background or occlusion scenes.
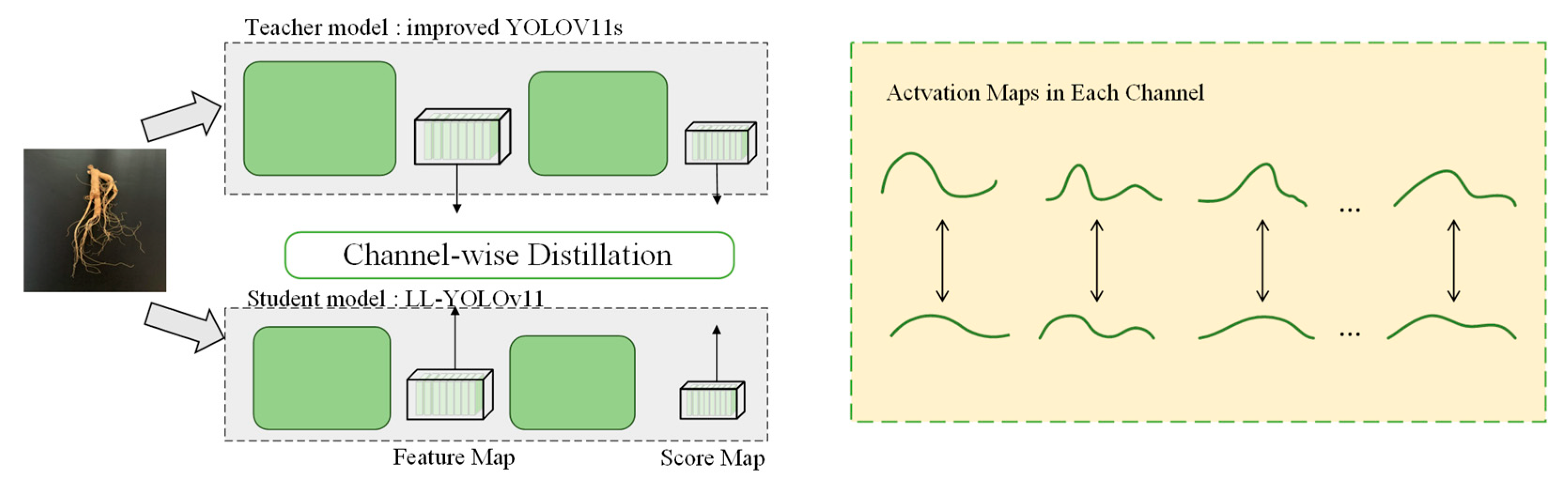
- Channel-wise knowledge distillation (CWD) techniques are used to enable the model to learn the properties in the teacher model, resulting in a lightweight model and achieving better detection performance.
2. Materials and Methods
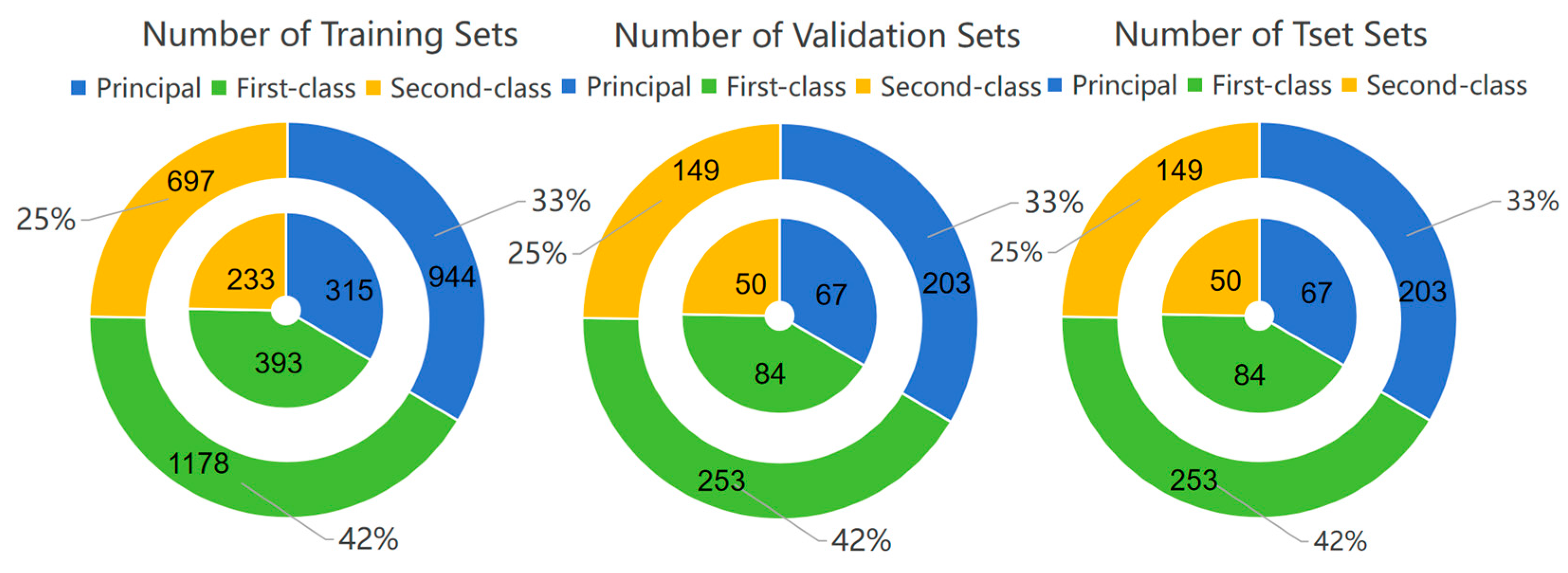
2.1. Data Set Construction
2.2. Data Augmentation
2.3. Novel Network Construction
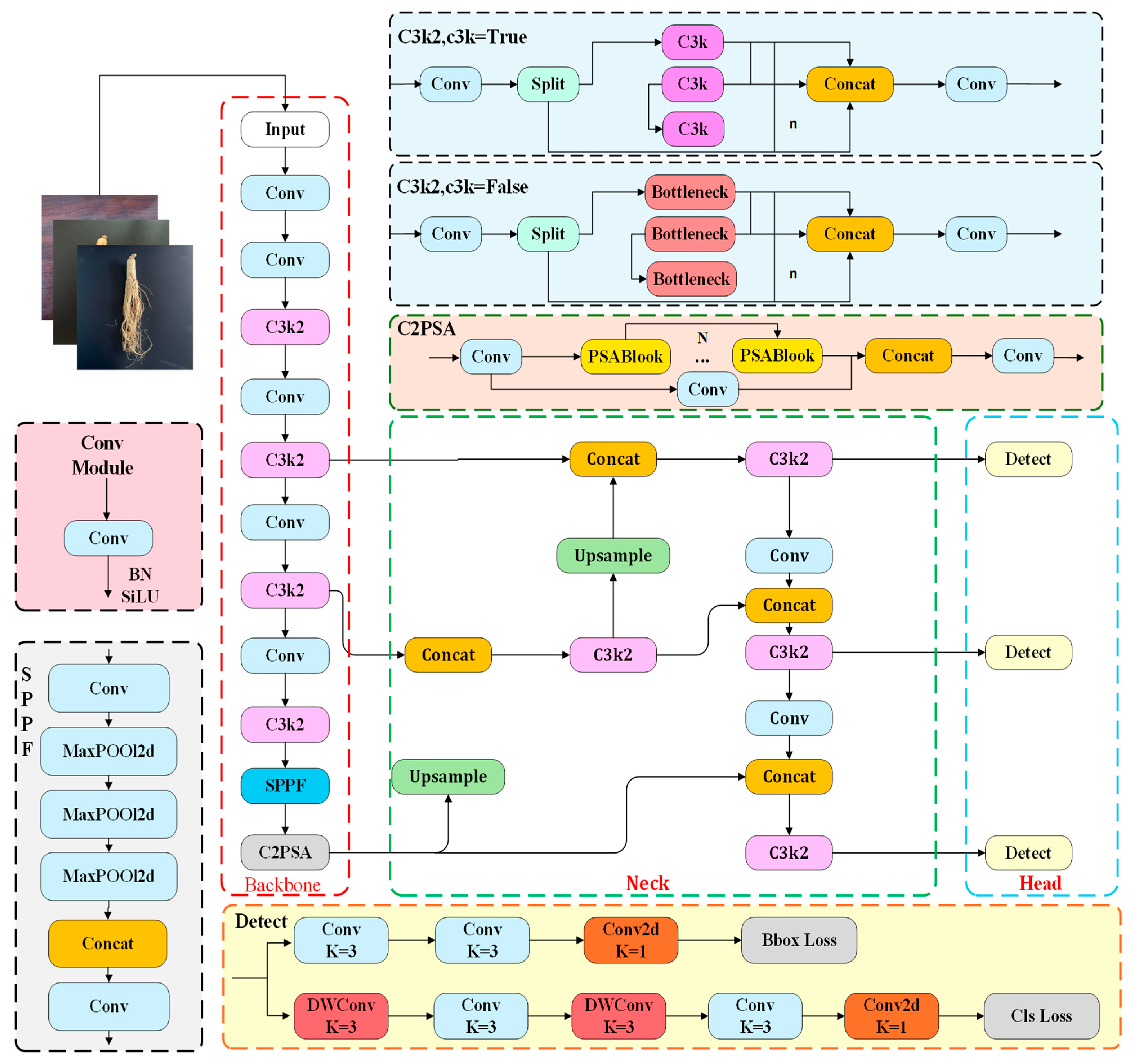
2.3.1. LLT-YOLOV11 Network Structure
- The DCA module is added as a module for feature extraction that is lightweight, integrated into the YOLOv11n network, replacing the existing c3k2 module for feature extraction. This significantly enhances the model’s training speed while more accurately capturing the shape and features of target objects.
- In the backbone network and neck network, deep separable convolutions (DWSConv) are used instead of conventional convolutions for downsampling, significantly reducing computational complexity and parameter counts.
- To address the complexity of target features and help the model focus on small target regions, an efficient multi-scale attention mechanism (EMA) is introduced.
- Knowledge distillation techniques are applied to increase the accuracy of the model.
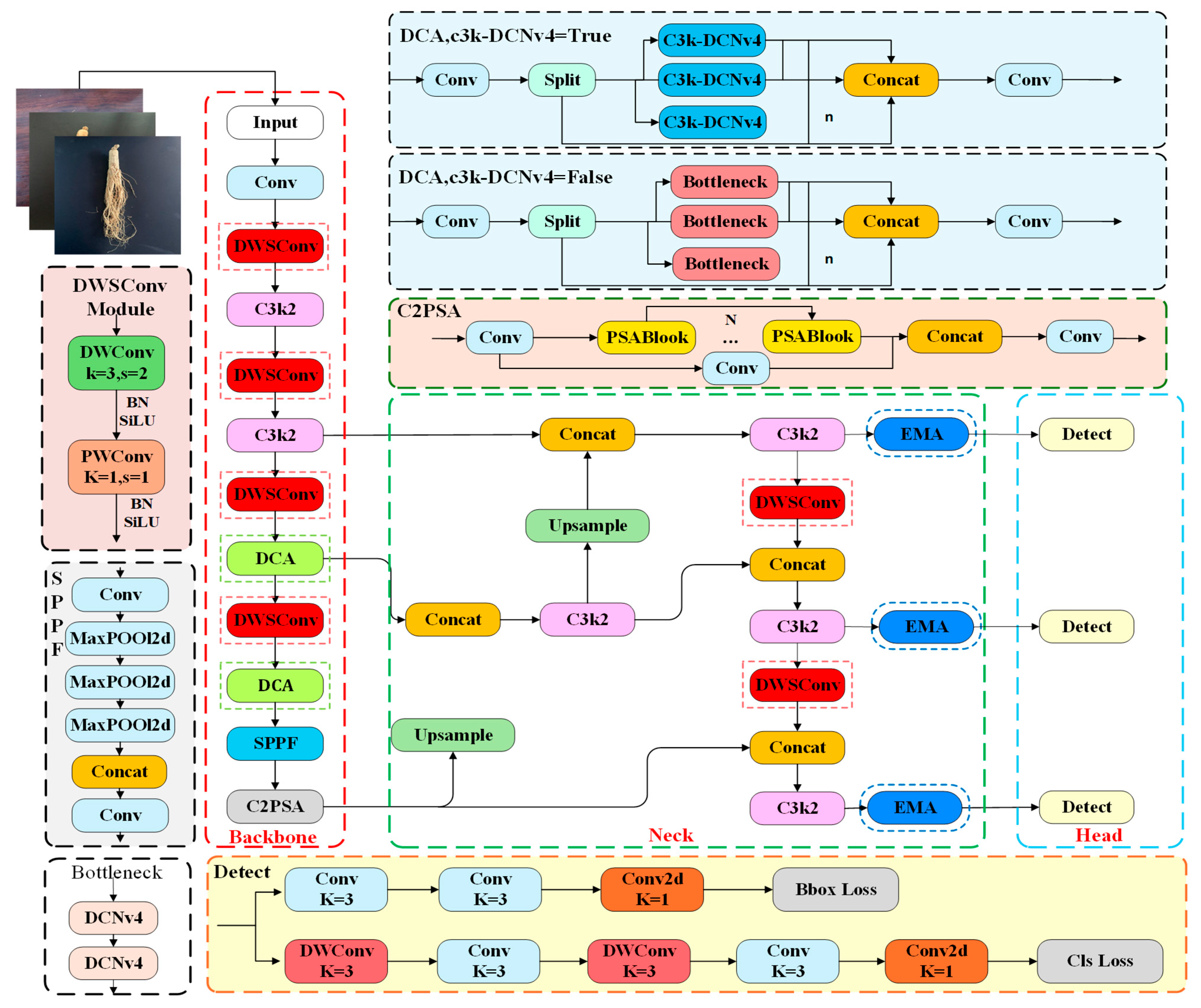
2.3.2. DCA Module
2.3.3. Depth-Wise Separable Convolution
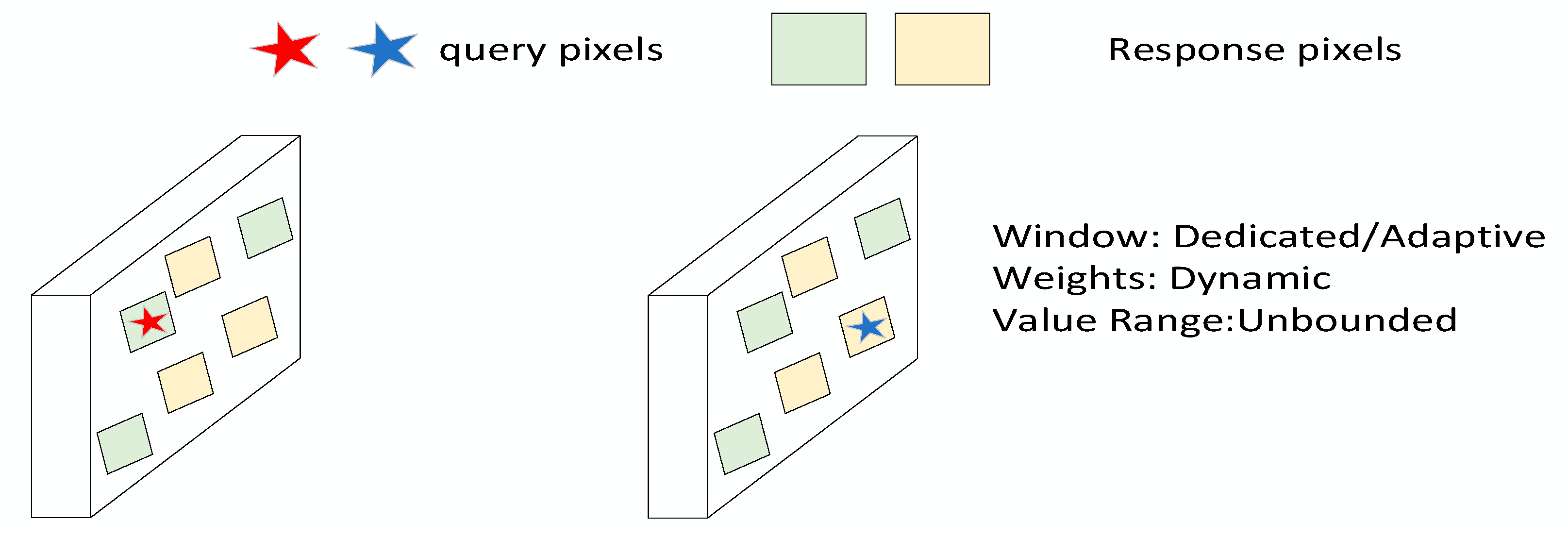
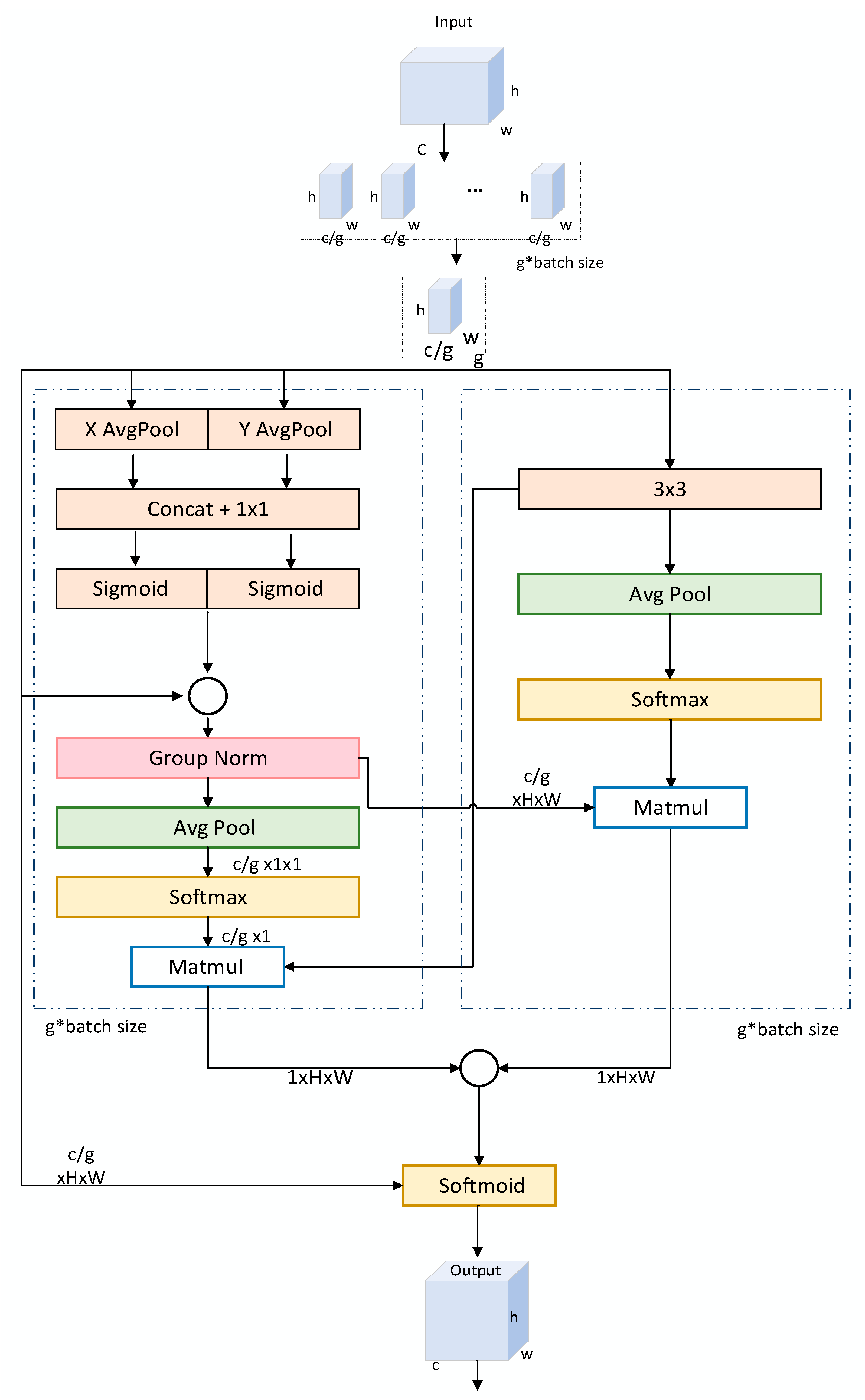
2.3.4. Efficient Multi-Scale Attention
2.3.5. Knowledge Distillation
3. Result and Analysis
3.1. Experimental Environment
3.2. Evaluation Criteria
3.3. Experiments and Analysis of Results
3.4. Before and After Data Enhancement Comparison
3.5. Ablation Experiment
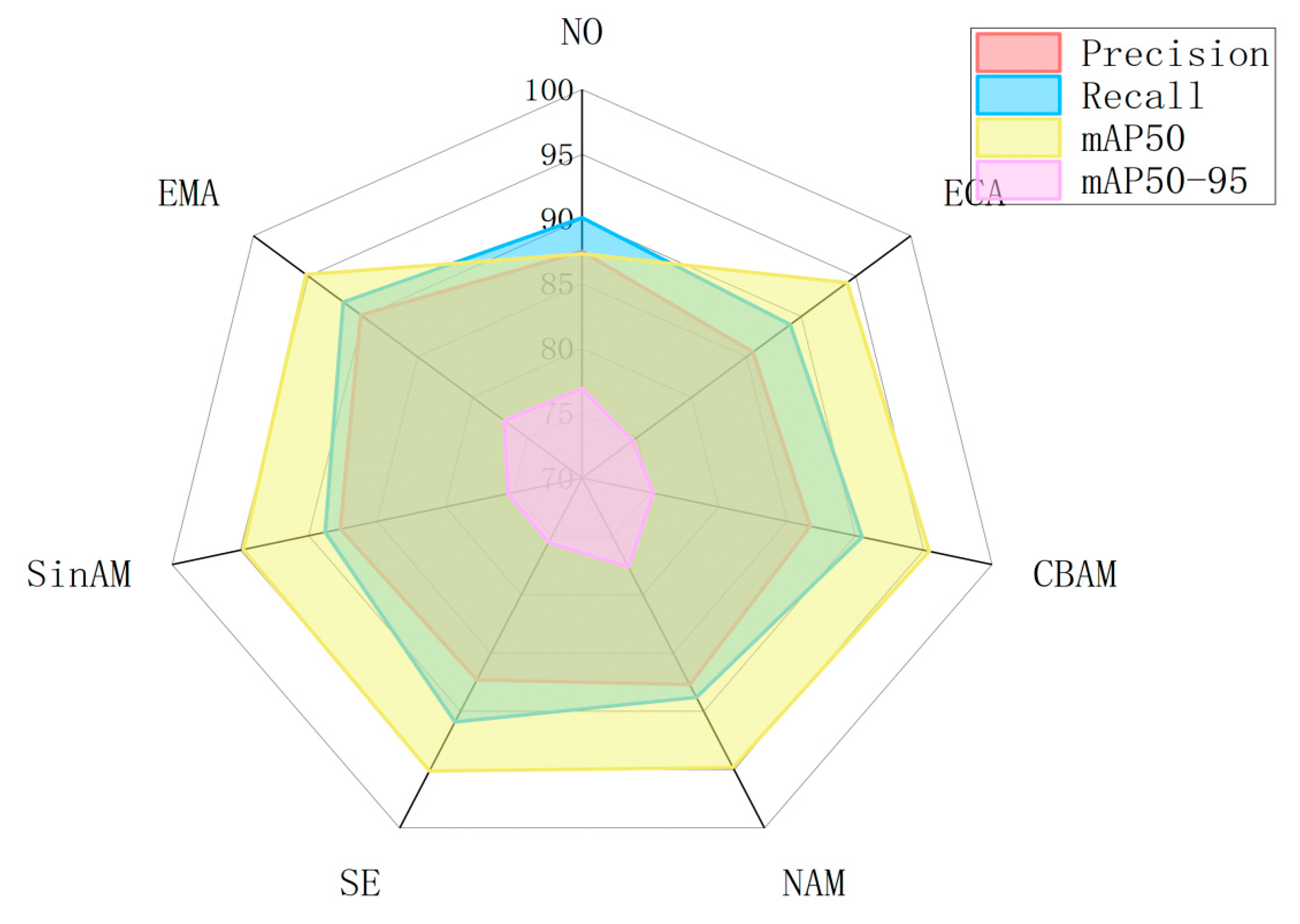
3.6. The Attention Mechanism on Model Performance
3.7. Experiments on Knowledge Distillation
3.7.1. Selection of Teacher Models
3.7.2. Analysis of Knowledge Distillation Scheme Selection
3.8. Comparison Experiment
3.9. Interference Robustness Test
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Coon, J.T.; Ernst, E. Panax ginseng. Drug Saf. 2002, 25, 323–344. [Google Scholar] [CrossRef] [PubMed]
