Structural Variation and Evolutionary Dynamics of Orobanchaceae from the Perspective of the Mitochondrial Genomes Pedicularis kansuensis and Pedicularis chinensis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant DNA Extraction and Sequencing
2.2. Mitochondrial Genome Assembly and Annotation
2.3. Codon Usage Bias Analysis
2.4. Repeat Sequence Analysis
2.5. Structural Analysis of the Mitochondrial Genome
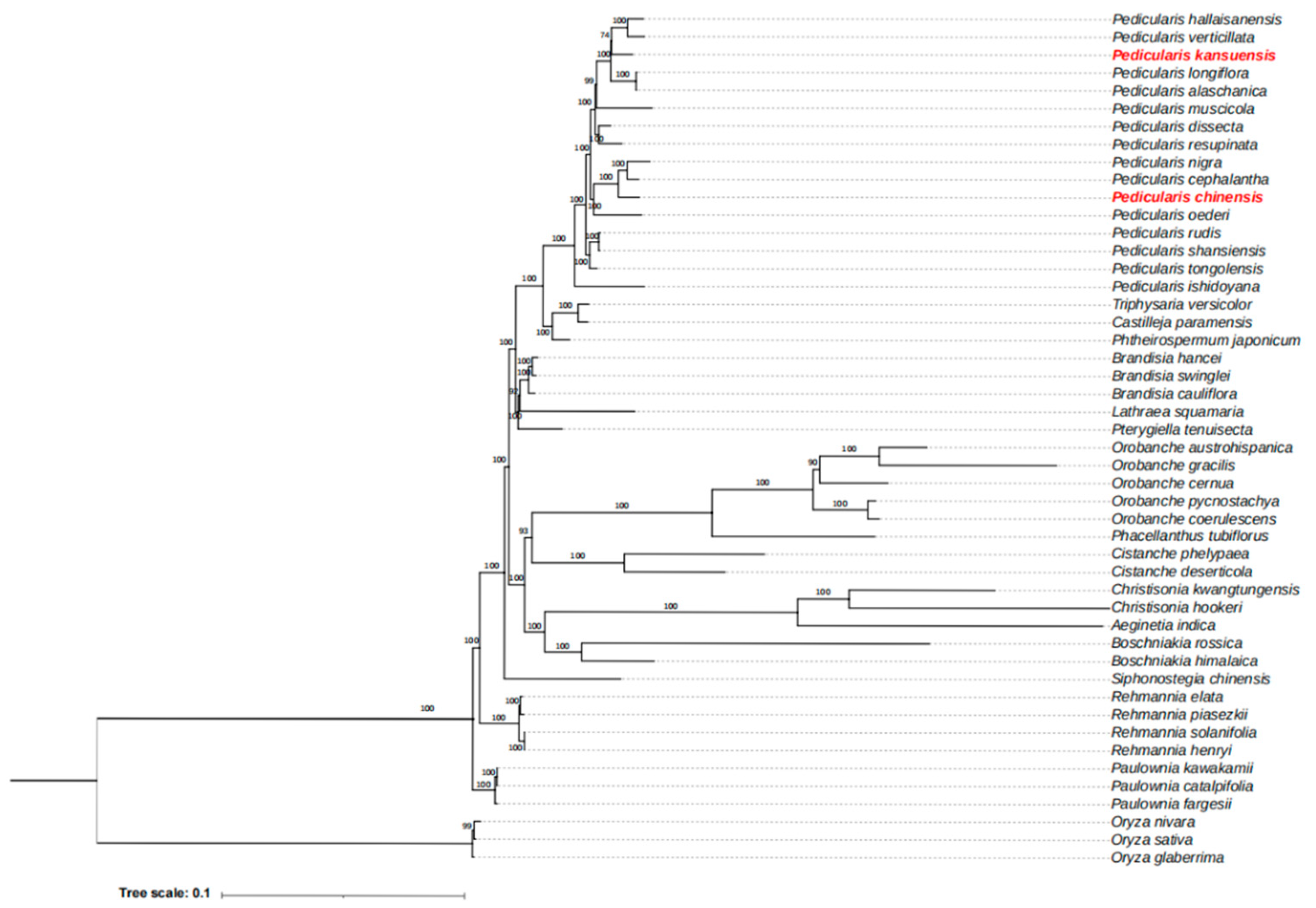
2.6. Phylogenetic Analysis
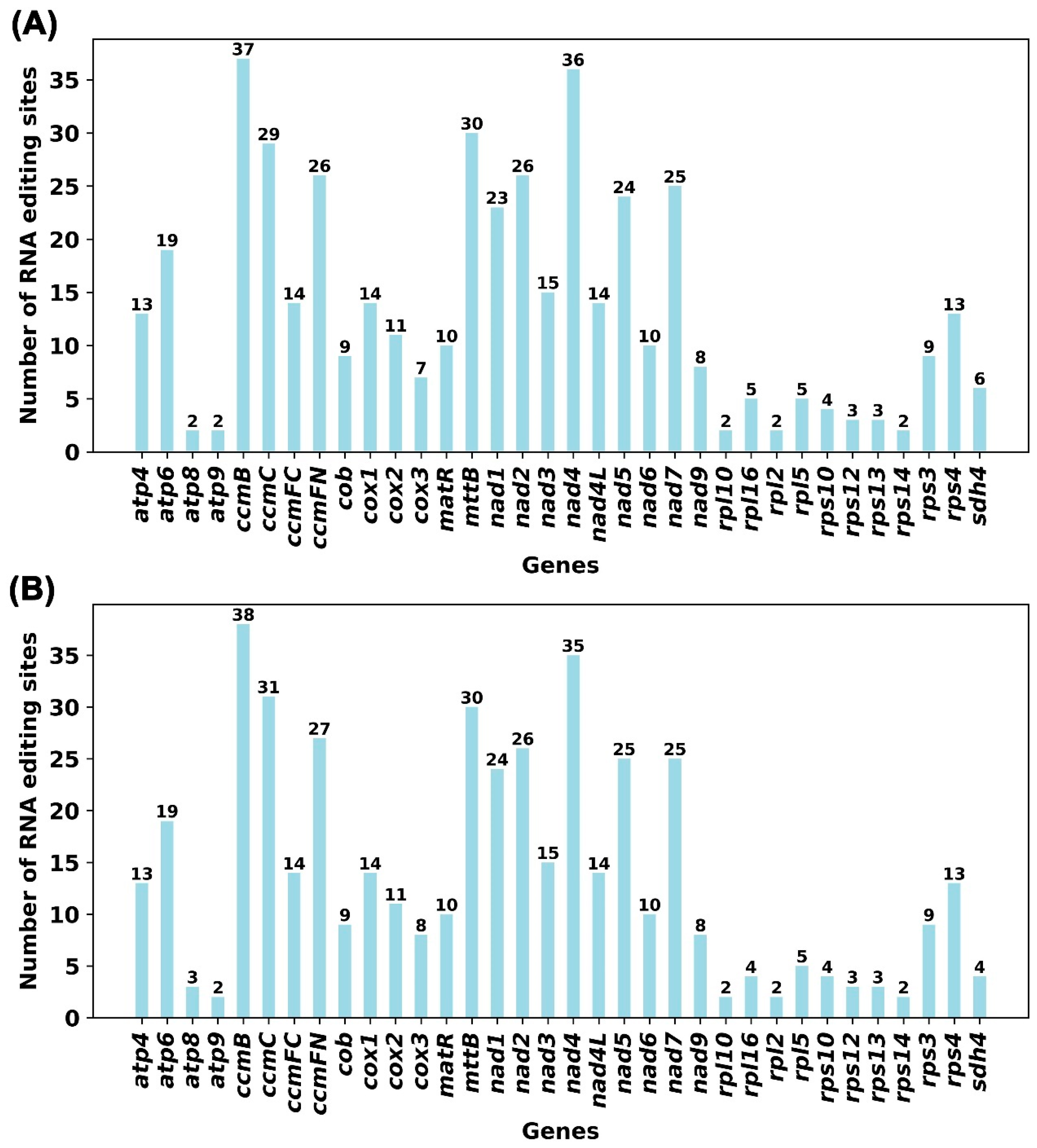
2.7. RNA Editing Event Analysis
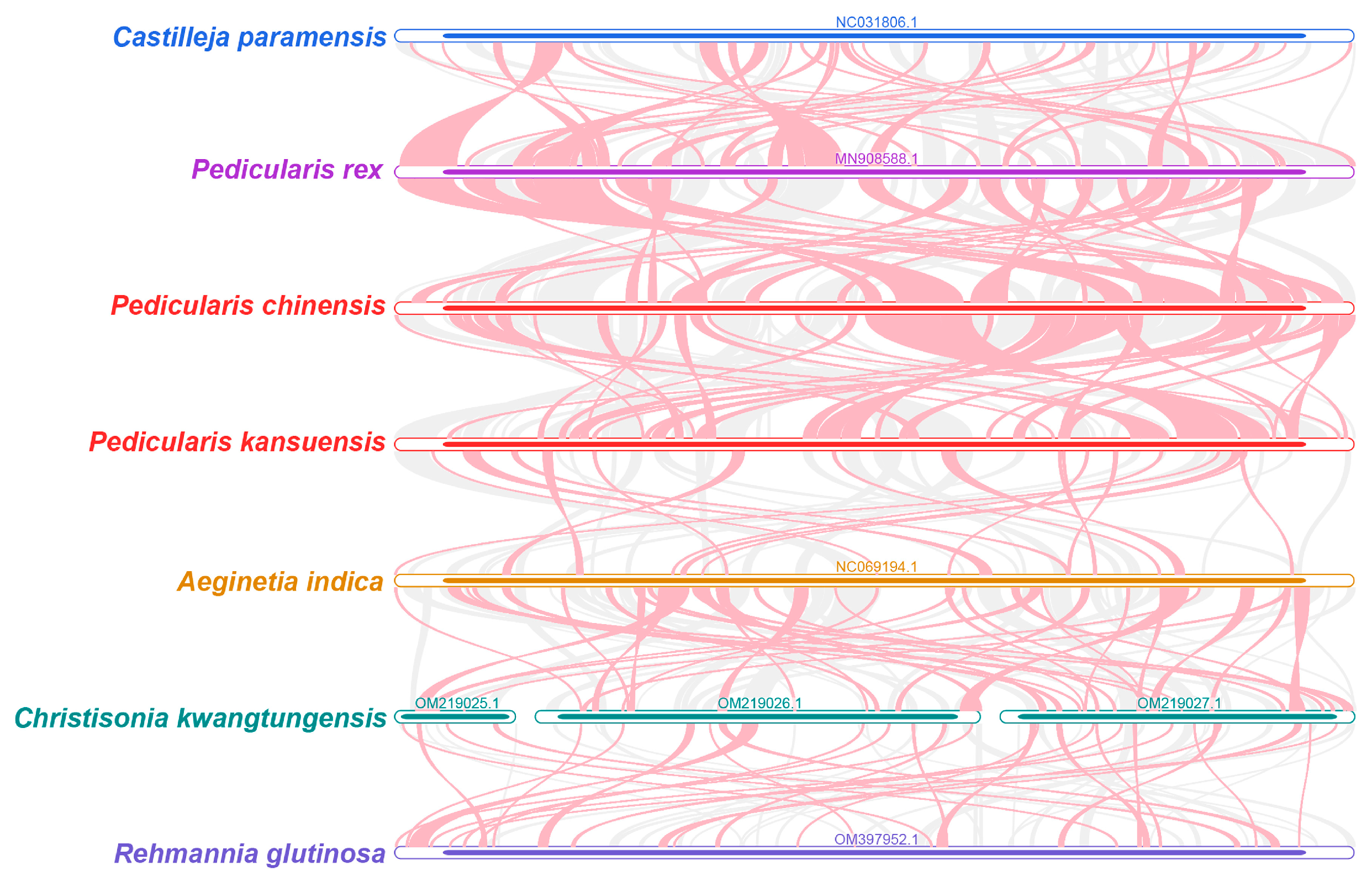
2.8. Collinearity Analysis
3. Results
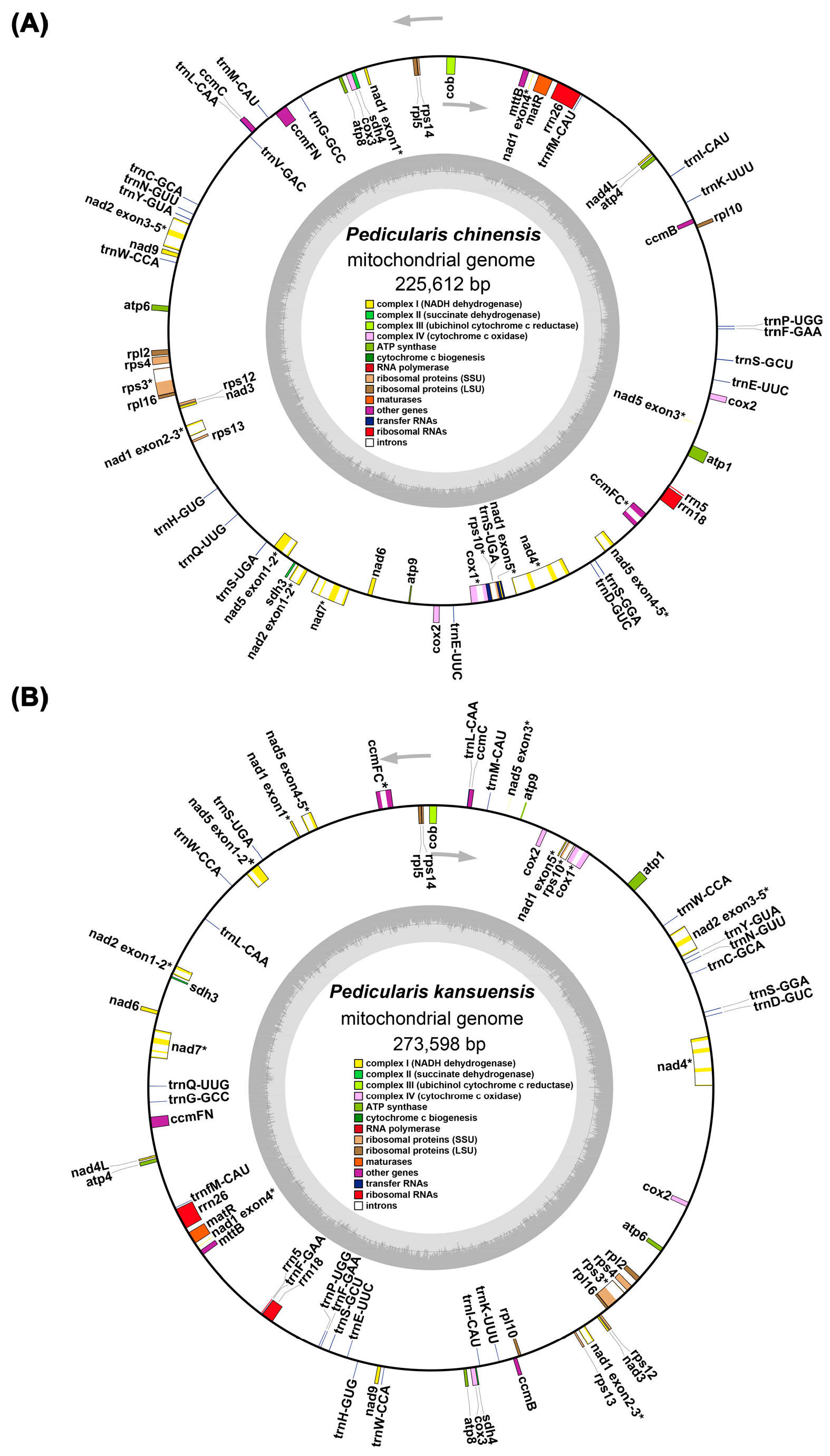
3.1. Structural Characteristics of the Mitochondrial Genomes of Pedicularis kansuensis and Pedicularis chinensis
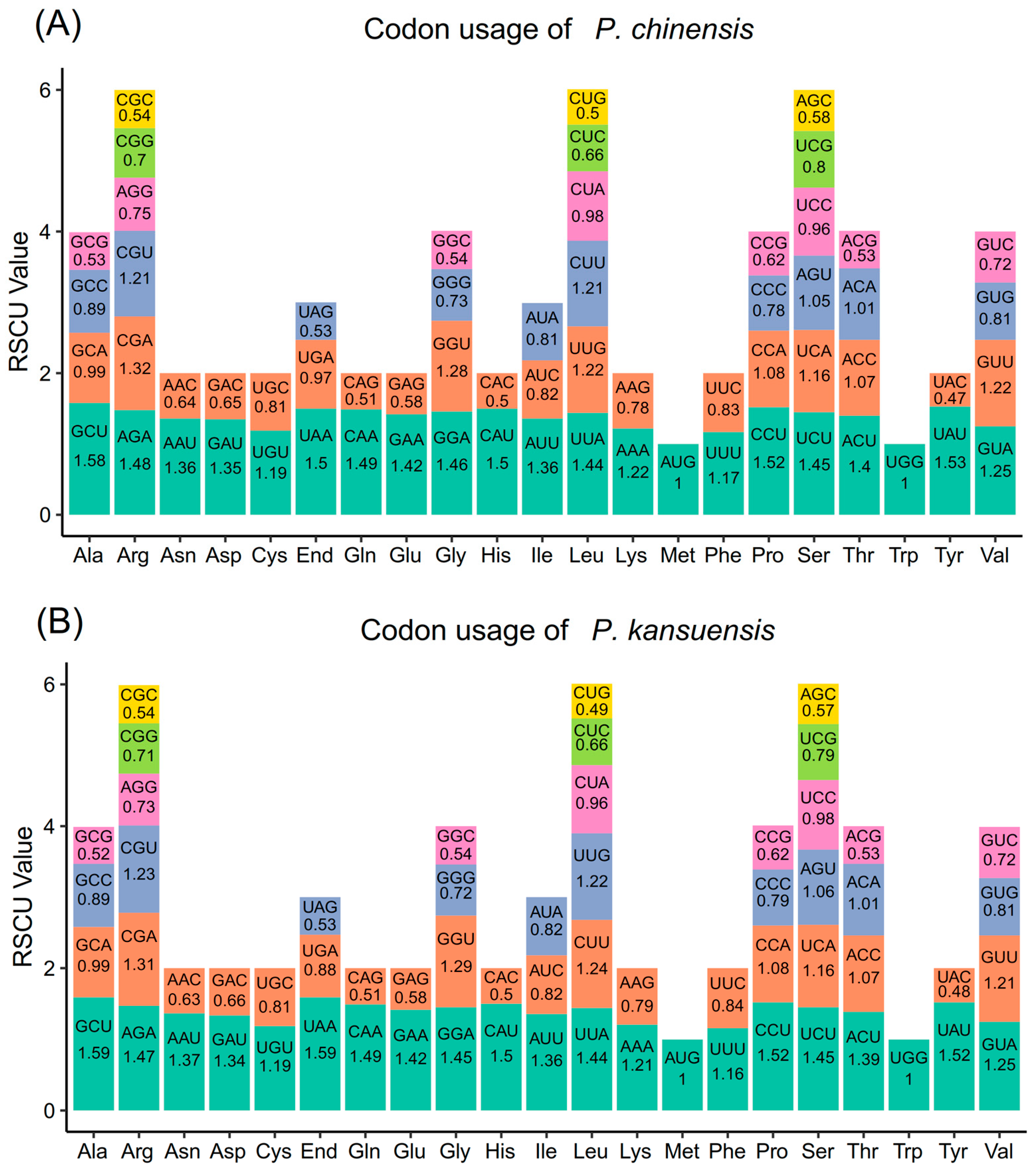
3.2. Codon Usage Bias Analysis of Mitochondrial Genes
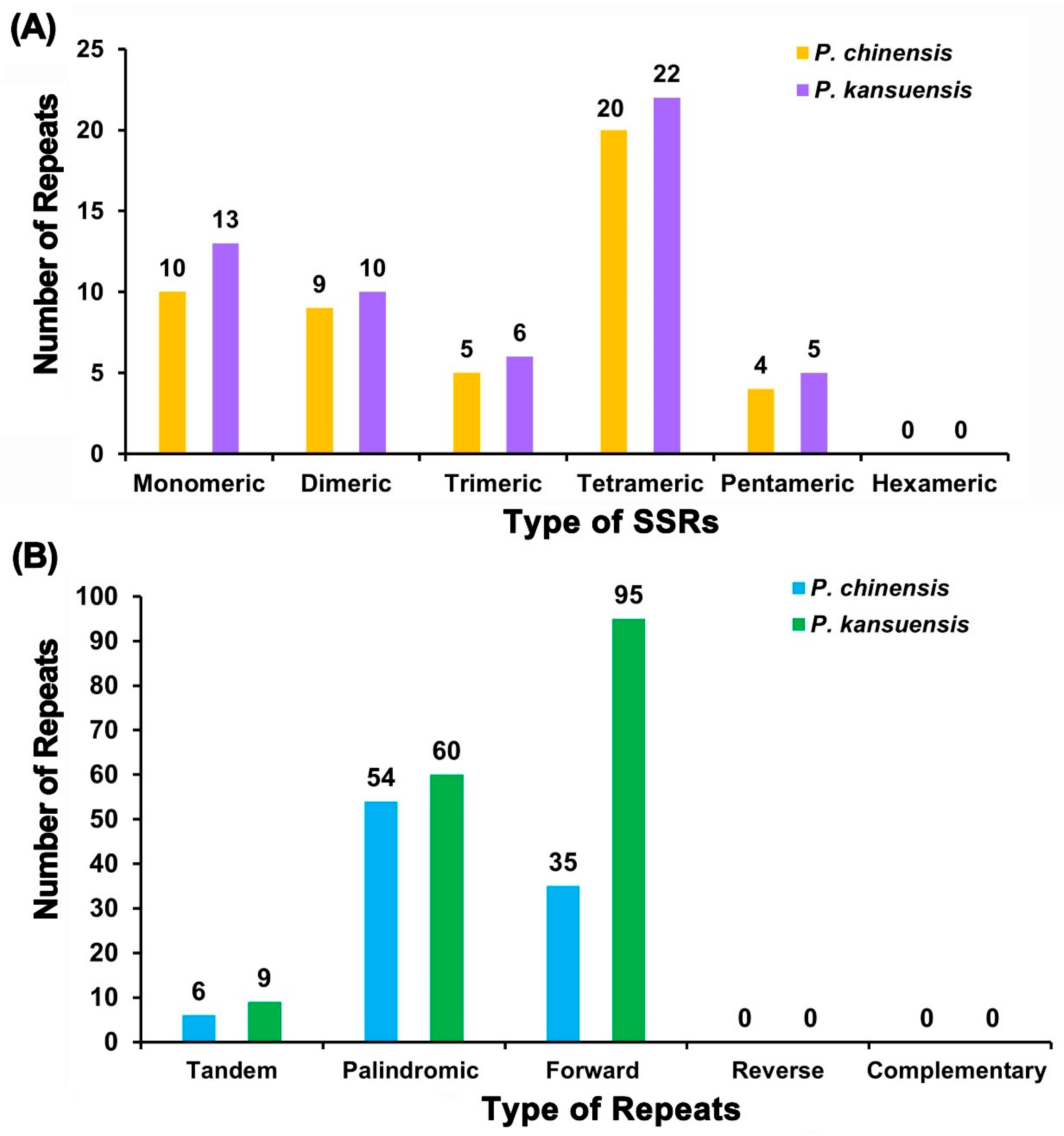
3.3. Divergent Repeat Sequence Profiles of the Mitochondrial Genomes in Pedicularis kansuensis and Pedicularis chinensis
