Mapping of Leaf Pigments in Lettuce via Hyperspectral Imaging and Machine Learning
Abstract
1. Introduction
2. Materials and Methods
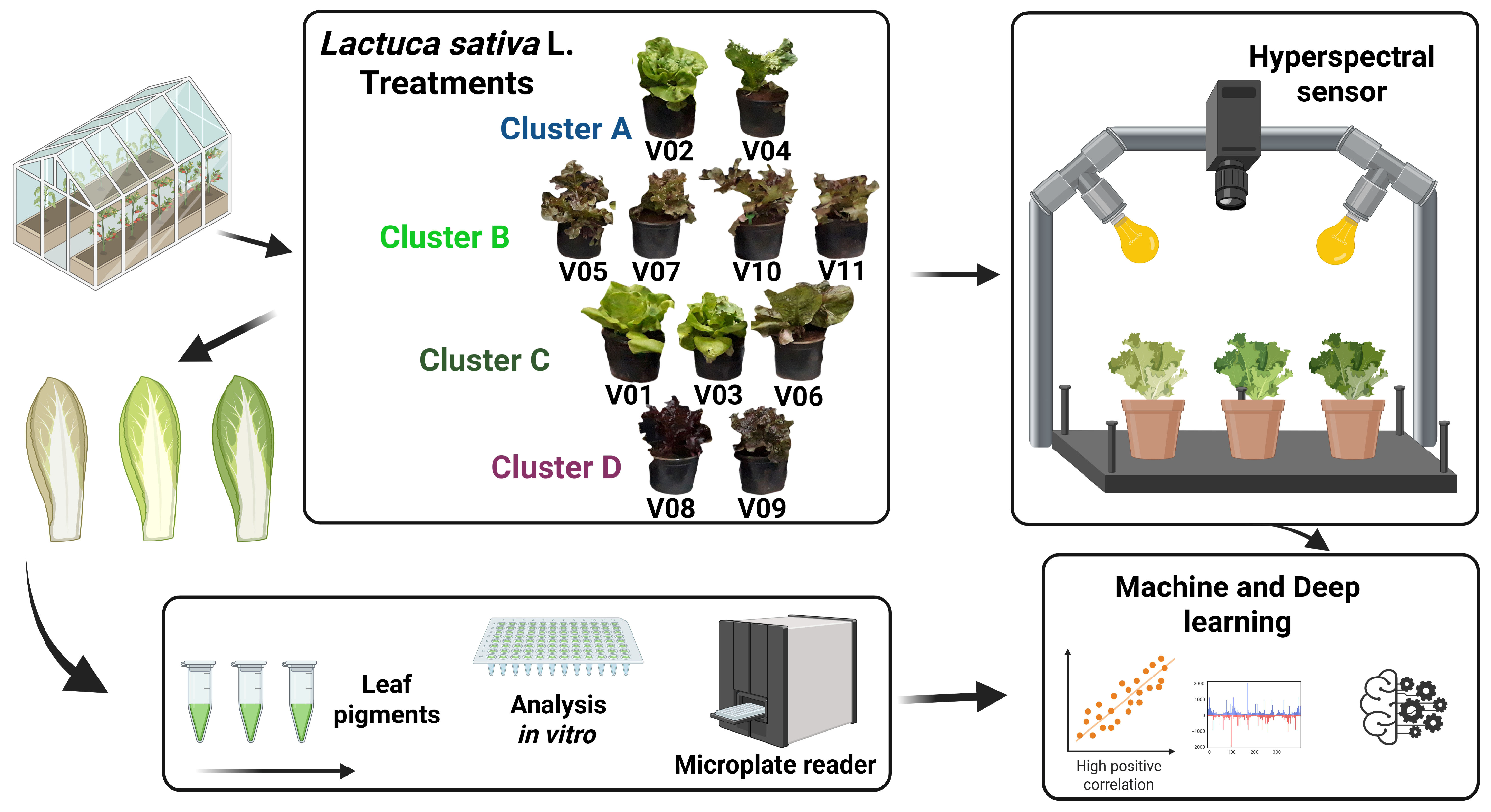
2.1. Plant Materials, Growth Conditions and Experimental Design
2.2. Extraction and Quantification of the Pigment Profile
Chlb = 34.09 × A652 − 15.28 × A665
Carotenoids = (1000 × A470 − 1.63 × Chla − 104.96 × Chlb)/221
2.3. Quantification of Total Soluble Phenolic Compounds
2.4. Spectral Collection of Leaf Reflectance Data
2.5. Vegetation Indices
2.6. Correlation and Heatmap Analyses
2.7. Preprocessing of Hyperspectral Data and Dataset Division
2.8. Machine Learning Algorithms
2.9. Statistical Analyses
3. Results
3.1. Descriptive Analyses
3.2. Effects of Cultivars on Biochemical Composition
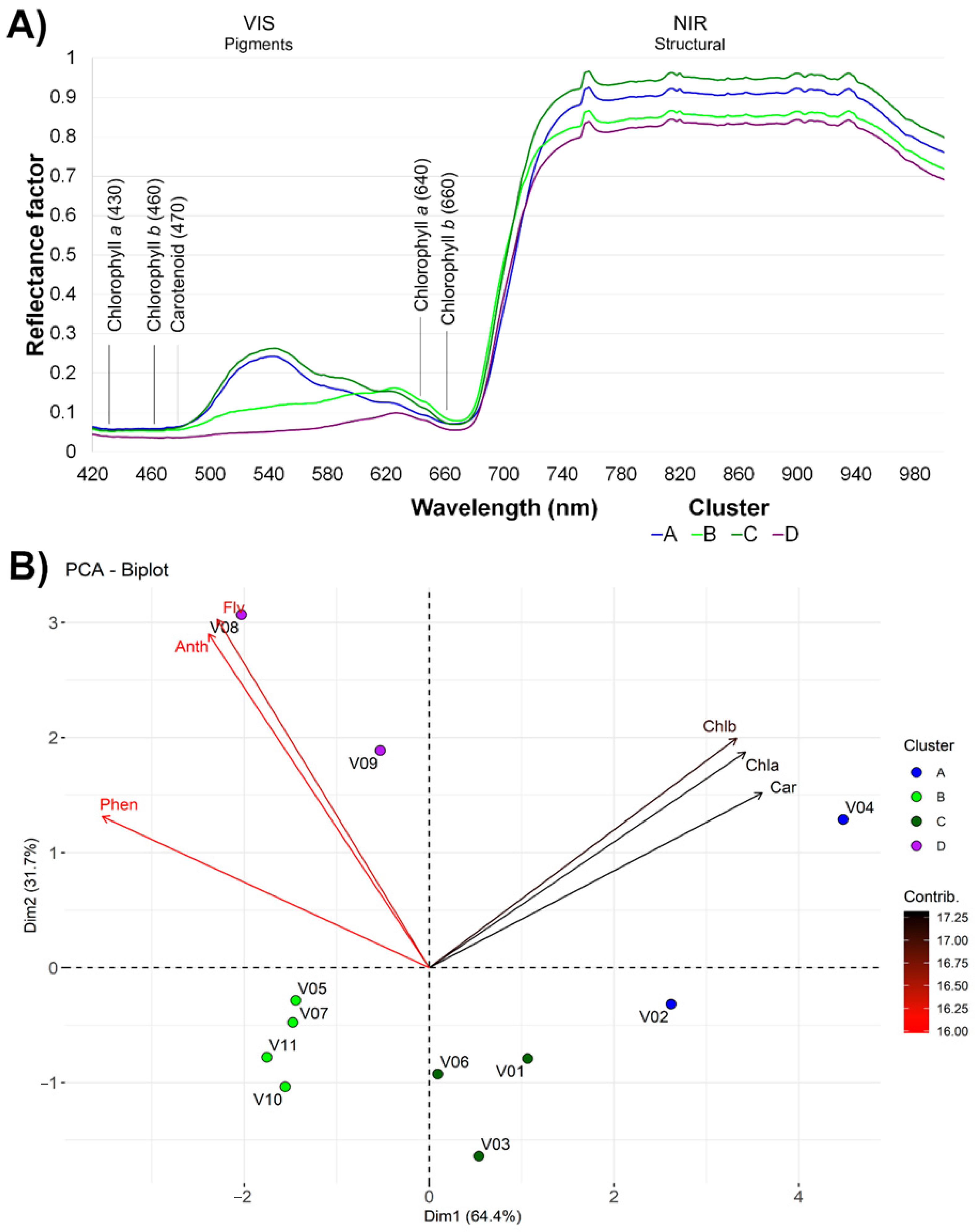
3.3. Cluster, PCA, and Spectral Analysis
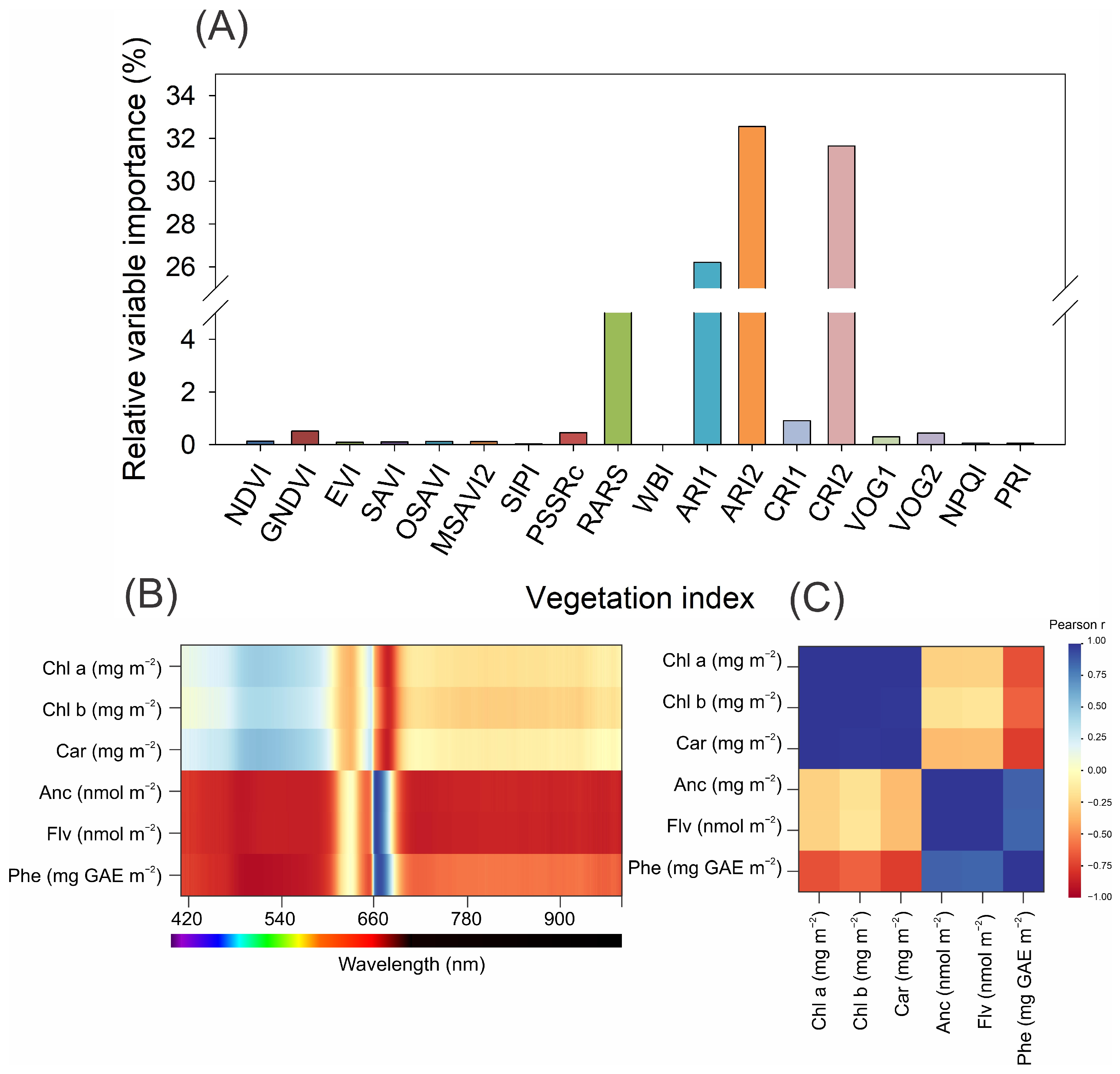
3.4. Vegetation and Cluster Heatmaps
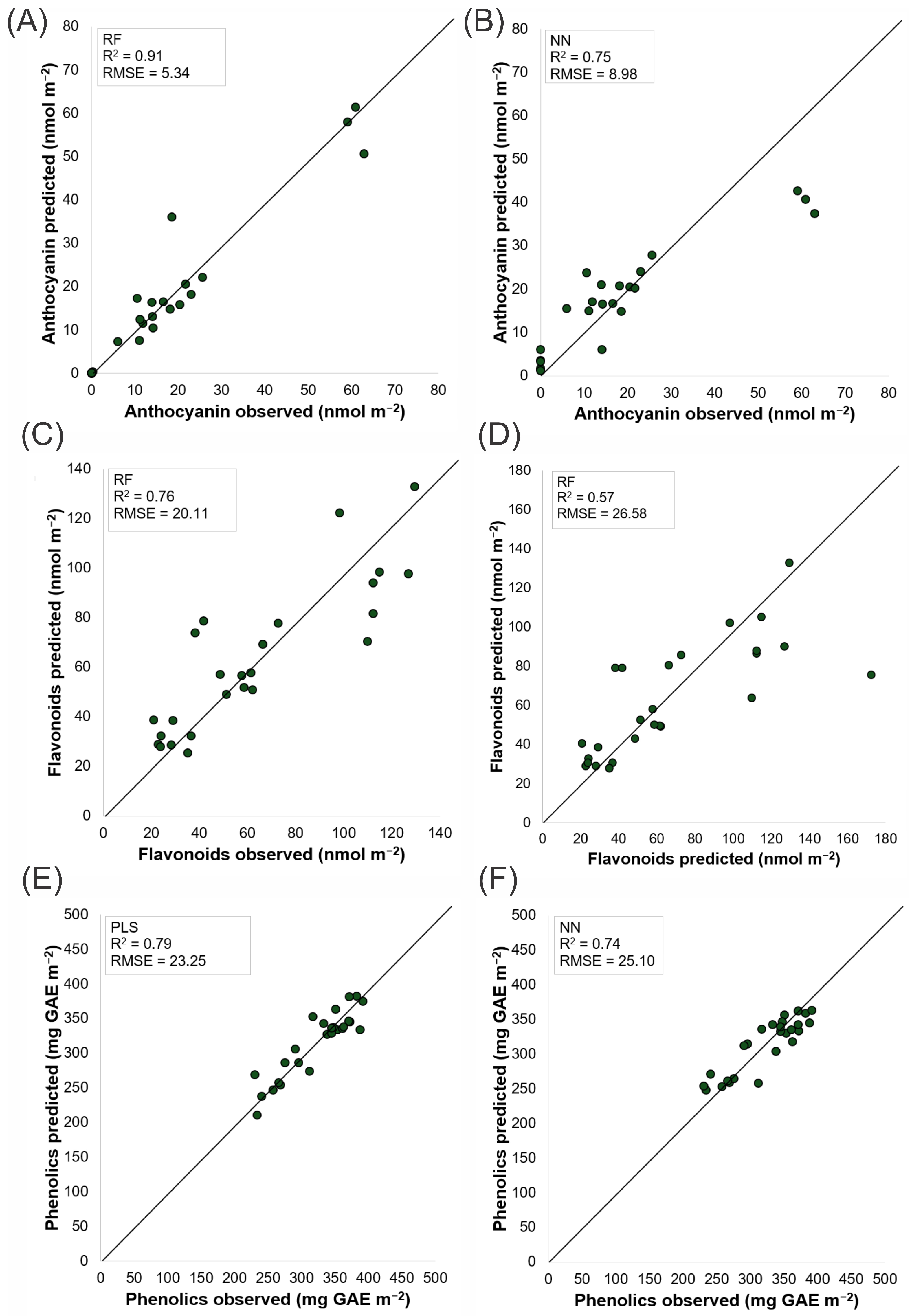
3.5. Machine Learning Models
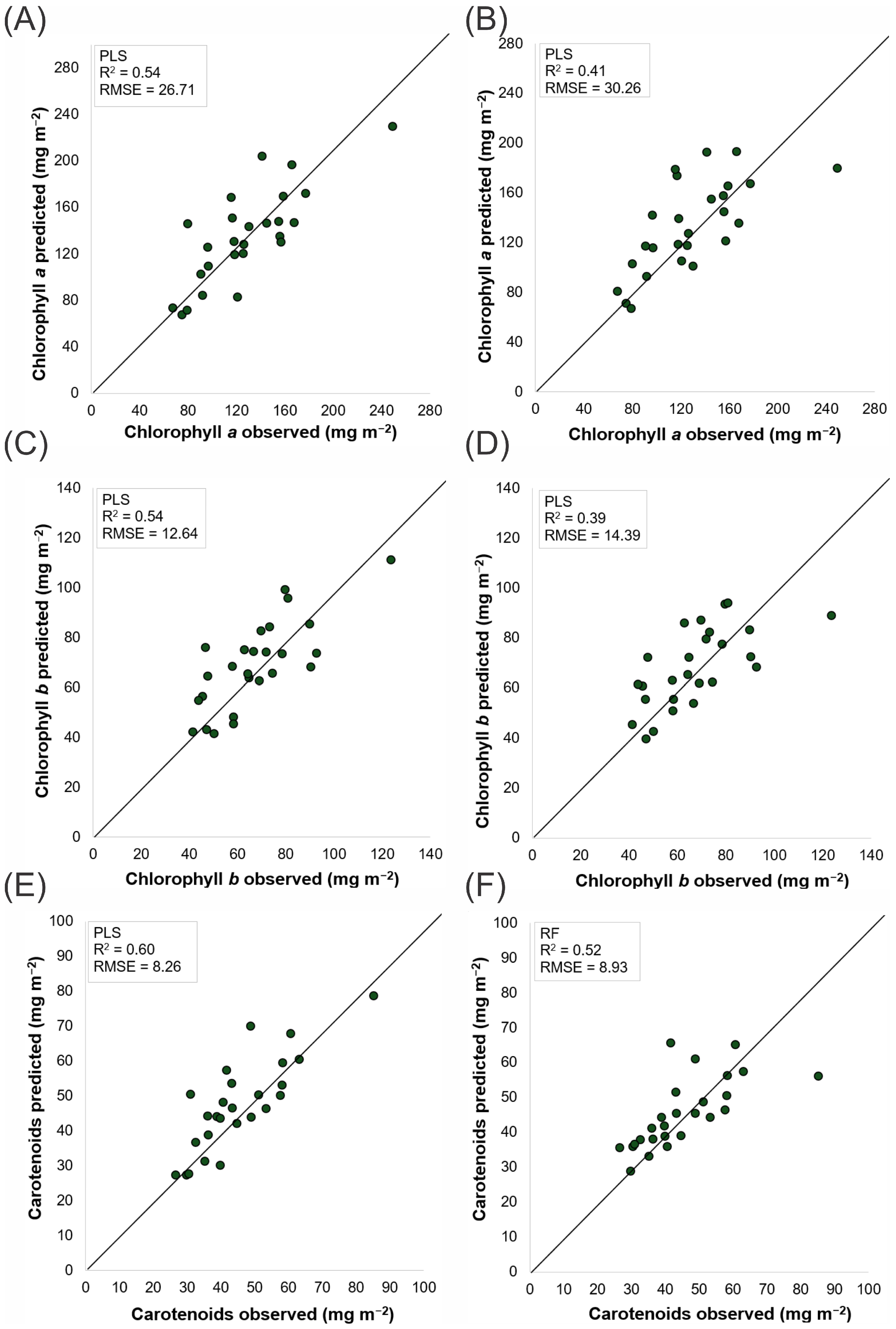
3.6. Performance of Regression Models in Predicting Phytochemical Compounds
3.7. Spatial Mapping of Lettuce Biochemical Traits
4. Discussion
4.1. Impact of Cultivars on Biochemical Composition
4.2. Spectral Reflectance Pattern and Pigment Content
4.3. Predictive Model Performance Across Sensors
4.4. Spatial Mapping of Predicted Traits
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Moura, L.d.O.; Lopes, D.d.C.; Neto, A.J.S.; Ferraz, L.d.C.L.; Carlos, L.d.A.; Martins, L.M. Evaluation of Techniques for Automatic Classification of Lettuce Based on Spectral Reflectance. Food Anal. Methods 2016, 9, 1799–1806. [Google Scholar] [CrossRef]
- Lopes, D.d.C.; Moura, L.d.O.; Neto, A.J.S.; Ferraz, L.d.C.L.; Carlos, L.d.A.; Martins, L.M. Spectral Indices for Non-Destructive Determination of Lettuce Pigments. Food Anal. Methods 2017, 10, 2807–2814. [Google Scholar] [CrossRef]
- Steidle Neto, A.J.; Moura, L.O.; Lopes, D.C.; Carlos, L.A.; Martins, L.M.; Ferraz, L.C. Non-Destructive Prediction of Pigment Content in Lettuce Based on Visible-NIR Spectroscopy. J. Sci. Food Agric. 2017, 97, 2015–2022. [Google Scholar] [CrossRef]
- Falcioni, R.; Gonçalves, J.V.F.; de Oliveira, K.M.; de Oliveira, C.A.; Demattê, J.A.M.; Antunes, W.C.; Nanni, M.R. Enhancing Pigment Phenotyping and Classification in Lettuce through the Integration of Reflectance Spectroscopy and AI Algorithms. Plants 2023, 12, 1333. [Google Scholar] [CrossRef]
- Agnolucci, M.; Avio, L.; Palla, M.; Sbrana, C.; Turrini, A.; Giovannetti, M. Health-Promoting Properties of Plant Products: The Role of Mycorrhizal Fungi and Associated Bacteria. Agronomy 2020, 10, 1864. [Google Scholar] [CrossRef]