- Irfan, M.; Kwak, Y.-S.; Han, C.-K.; Hyun, S.H.; Rhee, M.H. Adaptogenic effects of Panax ginseng on modulation of cardiovascular functions. J. Ginseng Res. 2020, 44, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Kitts, D.D.; Hu, C. Efficacy and safety of Ginseng. Public Health Nutr. 2000, 3, 473–485. [Google Scholar] [CrossRef]
- Liu, S.; Ai, Z.; Hu, Y.; Ren, G.; Zhang, J.; Tang, P.; Zou, H.; Li, X.; Wang, Y.; Nan, B.; et al. Ginseng glucosyl oleanolate inhibits cervical cancer cell proliferation and angiogenesis via PI3K/AKT/HIF-1α pathway. npj Sci. Food 2024, 8, 105. [Google Scholar] [CrossRef]
- Lee, K.-Y.; Shim, S.-L.; Jang, E.-S.; Choi, S.-G. Ginsenoside stability and antioxidant activity of Korean red ginseng (Panax ginseng CA Meyer) extract as affected by temperature and time. LWT 2024, 200, 116205. [Google Scholar] [CrossRef]
- Mancuso, C.; Santangelo, R. Panax ginseng and Panax quinquefolius: From pharmacology to toxicology. Food Chem. Toxicol. 2017, 107, 362–372. [Google Scholar] [CrossRef]
- Zhou, Z.; Li, M.; Zhang, Z.; Song, Z.; Xu, J.; Zhang, M.; Gong, M. Overview of Panax ginseng and its active ingredients’ protective mechanism on cardiovascular diseases. J. Ethnopharmacol. 2024, 334, 118506. [Google Scholar] [CrossRef]
- Fang, J.; Xu, Z.-F.; Zhang, T.; Chen, C.-B.; Liu, C.-S.; Liu, R.; Chen, Y.-Q. Effects of soil microbial ecology on ginsenoside accumulation in Panax ginseng across different cultivation years. Ind. Crops Prod. 2024, 215, 118637. [Google Scholar] [CrossRef]
- Ye, X.-W.; Li, C.-S.; Zhang, H.-X.; Li, Q.; Cheng, S.-Q.; Wen, J.; Wang, X.; Ren, H.-M.; Xia, L.-J.; Wang, X.-X.; et al. Saponins of ginseng products: A review of their transformation in processing. Front. Pharmacol. 2023, 14, 1177819. [Google Scholar] [CrossRef]
- Dong, S.; Wang, P.; Abbas, K. A survey on deep learning and its applications. Comput. Sci. Rev. 2021, 40, 100379. [Google Scholar] [CrossRef]
- Chen, H.; Engkvist, O.; Wang, Y.; Olivecrona, M.; Blaschke, T. The rise of deep learning in drug discovery. Drug Discov. Today 2018, 23, 1241–1250. [Google Scholar] [CrossRef]