3.4. Analysis of Mitochondrial Genomic Sequence Variations of Pedicularis kansuensis and Pedicularis chinensis
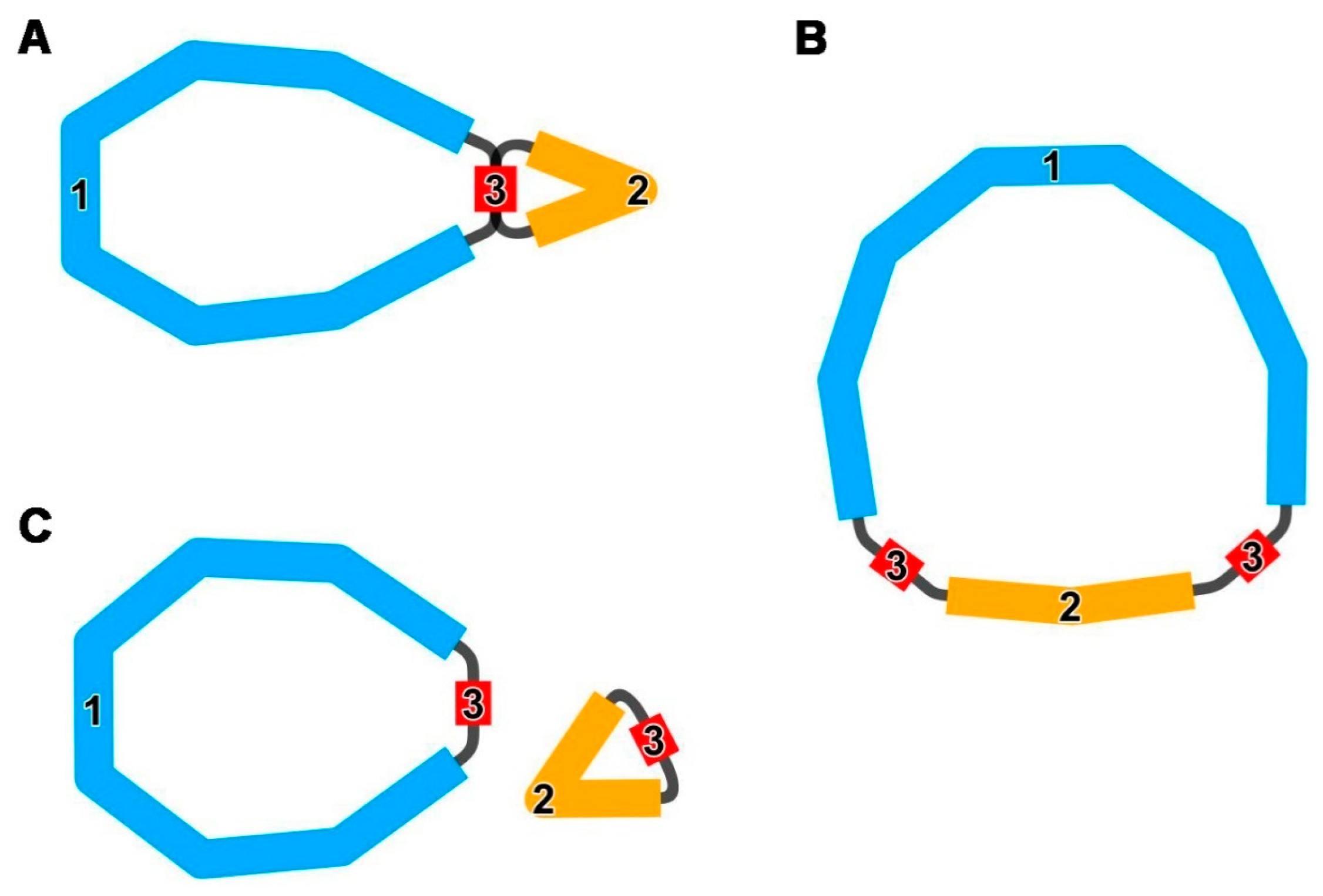
3.4.1. Synteny Analysis
3.4.2. RNA Editing Events in Mitochondrial Genes
3.5. Phylogenetic Analysis Based on Mitochondrial Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gray, M.W.; Burger, G.; Lang, B.F. Mitochondrial evolution. Science 1999, 283, 1476–1481. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kan, S.L.; Liao, X.Z.; Zhou, J.W.; Tembrock, L.R.; Daniell, H.; Jin, S.X.; Wu, Z.Q. Plant organellar genomes: Much done, much more to do. Trends Plant Sci. 2024, 29, 754–769. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Newton, K.J. Plant mitochondrial genomes: Dynamics and mechanisms of mutation. Annu. Rev. Plant Biol. 2017, 68, 225–252. [Google Scholar] [CrossRef]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 2015, 112, E3515–E3524. [Google Scholar] [CrossRef]