- Matysiak, B.; Ropelewska, E.; Wrzodak, A.; Kowalski, A.; Kaniszewski, S. Yield and Quality of Romaine Lettuce at Different Daily Light Integral in an Indoor Controlled Environment. Agronomy 2022, 12, 1026. [Google Scholar] [CrossRef]
- Llorach, R.; Martínez-Sánchez, A.; Tomás-Barberán, F.A.; Gil, M.I.; Ferreres, F. Characterisation of Polyphenols and Antioxidant Properties of Five Lettuce Varieties and Escarole. Food Chem. 2008, 108, 1028–1038. [Google Scholar] [CrossRef]
- Clemente, A.A.; Maciel, G.M.; Siquieroli, A.C.S.; Gallis, R.B.d.A.; Pereira, L.M.; Duarte, J.G. High-Throughput Phenotyping to Detect Anthocyanins, Chlorophylls, and Carotenoids in Red Lettuce Germplasm. Int. J. Appl. Earth Obs. Geoinf. 2021, 103, 102533. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Gibin, M.S.; Vollmann, A.; Pattaro, M.C.; Giacomelli, M.E.; Sato, F.; Nanni, M.R.; Antunes, W.C. Classification and Prediction by Pigment Content in Lettuce (Lactuca sativa L.) Varieties Using Machine Learning and ATR-FTIR Spectroscopy. Plants 2022, 11, 3413. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, J.K.; Moura-Bueno, J.M.; Ramon, R.; Almeida, T.F.; Naibo, G.; Martins, A.P.; Santos, L.S.; Gianello, C.; Tiecher, T. Combining Different Pre-Processing and Multivariate Methods for Prediction of Soil Organic Matter by near Infrared Spectroscopy (NIRS) in Southern Brazil. Geoderma Reg. 2022, 29, e00530. [Google Scholar] [CrossRef]
- Crusiol, L.G.T.; Sun, L.; Sibaldelli, R.N.R.; Felipe, V., Jr.; Furlaneti, W.X.; Chen, R.; Sun, Z.; Wuyun, D.; Chen, Z.; Nanni, M.R.; et al. Strategies for Monitoring Within-Field Soybean Yield Using Sentinel-2 Vis-NIR-SWIR Spectral Bands and Machine Learning Regression Methods. Precis. Agric. 2022, 23, 1093–1123. [Google Scholar] [CrossRef]
- Wang, D.; Cao, W.; Zhang, F.; Li, Z.; Xu, S.; Wu, X. A Review of Deep Learning in Multiscale Agricultural Sensing. Remote Sens. 2022, 14, 559. [Google Scholar] [CrossRef]
- Huang, C.Y.; Asner, G.P. Applications of Remote Sensing to Alien Invasive Plant Studies. Sensors 2009, 9, 4869–4889. [Google Scholar] [CrossRef] [PubMed]
- Falcioni, R.; de Oliveira, R.B.; Chicati, M.L.; Antunes, W.C.; Demattê, J.A.M.; Nanni, M.R. Estimation of Biochemical Compounds in Tradescantia Leaves Using VIS-NIR-SWIR Hyperspectral and Chlorophyll a Fluorescence Sensors. Remote Sens. 2024, 16, 1910. [Google Scholar] [CrossRef]
- Chicati, M.S.; Nanni, M.R.; Chicati, M.L.; Furlanetto, R.H.; Cezar, E.; De Oliveira, R.B. Hyperspectral Remote Detection as an Alternative to Correlate Data of Soil Constituent. Remote Sens. Appl. Soc. Environ. 2019, 16, 100270. [Google Scholar] [CrossRef]