- Li, D.; Yang, C.; Yao, R.; Ma, L. Origin identification of Saposhnikovia divaricata by CNN Embedded with the hierarchical residual connection block. Agronomy 2023, 13, 1199. [Google Scholar] [CrossRef]
- Li, D.; Piao, X.; Lei, Y.; Li, W.; Zhang, L.; Ma, L. A Grading Method of Ginseng (Panax ginseng C. A. Meyer) Appearance Quality Based on an Improved ResNet50 Model. Agronomy 2022, 12, 2925. [Google Scholar] [CrossRef]
- Li, D.; Zhai, M.; Piao, X.; Li, W.; Zhang, L. A Ginseng Appearance Quality Grading Method Based on an Improved ConvNeXt Model. Agronomy 2023, 13, 1770. [Google Scholar] [CrossRef]
- Yang, H.; Yang, L.; Wu, T.; Yuan, Y.; Li, J.; Li, P. MFD-YOLO: A fast and lightweight model for strawberry growth state detection. Comput. Electron. Agric. 2025, 234, 110177. [Google Scholar] [CrossRef]
- Li, H.; Gu, Z.; He, D.; Wang, X.; Huang, J.; Mo, Y.; Li, P.; Huang, Z.; Wu, F. A lightweight improved YOLOv5s model and its deployment for detecting pitaya fruits in daytime and nighttime light-supplement environments. Comput. Electron. Agric. 2024, 220, 108914. [Google Scholar] [CrossRef]
- Gai, R.; Chen, N.; Yuan, H. A detection algorithm for cherry fruits based on the improved YOLO-v4 model. Neural Comput. Appl. 2023, 35, 13895–13906. [Google Scholar] [CrossRef]
- Ren, R.; Zhang, S.; Sun, H.; Wang, N.; Yang, S.; Zhao, H.; Xin, M. YOLO-RCS: A method for detecting phenological period of ’Yuluxiang’ pear in an unstructured environment. Comput. Electron. Agric. 2025, 229, 109819. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, J.; Zhang, F.; Gao, J. LACTA: A lightweight and accurate algorithm for cherry tomato detection in unstructured environments. Expert Syst. Appl. 2024, 238, 122073. [Google Scholar] [CrossRef]
- Jrondi, Z.; Moussaid, A.; Hadi, M.Y. Exploring End-to-End object detection with transformers versus YOLOv8 for enhanced citrus fruit detection within trees. Syst. Soft Comput. 2024, 6, 200103. [Google Scholar] [CrossRef]