- Putintseva, Y.A.; Bondar, E.I.; Simonov, E.P.; Sharov, V.V.; Oreshkova, N.V.; Kuzmin, D.A.; Konstantinov, Y.M.; Shmakov, V.N.; Belkov, V.I.; Sadovsky, M.G.; et al. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genom. 2020, 21, 654. [Google Scholar] [CrossRef]
- Frezza, C.; Venditti, A.; Toniolo, C.; Vita, D.; Serafini, I.; Ciccòla, A.; Franceschin, M.; Ventrone, A.; Tomassini, L.; Foddai, S.; et al. Pedicularis L. Genus: Systematics, botany, phytochemistry, chemotaxonomy, ethnopharmacology, and other. Plants 2019, 8, 306. [Google Scholar] [CrossRef]
- Yatoo, M.I.; Dimri, U.; Gopalakrishnan, A.; Karthik, K.; Gopi, M.; Khandia, R.; Saminathan, M.; Saxena, A.; Alagawany, M.; Farag, M.R.; et al. Beneficial health applications and medicinal values of Pedicularis plants: A review. Biomed. Pharmacother. 2017, 95, 1301–1313. [Google Scholar] [CrossRef]
- Li, X.; Lin, C.Y.; Yang, J.B.; Yu, W.B. De novo assembling a complete mitochondrial genome of Pedicularis rex (Orobanchaceae) using GetOrganelle toolkit. Mitochondrial DNA Part B 2020, 5, 1056–1057. [Google Scholar] [CrossRef]