- Vigneault, P.; Lafond-Lapalme, J.; Deshaies, A.; Khun, K.; de la Sablonnière, S.; Filion, M.; Longchamps, L.; Mimee, B. An Integrated Data-Driven Approach to Monitor and Estimate Plant-Scale Growth Using UAV. ISPRS Open J. Photogramm. Remote Sens. 2024, 11, 100052. [Google Scholar] [CrossRef]
- Zsebő, S.; Bede, L.; Kukorelli, G.; Kulmány, I.M.; Milics, G.; Stencinger, D.; Teschner, G.; Varga, Z.; Vona, V.; Kovács, A.J. Yield Prediction Using NDVI Values from GreenSeeker and MicaSense Cameras at Different Stages of Winter Wheat Phenology. Drones 2024, 8, 88. [Google Scholar] [CrossRef]
- Furlanetto, R.H.; Moriwaki, T.; Falcioni, R.; Pattaro, M.; Vollmann, A.; Sturion, A.C., Jr.; Antunes, W.C.; Nanni, M.R. Hyperspectral Reflectance Imaging to Classify Lettuce Varieties by Optimum Selected Wavelengths and Linear Discriminant Analysis. Remote Sens. Appl. Soc. Environ. 2020, 20, 100400. [Google Scholar] [CrossRef]
- Shurygin, B.; Chivkunova, O.; Solovchenko, O.; Solovchenko, A.; Dorokhov, A.; Smirnov, I.; Astashev, M.E.; Khort, D. Comparison of the Non-Invasive Monitoring of Fresh-Cut Lettuce Condition with Imaging Reflectance Hyperspectrometer and Imaging PAM-Fluorimeter. Photonics 2021, 8, 425. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, X.; Kang, M.; Hu, B.-G.; Heuvelink, E.; Marcelis, L.F.M. Machine Learning versus Crop Growth Models: An Ally, Not a Rival. AoB PLANTS 2022, 15, plac061. [Google Scholar] [CrossRef]
- Habibi, L.N.; Watanabe, T.; Matsui, T.; Tanaka, T.S.T. Machine Learning Techniques to Predict Soybean Plant Density Using UAV and Satellite-Based Remote Sensing. Remote Sens. 2021, 13, 2548. [Google Scholar] [CrossRef]
- Ropelewska, E. Application of Imaging and Artificial Intelligence for Quality Monitoring of Stored Black Currant (Ribes nigrum L.). Foods 2022, 11, 3589. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart Breeding Driven by Big Data, Artificial Intelligence, and Integrated Genomic-Enviromic Prediction. Mol. Plant 2022, 15, 1664–1695. [Google Scholar] [CrossRef]
- Iqbal, I.M.; Balzter, H.; Firdaus-e-Bareen; Shabbir, A. Identifying the Spectral Signatures of Invasive and Native Plant Species in Two Protected Areas of Pakistan through Field Spectroscopy. Remote Sens. 2021, 13, 4009. [Google Scholar] [CrossRef]
- Hoeppner, J.M.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M.; Chang, H.-C.; Gara, T.W. Mapping Canopy Chlorophyll Content in a Temperate Forest Using Airborne Hyperspectral Data. Remote Sens. 2020, 12, 3573. [Google Scholar] [CrossRef]
- Wang, L.; Gao, R.; Li, C.; Wang, J.; Liu, Y.; Hu, J.; Li, B.; Qiao, H.; Feng, H.; Yue, J. Mapping Soybean Maturity and Biochemical Traits Using UAV-Based Hyperspectral Images. Remote Sens. 2023, 15, 4807. [Google Scholar] [CrossRef]