- Wang, J.; Liu, M.; Du, Y.; Zhao, M.; Jia, H.; Guo, Z.; Su, Y.; Lu, D.; Liu, Y. PG-YOLO: An efficient detection algorithm for pomegranate before fruit thinning. Eng. Appl. Artif. Intell. 2024, 134, 108700. [Google Scholar] [CrossRef]
- Jin, S.; Zhou, L.; Zhou, H. CO-YOLO: A lightweight and efficient model for Camellia oleifera fruit object detection and posture determination. Comput. Electron. Agric. 2025, 235, 110394. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, X.; Zhang, K.; Zhang, R.; Wang, Y. Research on the current situation of ginseng industry and development counter-measures in Jilin Province. J. Jilin Agric. Univ. 2023, 45, 649–655. [Google Scholar]
- Jiang, M.; Liang, Y.; Pei, Z.; Wang, X.; Zhou, F.; Wei, C.; Feng, X. Diagnosis of breast hyperplasia and evaluation of RuXian-I based on metabolomics deep belief networks. Int. J. Mol. Sci. 2019, 20, 2620. [Google Scholar] [CrossRef]
- Zhou, F.; Jin, L.; Dong, J. A Survey on Convolutional Neural Networks. Chin. J. Comput. 2017, 40, 1229–1251. [Google Scholar]
- Zheng, Y.; Li, G.; Li, Y. A Survey on the Application of Deep Learning in Image Recognition. Comput. Eng. Appl. 2019, 55, 20–36. [Google Scholar]
- Terven, J.; Córdova-Esparza, D.-M.; Romero-González, J.-A. A comprehensive review of yolo architectures in computer vision: From YOLOv1 to YOLOv8 and YOLO-NAS. Mach. Learn. Knowl. Extr. 2023, 5, 1680–1716. [Google Scholar] [CrossRef]
- Khanam, R.; Hussain, M. Yolov11: An overview of the key architectural enhancements. arXiv 2024, arXiv:2410.17725. [Google Scholar] [CrossRef]
- Shao, Y.; Zhang, D.; Chu, H.; Zhang, X.; Rao, Y. A Survey on YOLO Object Detection Based on Deep Learning. J. Electron. Inf. Technol. 2022, 44, 3697–3708. [Google Scholar]
- Xiong, Y.; Li, Z.; Chen, Y.; Wang, F.; Zhu, X.; Luo, J.; Wang, W.; Lu, T.; Li, H.; Qiao, Y.; et al. Efficient deformable convnets: Rethinking dynamic and sparse operators for vision applications. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–22 June 2024; pp. 5652–5661. [Google Scholar]
- Chollet, F. Xception: Deep Learning with Depthwise Separable Convolutions. In Proceedings of the 30th IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1251–1258. [Google Scholar]