- Sui, X.L.; Guan, K.Y.; Chen, Y.; Xue, R.J.; Li, A.R. A legume host benefits more from Arbuscular mycorrhizal fungi than a grass host in the presence of a root hemiparasitic plant. Microorganisms 2022, 10, 440. [Google Scholar] [CrossRef]
- Wei, W.R.; Zhen, Q.Y.; Deng, J.; Yue, H.L.; Qin, M.S.; Oosthuizen, M.K. Grazing during the grassland greenup period promotes plant species richness in alpine grassland in winter pastures. Front. Plant Sci. 2022, 13, 973662. [Google Scholar] [CrossRef]
- Wang, D.; Cui, B.C.; Duan, S.S.; Chen, J.J.; Fan, H.; Lu, B.B.; Zheng, J.H. Moving north in China: The habitat of Pedicularis kansuensis in the context of climate change. Sci. Total Environ. 2019, 697, 133979. [Google Scholar] [CrossRef]
- Doyle, J. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef]
- Wick, R.R.; Schultz, M.B.; Justin, Z.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Ye, W.C.; Zhang, Y.D.; Xu, Y.S. High speed BLASTN: An accelerated MegaBLAST search tool. Nucleic Acids Res. 2015, 43, 7762. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef]
- Michael, T.; Pascal, L.; Tommaso, P.; Ulbricht-Jones, E.S.; Axel, F.; Ralph, B.; Stephan, G. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef]
- Lewis, S.E.; Searle, S.M.J.; Harris, N.; Gibson, M.; Lyer, V.; Richter, J.; Wiel, C.; Bayraktarogli, L.; Birney, E.; Crosby, M.A.; et al. Apollo: A sequence annotation editor. Genome Biol. 2002, 3, 1–14. [Google Scholar] [CrossRef]
- Li, J.L.; Ni, Y.; Lu, Q.Q.; Chen, H.M.; Liu, C. PMGA: A plant mitochondrial genome annotator. Plant Commun. 2025, 6, 101191. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.L.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Sebastian, B.; Thomas, T.; Thomas, M.; Uwe, S.; Martin, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Gary, B. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef]