- dos Santos, G.L.A.A.; Reis, A.S.; Besen, M.R.; Furlanetto, R.H.; Rodrigues, M.; Crusiol, L.G.T.; de Oliveira, K.M.; Falcioni, R.; de Oliveira, R.B.; Batista, M.A.; et al. Spectral Method for Macro and Micronutrient Prediction in Soybean Leaves Using Interval Partial Least Squares Regression. Eur. J. Agron. 2023, 143, 126717. [Google Scholar] [CrossRef]
- Falcioni, R.; Moriwaki, T.; Rodrigues, M.; de Oliveira, K.M.; Furlanetto, R.H.; dos Reis, A.S.; dos Santos, G.L.A.A.; Mendonça, W.A.; Crusiol, L.G.T.; Gonçalves, J.V.F.; et al. Nutrient Deficiency Lowers Photochemical and Carboxylation Efficiency in Tobacco. Theor. Exp. Plant Physiol. 2023, 35, 81–97. [Google Scholar] [CrossRef]
- Braga, P.; Crusiol, L.G.T.; Nanni, M.R.; Caranhato, A.L.H.; Fuhrmann, M.B.; Nepomuceno, A.L.; Neumaier, N.; Farias, J.R.B.; Koltun, A.; Gonçalves, L.S.A.; et al. Vegetation Indices and NIR-SWIR Spectral Bands as a Phenotyping Tool for Water Status Determination in Soybean. Precis. Agric. 2021, 22, 249–266. [Google Scholar] [CrossRef]
- Li, X.; Zhou, R.; Xu, K.; Xu, J.; Jin, J.; Fang, H.; He, Y. Rapid Determination of Chlorophyll and Pheophytin in Green Tea Using Fourier Transform Infrared Spectroscopy. Molecules 2018, 23, 1010. [Google Scholar] [CrossRef]
- Overbeck, V.; Schmitz, M.; Blanke, M. Non-Destructive Sensor-Based Prediction of Maturity and Optimum Harvest Date of Sweet Cherry Fruit. Sensors 2017, 17, 277. [Google Scholar] [CrossRef] [PubMed]
- Gitelson, A.; Solovchenko, A.; Viña, A. Foliar Absorption Coefficient Derived from Reflectance Spectra: A Gauge of the Efficiency of in situ Light-Capture by Different Pigment Groups. J. Plant Physiol. 2020, 254, 153277. [Google Scholar] [CrossRef]
- Ragaee, S. Antioxidant Activity and Nutrient Composition of Selected Cereals for Food Use. Food Chem. 2006, 98, 32–38. [Google Scholar] [CrossRef]
- Luz, R.B. Attenuated Total Reflectance Spectroscopy of Plant Leaves: A Tool for Ecological and Botanical Studies. New Phytol. 2006, 172, 305–318. [Google Scholar] [CrossRef] [PubMed]
- Falcioni, R.; Gonçalves, J.V.F.; de Oliveira, K.M.; Antunes, W.C.; Nanni, M.R. VIS-NIR-SWIR Hyperspectroscopy Combined with Data Mining and Machine Learning for Classification of Predicted Chemometrics of Green Lettuce. Remote Sens. 2022, 14, 6330. [Google Scholar] [CrossRef]
- Asner, G.P.; Jones, M.O.; Martin, R.E.; Knapp, D.E.; Hughes, R.F. Remote Sensing of Native and Invasive Species in Hawaiian Forests. Remote Sens. Environ. 2008, 112, 1912–1926. [Google Scholar] [CrossRef]
- Ling, B.; Goodin, D.G.; Raynor, E.J.; Joern, A. Hyperspectral Analysis of Leaf Pigments and Nutritional Elements in Tallgrass Prairie Vegetation. Front. Plant Sci. 2019, 10, 142. [Google Scholar] [CrossRef]