- Ouyang, D.; He, S.; Zhang, G.; Luo, M.; Guo, H.; Zhan, J.; Huang, Z. Efficient multi-scale attention module with cross-spatial learning. In Proceedings of the ICASSP 2023—2023 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Rhodes Island, Greece, 4–10 June 2023; IEEE: New York, NY, USA, 2023; pp. 1–5. [Google Scholar]
- Shu, C.; Liu, Y.; Gao, J.; Yan, Z.; Shen, C. Channel-wise knowledge distillation for dense prediction. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 5311–5320. [Google Scholar]
- Selvaraju, R.R.; Cogswell, M.; Das, A.; Vedantam, R.; Parikh, D.; Batra, D. Grad-cam: Visual explanations from deep networks via gradient-based localization. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 618–626. [Google Scholar]
- Woo, S.; Park, J.; Lee, J.-Y.; Kweon, I.S. Cbam: Convolutional block attention module. In Proceedings of the European Conference on Computer Vision (ECCV), Munich, Germany, 8–14 September 2018; pp. 3–19. [Google Scholar]
- Hou, Q.; Zhou, D.; Feng, J. Coordinate attention for efficient mobile network design. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2021; pp. 13713–13722. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-and-excitation networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Liu, Y.; Shao, Z.; Hoffmann, N. Global attention mechanism: Retain information to enhance channel-spatial interactions. arXiv 2021, arXiv:2112.05561. [Google Scholar] [CrossRef]
- Liu, Y.; Shao, Z.; Teng, Y.; Hoffmann, N. NAM: Normalization-based attention module. arXiv 2021, arXiv:2111.12419. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, R.Y.; Li, L.; Xie, L. Simam: A simple, parameter-free attention module for convolutional neural networks. In Proceedings of the International Conference on Machine Learning, Online, 18–24 July 2021; Volume 139, pp. 11863–11874. [Google Scholar]
- Yang, Z.; Li, Z.; Shao, M.; Shi, D.; Yuan, Z.; Yuan, C. Masked generative distillation. In Proceedings of the European Conference on Computer Vision, Tel Aviv, Israel, 23–27 October 2022; Springer Nature: Cham, Switzerland, 2022; pp. 53–69. [Google Scholar]