- Yu, W.B.; Yang, J.B.; Song, Y.; DePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Edera, A.A.; Small, I.; Milone, D.H.; Sanchez-Puerta, M.V. Deepred-Mt: Deep representation learning for predicting C-to-U RNA editing in plant mitochondria. Comput. Biol. Med. 2021, 136, 104682. [Google Scholar] [CrossRef]
- Wang, Y.P.; Tang, H.B.; Debarry, J.D.; Tan, X.; Li, J.P.; Wang, X.Y.; Lee, T.H.; Jin, H.Z.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Alverson, A.J.; Xiaoxin, W.; Rice, D.W.; Stern, D.B.; Kerrie, B.; Palmer, J.D. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol. Biol. Evol. 2010, 27, 1436–1448. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef] [PubMed]
- Raven, R.H.; Hong, D.Y.; Wu, Z.Y.; Raven, R.H.; Hong, D.Y. History of the flora of China. Flora China 2013, 1, 1–21. [Google Scholar]
- Wicke, S.; Müller, K.F.; de Pamphilis, C.W.; Quandt, D.; Wickett, N.J.; Zhang, Y.; Renner, S.S.; Schneeweiss, G.M. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell 2013, 25, 3711–3725. [Google Scholar] [CrossRef] [PubMed]
- T.A.P. GROUP. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 141, 436. [Google Scholar]