- Blackburn, G.A. Hyperspectral Remote Sensing of Plant Pigments. J. Exp. Bot. 2007, 58, 855–867. [Google Scholar] [CrossRef]
- Shi, M.; Gu, J.; Wu, H.; Rauf, A.; Emran, T.B.; Khan, Z.; Mitra, S.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Al-Awthan, Y.S.; et al. Phytochemicals, Nutrition, Metabolism, Bioavailability, and Health Benefits in Lettuce: A Comprehensive Review. Antioxidants 2022, 11, 1158. [Google Scholar] [CrossRef]
- Sumi, M.J.; Jahan, N.; Thamid, S.S.; Tarik, M.E.I.; Hassannejad, S.; Rahimi, M.; Imran, S. LED Light Effect on Growth, Pigments, and Antioxidants of Lettuce (Lactuca sativa L.) Baby Greens. BMC Plant Biol. 2025, 25, 582. [Google Scholar] [CrossRef]
- Lee, M.; Kim, J.; Oh, M.-M.; Lee, J.-H.; Rajashekar, C.B. Effects of Supplemental UV-A LEDs on the Nutritional Quality of Lettuce: Accumulation of Protein and Other Essential Nutrients. Horticulturae 2022, 8, 680. [Google Scholar] [CrossRef]
- Sonobe, R.; Hirono, Y. Applying Variable Selection Methods and Preprocessing Techniques to Hyperspectral Reflectance Data to Estimate Tea Cultivar Chlorophyll Content. Remote Sens. 2023, 15, 19. [Google Scholar] [CrossRef]
- Sonobe, R.; Wang, Q. Hyperspectral Indices for Quantifying Leaf Chlorophyll Concentrations Performed Differently with Different Leaf Types in Deciduous Forests. Ecol. Inform. 2017, 37, 1–9. [Google Scholar] [CrossRef]
- Katuwal, K.B.; Yang, H.; Huang, B. Evaluation of Phenotypic and Photosynthetic Indices to Detect Water Stress in Perennial Grass Species Using Hyperspectral, Multispectral and Chlorophyll Fluorescence Imaging. Grass Res. 2023, 3, 16. [Google Scholar] [CrossRef]
- Zhang, Y.-W.; Wang, T.; Guo, Y.; Skidmore, A.; Zhang, Z.; Tang, R.; Song, S.; Tang, Z. Estimating Community-Level Plant Functional Traits in a Species-Rich Alpine Meadow Using UAV Image Spectroscopy. Remote Sens. 2022, 14, 3399. [Google Scholar] [CrossRef]
- Wang, S.; Guan, K.; Zhang, C.; Jiang, C.; Zhou, Q.; Li, K.; Qin, Z.; Ainsworth, E.A.; He, J.; Wu, J.; et al. Airborne Hyperspectral Imaging of Cover Crops through Radiative Transfer Process-Guided Machine Learning. Remote Sens. Environ. 2023, 285, 113386. [Google Scholar] [CrossRef]
- Coast, O.; Shah, S.; Ivakov, A.; Gaju, O.; Wilson, P.B.; Posch, B.C.; Bryant, C.J.; Negrini, A.C.A.; Evans, J.R.; Condon, A.G.; et al. Predicting Dark Respiration Rates of Wheat Leaves from Hyperspectral Reflectance. Plant Cell Environ. 2019, 42, 2133–2150. [Google Scholar] [CrossRef]
- Zhang, Y.; Kong, X.; Deng, L.; Liu, Y. Monitor Water Quality through Retrieving Water Quality Parameters from Hyperspectral Images Using Graph Convolution Network with Superposition of Multi-Point Effect: A Case Study in Maozhou River. J. Environ. Manag. 2023, 342, 118283. [Google Scholar] [CrossRef]