- Kim, J.; Park, S.U.; Kwak, N. Paraphrasing complex network: Network compression via factor transfer. arXiv 2018, arXiv:1802.04977. [Google Scholar]
- Romero, A.; Ballas, N.; Kahou, S.E.; Chassang, A.; Gatta, C.; Bengio, Y. Fitnets: Hints for thin deep nets. arXiv 2014, arXiv:1412.6550. [Google Scholar]




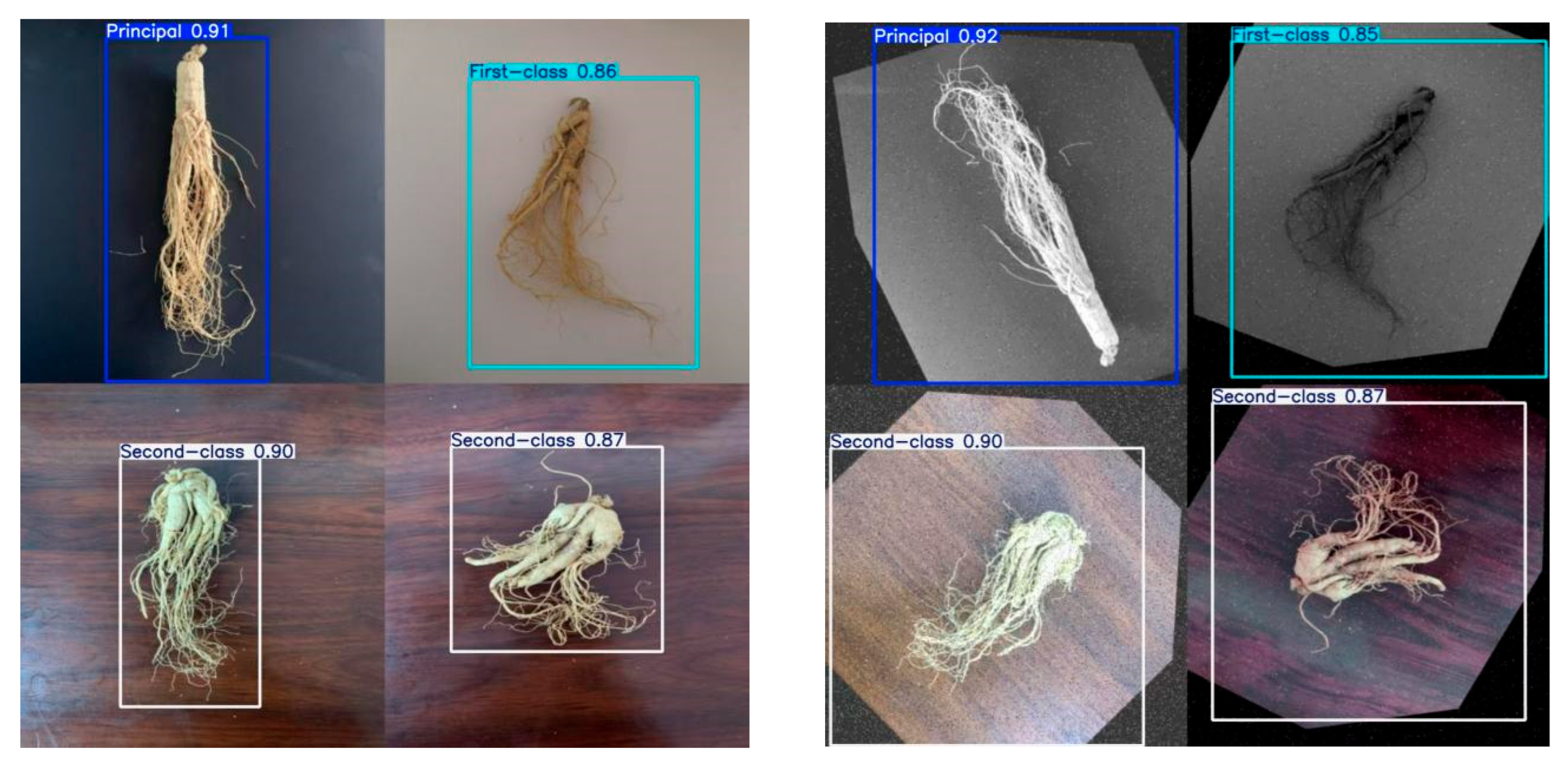
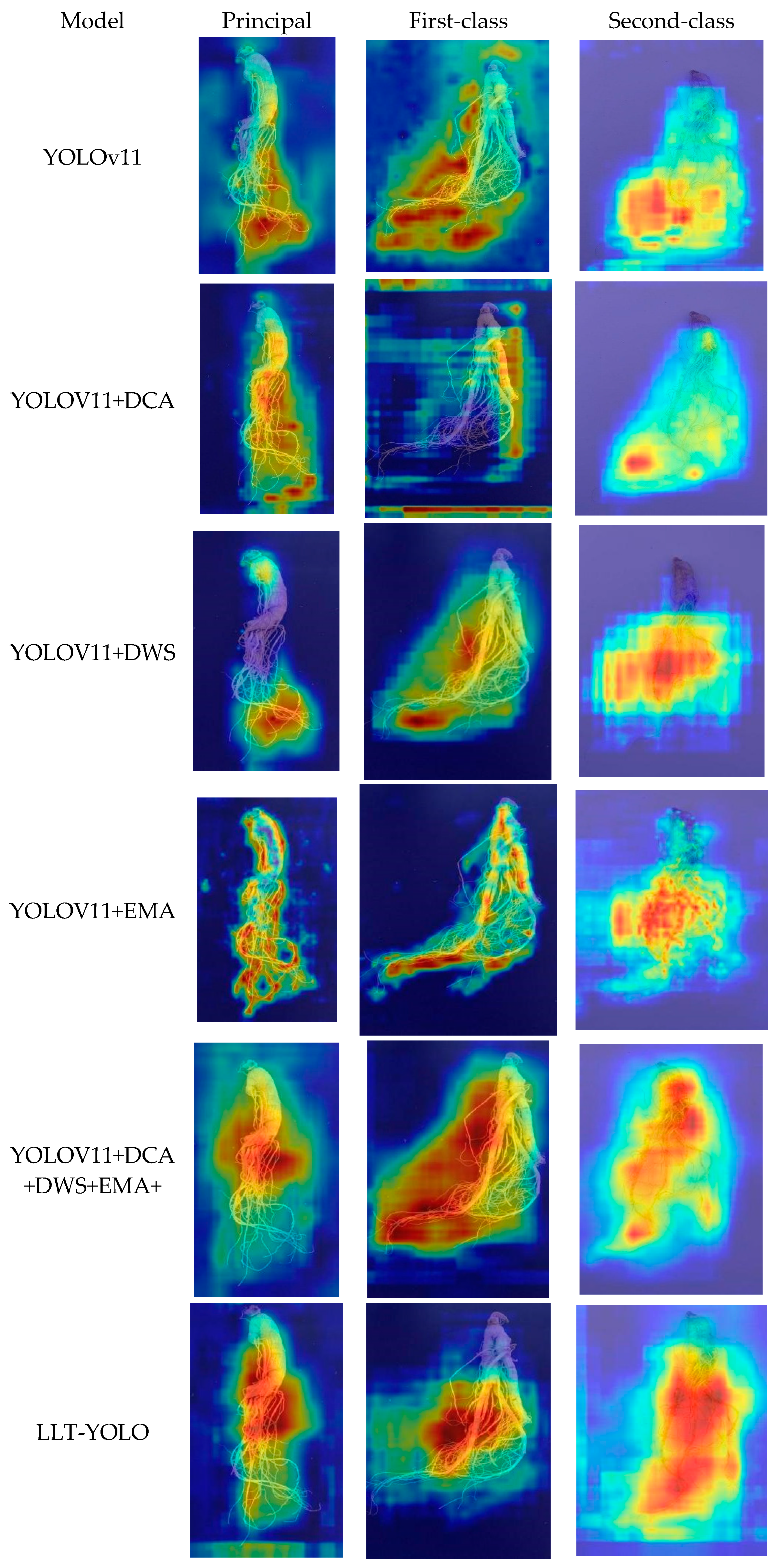


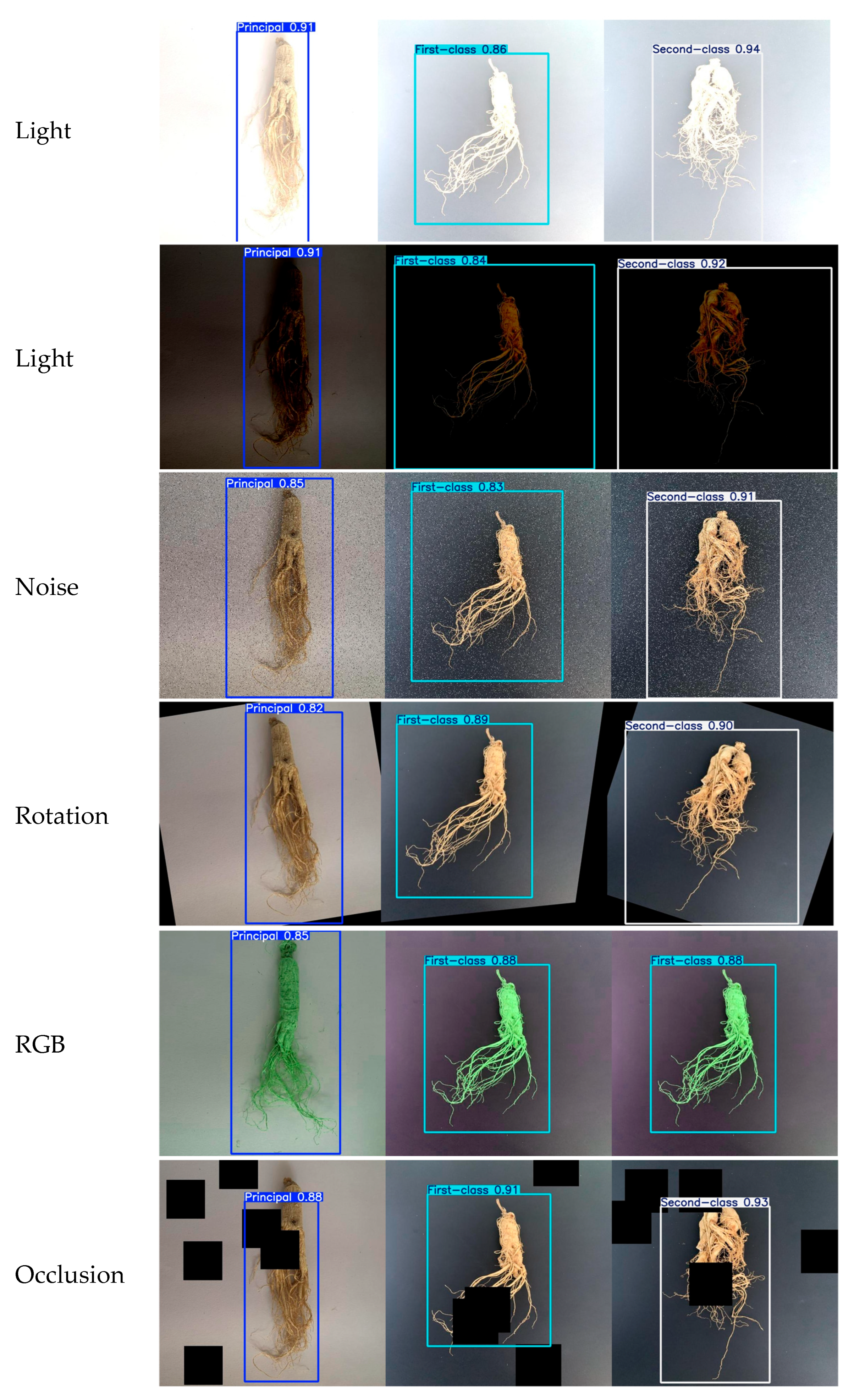
| Projects | Principal Ginseng | First-Class Ginseng | Second-Class Ginseng |
|---|---|---|---|
| Main Root | Like a cylindrical | ||
| Branch Root | There are two to three clear branching roots, and the thickness is more even | One to four branches, some rough and others smooth | |
| Rutabaga | With a reed head and ginseng fibrous roots | The reed head and ginseng’s fibrous roots are more complete | Rutabaga and ginseng with roots that are not fully fibrous |
| Groove | Grooves that are clear | Not a clear, distinct groove | Without grooves |
| Surface | Light yellow or pale yellow, no rust in water, and no draw lines | Grayish-yellow or yellowish-white, with light water rust, or with holes for pumps | yellowish-white or grayish-yellow, with a little more water rust and holes for pumps |
| Texture | Harder, powdery, non-hollow | ||
| Gross-section | Powdery and yellowish-white in color, with a resinous tract that may be seen | ||
| Diameter Length | ≥3.5 | 3.0–3.49 | 2.5–2.99 |
| Damage, Scars | No significant injury | Minor injury | More serious |
| Insects, Mildew, Impurities | None | Mild | Presence |
| Section | Section neat, clear | Segment is obvious | Segments are not obvious |
| Springtails | Square or rectangular | Made conical or cylindrical | Irregular shape |
| Weight | 500 g/root or more | 250–500 g/root | 100–250 g/root |
| Hyperparameters | Value |
|---|---|
| Image size | 640 × 640 |
| Epochs | 300 |
| Optimizer | SGD |
| Batch Size | 32 |
| Initial Learning Rate | 0.01 |
| Final Learning Rate | 0.01 |
| Close Mosaic | Last ten epochs |
| Workers | 8 |
| Weight-decay | 0.0005 |