- Mower, J.P.; Sloan, D.B.; Alverson, A.J. Plant mitochondrial genome diversity: The genomics revolution. In Plant Genome Diversity Volume 1: Plant Genomes, Their Residents, and Their Evolutionary Dynamics; Wendel, J.F., Greilhuber, J., Dolezel, J., Leitch, I.J., Eds.; Springer: Vienna, Austria, 2012; pp. 123–144. [Google Scholar]
- Morton, B.R. Selection on the codon bias of chloroplast and cyanelle genes in different plant and algal lineages. J. Mol. Evol. 1998, 46, 449–459. [Google Scholar] [CrossRef]
- Maier, R.M.; Zeltz, P.; Kössel, H.; Bonnard, G.; Gualberto, J.M.; Grienenberger, J.M. RNA editing in plant mitochondria and chloroplasts. Plant Mol. Biol. 1996, 32, 343–365. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Wu, M.; Palmer, J.D.; Taylor, D.R. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol. Evol. 2012, 4, 294–306. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Q.; Li, X.; Li, Y. Structural Variation and Evolutionary Dynamics of Orobanchaceae from the Perspective of the Mitochondrial Genomes Pedicularis kansuensis and Pedicularis chinensis. Horticulturae 2025, 11, 1095. https://doi.org/10.3390/horticulturae11091095
Shi Q, Li X, Li Y. Structural Variation and Evolutionary Dynamics of Orobanchaceae from the Perspective of the Mitochondrial Genomes Pedicularis kansuensis and Pedicularis chinensis. Horticulturae. 2025; 11(9):1095. https://doi.org/10.3390/horticulturae11091095
Chicago/Turabian StyleShi, Qian, Xiuzhang Li, and Yuling Li. 2025. "Structural Variation and Evolutionary Dynamics of Orobanchaceae from the Perspective of the Mitochondrial Genomes Pedicularis kansuensis and Pedicularis chinensis" Horticulturae 11, no. 9: 1095. https://doi.org/10.3390/horticulturae11091095
APA StyleShi, Q., Li, X., & Li, Y. (2025). Structural Variation and Evolutionary Dynamics of Orobanchaceae from the Perspective of the Mitochondrial Genomes Pedicularis kansuensis and Pedicularis chinensis. Horticulturae, 11(9), 1095. https://doi.org/10.3390/horticulturae11091095