- Bartsch, B.d.A.; Rosin, N.A.; dos Santos, U.J.; Coblinski, J.A.; Pelegrino, M.H.P.; Rosas, J.T.F.; Poppiel, R.R.; Ortiz, E.B.; Kochinki, V.C.V.; Gallo, P.; et al. A Step Forward in Hybrid Soil Laboratory Analysis: Merging Chemometric Corrections, Protocols and Data-Driven Methods. Remote Sens. 2024, 16, 4543. [Google Scholar] [CrossRef]
- Li, Y.; Xu, X.; Wu, W.; Zhu, Y.; Gao, L.; Jiang, X.; Meng, Y.; Yang, G.; Xue, H. Hyperspectral Estimation of Chlorophyll Content in Grapevine Based on Feature Selection and GA-BP. Sci. Rep. 2025, 15, 8029. [Google Scholar] [CrossRef]
- Isgró, M.A.; Basallote, M.D.; Caballero, I.; Barbero, L. Comparison of UAS and Sentinel-2 Multispectral Imagery for Water Quality Monitoring: A Case Study for Acid Mine Drainage Affected Areas (SW Spain). Remote Sens. 2022, 14, 4053. [Google Scholar] [CrossRef]
- Neri, I.; Caponi, S.; Bonacci, F.; Clementi, G.; Cottone, F.; Gammaitoni, L.; Figorilli, S.; Ortenzi, L.; Aisa, S.; Pallottino, F.; et al. Real-Time AI-Assisted Push-Broom Hyperspectral System for Precision Agriculture. Sensors 2024, 24, 344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yu, L.; Zhang, X.; Liu, Y.; Zhan, Z.; Ren, L.; Luo, Y. Fusion of UAV Hyperspectral Imaging and LiDAR for the Early Detection of EAB Stress in Ash and a New EAB Detection Index NDVI(776,678). Remote Sens. 2022, 14, 2428. [Google Scholar] [CrossRef]
- Jin, J.; Wang, Q. Selection of Informative Spectral Bands for PLS Models to Estimate Foliar Chlorophyll Content Using Hyperspectral Reflectance. IEEE Trans. Geosci. Remote Sens. 2019, 57, 3064–3072. [Google Scholar] [CrossRef]
- Calviño-Cancela, M.; Martín-Herrero, J. Spectral Discrimination of Vegetation Classes in Ice-Free Areas of Antarctica. Remote Sens. 2016, 8, 856. [Google Scholar] [CrossRef]
- Gierlinger, N.; Keplinger, T.; Harrington, M. Imaging of Plant Cell Walls by Confocal Raman Microscopy. Nat. Protoc. 2012, 7, 1694–1708. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Spectral Reflectance Changes Associated with Autumn Senescence of Aesculus hippocastanum L. and Acer platanoides L. Leaves. Spectral Features and Relation to Chlorophyll Estimation. J. Plant Physiol. 1994, 143, 286–292. [Google Scholar] [CrossRef]

| Parameters of Dataset | (n) | Mean | SD | Median | Minimum | Maximum | CV (%) |
|---|---|---|---|---|---|---|---|
| Chla | 132 | 128.89 | 50.49 | 118.95 | 63.15 | 306.40 | 39.18 |
| Chlb | 132 | 66.76 | 23.42 | 61.75 | 39.41 | 147.27 | 35.09 |
| Car | 132 | 45.70 | 16.21 | 42.41 | 25.56 | 101.92 | 35.47 |
| Anc | 132 | 13.43 | 16.98 | 11.46 | 0.00 | 68.57 | 126.53 |
| Flv | 132 | 59.55 | 36.12 | 49.75 | 11.77 | 178.28 | 60.66 |