| Momentum | 0.937 |
| Level | Model | Precision (%) | Recall (%) | mAP (%) | mAP50–95 (%) |
|---|---|---|---|---|---|
| ALL | YOLOV11 | 87.5 | 90.1 | 87.3 | 76.9 |
| LLT-YOLOv11 | 90.5 | 92.3 | 95.1 | 77.4 | |
| Principal | YOLOV11 | 91.3 | 91.6 | 92.4 | 72.9 |
| LLT-YOLOv11 | 95.7 | 91.6 | 96.7 | 78.8 | |
| First-class | YOLOV11 | 82.2 | 83.0 | 87.7 | 67.0 |
| LLT-YOLOv11 | 84.8 | 93.7 | 94.2 | 75.2 | |
| Second-class | YOLOV11 | 76.6 | 81.1 | 85.7 | 72.6 |
| LLT-YOLOv11 | 90.9 | 86.9 | 94.4 | 78.3 |
| Augmentation Strategy | Precision (%) | Recall (%) | mAP50 (%) | mAP50–95 (%) |
|---|---|---|---|---|
| original | 87.1 | 86.4 | 91.89 | 71.77 |
| Data enhancement | 90.5 | 92.3 | 95.1 | 77.4 |
| Model | Precision (%) | Recall (%) | mAP50 (%) | mAP50–95 (%) | GFlops | FPS | Model Size (MB) | Parameters (M) |
|---|---|---|---|---|---|---|---|---|
| YOLOV11 | 87.5 | 90.1 | 87.3 | 76.9 | 6.3 | 111.90 | 5.24 | 2.58 |
| A | 79.4 | 89.7 | 92.4 | 72.2 | 6.6 | 104.11 | 5.26 | 2.24 |
| B | 84.4 | 87.2 | 93.8 | 72.9 | 5.3 | 608.27 | 4.10 | 2.15 |
| C | 90.2 | 91.8 | 95.2 | 77.1 | 6.5 | 477.07 | 5.19 | 2.59 |
| D | 85.7 | 93.4 | 96.3 | 77.7 | 6.7 | 106.18 | 4.15 | 2.32 |
| E | 89.1 | 90.3 | 96.1 | 77.6 | 6.9 | 85.65 | 5.23 | 2.47 |
| F | 87.2 | 90.8 | 95.7 | 75.6 | 6.7 | 88.93 | 4.07 | 2.36 |
| G | 88.1 | 92.7 | 96.1 | 79.0 | 7.0 | 87.00 | 4.12 | 2.00 |
| LLT-YOLOv11 | 90.5 | 92.3 | 95.1 | 77.4 | 7.2 | 114.45 | 4.12 | 2.00 |
| Attention Mechanisms | Precision (%) | Recall (%) | mAP50 (%) | mAP50–95 (%) | FPS | Gflops | Weight |
|---|---|---|---|---|---|---|---|
| NO | 87.5 | 90.1 | 87.3 | 76.9 | 111.90 | 6.3 | 5.24 |
| ECA | 85.6 | 89.0 | 94.2 | 74.6 | 146.32 | 6.3 | 5.35 |
| CBAM | 86.7 | 90.5 | 95.4 | 75.2 | 111.29 | 6.4 | 5.52 |
| NAM | 87.7 | 88.8 | 94.8 | 77.6 | 126.47 | 6.3 | 7.03 |
| SE | 87.3 | 90.9 | 95.1 | 75.5 | 105.38 | 6.5 | 5.36 |
| SimAM | 87.7 | 88.8 | 94.8 | 75.4 | 130.49 | 6.3 | 5.35 |
| EMA | 90.2 | 91.8 | 95.2 | 77.1 | 114.21 | 6.5 | 5.19 |
| Model | Precision/% | Recall/% | mAP50/% | mAP50–95/% | Model Size/MB |
|---|---|---|---|---|---|