| Phe | 132 | 312.95 | 52.55 | 331.01 | 213.70 | 393.30 | 16.79 |
| Parameters Training | (n) | Mean | SD | Median | Minimum | Maximum | CV (%) |
| Chla | 106 | 129.18 | 52.89 | 117.76 | 63.14 | 306.40 | 40.94 |
| Chlb | 106 | 66.64 | 24.48 | 60.32 | 39.40 | 147.26 | 36.74 |
| Car | 106 | 45.82 | 16.92 | 42.41 | 25.55 | 101.91 | 36.92 |
| Anc | 106 | 12.76 | 16.63 | 11.01 | 0.00 | 68.57 | 130.03 |
| Flv | 106 | 57.59 | 34.64 | 48.26 | 11.76 | 178.28 | 60.15 |
| Phe | 106 | 310.40 | 52.90 | 325.20 | 213.65 | 393.30 | 17.05 |
| Parameters Test | (n) | Mean | SD | Median | Minimum | Maximum | CV (%) |
| Chla | 26 | 127.69 | 40.14 | 123.08 | 67.38 | 249.10 | 31.43 |
| Chlb | 26 | 67.25 | 18.91 | 65.65 | 41.33 | 123.60 | 28.11 |
| Car | 26 | 45.16 | 13.19 | 42.37 | 26.51 | 85.18 | 29.21 |
| Anc | 26 | 16.15 | 18.45 | 12.89 | 0.00 | 62.90 | 114.22 |
| Flv | 26 | 67.53 | 41.40 | 58.13 | 20.83 | 172.50 | 61.31 |
| Phe | 26 | 323.20 | 50.60 | 341.45 | 230.45 | 391.55 | 15.65 |
| Effects | Chla | Chlb | Car | Anc | Flv | Phe | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Varieties | mg m−2 | mg m−2 | mg m−2 | nmol m−2 | nmol m−2 | mg GAE m−2 | ||||||||||||||||||
| V01 | 138.16 | ± | 5.19 | c † | 73.35 | ± | 3.16 | b | 48.81 | ± | 2.10 | c | 0.00 | ± | 0.00 | f | 31.73 | ± | 2.30 | ef | 287.10 | ± | 10.20 | c |
| V02 | 177.29 | ± | 12.01 | a | 85.10 | ± | 5.56 | b | 63.62 | ± | 3.89 | b | 0.00 | ± | 0.00 | f | 26.75 | ± | 2.22 | f | 241.65 | ± | 6.50 | d |
| V03 | 110.15 | ± | 5.25 | d | 57.00 | ± | 3.07 | cde | 41.84 | ± | 1.81 | cd | 0.03 | ± | 0.01 | f | 26.92 | ± | 3.18 | f | 259.40 | ± | 5.90 | cd |
| V04 | 243.96 | ± | 19.31 | a | 118.80 | ± | 9.24 | a | 82.21 | ± | 6.42 | a | 0.45 | ± | 0.07 | f | 35.88 | ± | 0.94 | def | 238.60 | ± | 3.80 | d |
| V05 | 102.39 | ± | 3.47 | d | 51.82 | ± | 1.43 | de | 35.29 | ± | 0.97 | de | 16.01 | ± | 0.35 | c | 61.71 | ± | 5.65 | bc | 365.30 | ± | 8.65 | ab |
| V06 | 115.75 | ± | 3.76 | cd | 57.82 | ± | 1.91 | cd | 42.08 | ± | 1.39 | cd | 6.71 | ± | 0.47 | e | 45.38 | ± | 2.11 | cde | 284.30 | ± | 4.75 | c |
| V07 | 95.15 | ± | 2.19 | de | 45.64 | ± | 1.02 | e | 33.78 | ± | 0.44 | e | 15.86 | ± | 0.94 | c | 71.06 | ± | 5.25 | bc | 333.20 | ± | 6.35 | b |
| V08 | 137.46 | ± | 4.38 | c | 69.55 | ± | 1.94 | bc | 44.23 | ± | 1.30 | c | 60.88 | ± | 1.67 | a | 124.36 | ± | 16.46 | a | 383.75 | ± | 2.90 | ab |
| V09 | 141.41 | ± | 6.237 | bc | 79.85 | ± | 4.70 | b | 49.42 | ± | 1.71 | c | 22.39 | ± | 0.72 | b | 119.46 | ± | 4.82 | a | 347.90 | ± | 2.35 | b |
| V10 | 76.12 | ± | 3.72 | f | 49.22 | ± | 1.91 | de | 30.42 | ± | 1.18 | e | 11.22 | ± | 0.25 | d | 51.49 | ± | 2.27 | bcd | 347.50 | ± | 8.50 | b |