| YOLOv11m | 93.8 | 95.3 | 95.0 | 83.9 | 38.6 |
| YOLOv11l | 93.1 | 96.0 | 95.1 | 85.4 | 48.8 |
| YOLOv11x | 92.4 | 94.3 | 95.3 | 82.5 | 71.5 |
| YOLOv11s | 93.9 | 95.7 | 95.7 | 85.7 | 14.0 |
| Method | Precision/% | Recall/% | mAP50/% | mAP50–95/% | Model Size/MB |
|---|---|---|---|---|---|
| MGD | 88.1 | 90.7 | 91.5 | 77.0 | 4.22 |
| L1 | 91.2 | 90.3 | 91.7 | 77.6 | 4.63 |
| L2 | 89.7 | 90.8 | 92.4 | 78.2 | 4.47 |
| CWD | 90.5 | 92.3 | 95.1 | 77.4 | 4.12 |
| Model | Precision/% | Recall/% | mAP50/% | mAP50–95/% | Model Size/MB | Parameters/% | FPS |
|---|---|---|---|---|---|---|---|
| YOLOV5n | 86.5 | 90.6 | 91.2 | 71.8 | 4.43 | 2.18 | 163.38 |
| YOLOv7 | 71.63 | 75.91 | 81.77 | 51.6 | 74.7 | 37.21 | 66.31 |
| YOLOv7-tiny | 63.6 | 78.6 | 72 | 48.7 | 12.3 | 6.03 | 57.91 |
| YOLOv8n | 87.6 | 88.2 | 91.9 | 74.6 | 5.36 | 2.68 | 151.24 |
| YOLOv9s | 85.8 | 94.6 | 95.0 | 78.2 | 12.6 | 6.19 | 84.79 |
| YOLOv10n | 77.5 | 89.8 | 89.3 | 71.4 | 5.49 | 2.69 | 135.70 |
| YOLOv11n | 87.5 | 90.1 | 87.3 | 76.9 | 5.24 | 2.58 | 111.90 |
| YOLOv12n | 88.2 | 83.3 | 87.8 | 75.2 | 5.32 | 2.50 | 87.22 |
| LLT-YOLOv11 | 90.5 | 92.3 | 95.1 | 77.4 | 4.12 | 2.00 | 114.45 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Li, Y.; You, H.; Zhang, L. Ginseng Quality Identification Based on Multi-Scale Feature Extraction and Knowledge Distillation. Horticulturae 2025, 11, 1120. https://doi.org/10.3390/horticulturae11091120
Li J, Li Y, You H, Zhang L. Ginseng Quality Identification Based on Multi-Scale Feature Extraction and Knowledge Distillation. Horticulturae. 2025; 11(9):1120. https://doi.org/10.3390/horticulturae11091120
Chicago/Turabian StyleLi, Jian, Yuting Li, Haohai You, and Lijuan Zhang. 2025. "Ginseng Quality Identification Based on Multi-Scale Feature Extraction and Knowledge Distillation" Horticulturae 11, no. 9: 1120. https://doi.org/10.3390/horticulturae11091120
APA StyleLi, J., Li, Y., You, H., & Zhang, L. (2025). Ginseng Quality Identification Based on Multi-Scale Feature Extraction and Knowledge Distillation. Horticulturae, 11(9), 1120. https://doi.org/10.3390/horticulturae11091120