| V11 | 79.90 | ± | 2.68 | ef | 46.18 | ± | 1.02 | e | 30.94 | ± | 0.96 | e | 14.10 | ± | 0.09 | cd | 60.32 | ± | 0.65 | bc | 353.30 | ± | 6.60 | b |
| p value | *** | *** | *** | *** | *** | *** | ||||||||||||||||||
| <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | <0.001 | |||||||||||||||||||
| RMSE | R2 | ||||||
|---|---|---|---|---|---|---|---|
| Attribute | Model | Train | Test | Train | Test | AIC | BIC |
| Chla (mg m−2) | LR | 34.58 | 27.22 | 0.57 | 0.52 | 975.36 | 1263.64 |
| PLS | 32.98 | 26.71 | 0.61 | 0.54 | 774.75 | 789.16 | |
| Gboost | 39.30 | 41.28 | 0.44 | 0.00 | 1002.18 | 1031.00 | |
| RF | 37.66 | 27.65 | 0.49 | 0.51 | 792.22 | 806.63 | |
| NN | 34.46 | 28.09 | 0.57 | 0.49 | 801.66 | 816.08 | |
| SVM | 50.93 | 37.50 | 0.06 | 0.09 | 954.37 | 968.79 | |
| Cubist | 18.50 | 46.44 | 0.85 | 0.32 | 972.36 | 986.78 | |
| Chlb (mg m−2) | LR | 17.36 | 13.04 | 0.49 | 0.51 | 783.79 | 1072.07 |
| PLS | 16.63 | 12.64 | 0.53 | 0.54 | 593.79 | 608.21 | |
| Gboost | 19.02 | 16.87 | 0.39 | 0.17 | 731.35 | 745.76 | |
| RF | 18.70 | 13.46 | 0.41 | 0.47 | 612.52 | 626.94 | |
| NN | 17.11 | 15.36 | 0.51 | 0.31 | 682.21 | 696.62 | |
| SVM | 22.95 | 16.93 | 0.11 | 0.16 | 733.86 | 748.27 | |
| Cubist | 12.91 | 16.31 | 0.66 | 0.33 | 694.16 | 708.58 | |
| Car (mg m−2) | LR | 10.67 | 8.40 | 0.60 | 0.58 | 547.34 | 691.48 |
| PLS | 10.25 | 8.26 | 0.63 | 0.60 | 446.46 | 460.88 | |
| Gboost | 12.68 | 12.05 | 0.43 | 0.13 | 648.73 | 663.14 | |
| RF | 12.03 | 8.33 | 0.49 | 0.59 | 451.95 | 466.37 | |
| NN | 10.53 | 9.66 | 0.61 | 0.44 | 532.21 | 546.63 | |
| SVM | 14.73 | 11.13 | 0.23 | 0.26 | 606.40 | 620.82 | |
| Cubist | 7.21 | 9.42 | 0.77 | 0.43 | 527.90 | 542.32 | |
| Anc (nmol m−2) | LR | 12.44 | 13.01 | 0.43 | 0.48 | 691.03 | 835.17 |
| PLS | 11.07 | 9.35 | 0.55 | 0.73 | 427.31 | 441.72 | |
| Gboost | 6.93 | 7.97 | 0.82 | 0.81 | 338.76 | 353.17 | |
| RF | 4.44 | 5.34 | 0.93 | 0.91 | 134.41 | 148.82 | |
| NN | 10.86 | 10.10 | 0.57 | 0.69 | 465.91 | 480.33 | |
| SVM | 11.69 | 12.93 | 0.50 | 0.49 | 596.83 | 611.25 | |
| Cubist | 3.14 | 11.27 | 0.96 | 0.63 | 518.20 | 532.61 | |
| Flv (nmol m−2) | LR | 24.28 | 28.05 | 0.50 | 0.52 | 883.29 | 1027.43 |
| PLS | 24.82 | 27.34 | 0.48 | 0.55 | 778.01 | 792.41 | |
| Gboost | 28.27 | 26.10 | 0.33 | 0.59 | 753.46 | 767.87 | |
| RF | 24.34 | 20.11 | 0.50 | 0.76 | 613.94 | 628.35 | |
| NN | 27.05 | 27.94 | 0.38 | 0.53 | 789.47 | 803.88 | |
| SVM | 29.63 | 35.69 | 0.26 | 0.23 | 919.26 | 933.68 | |
| Cubist | 14.87 | 24.72 | 0.80 | 0.49 | 767.92 | 782.34 | |
| Phe (mg GAE m−2) | LR | 26.20 | 22.05 | 0.75 | 0.80 | 279.29 | 423.43 |
| PLS | 25.30 | 23.25 | 0.77 | 0.79 | 209.72 | 224.14 | |
| Gboost | 28.70 | 32.40 | 0.70 | 0.57 | 391.93 | 406.35 | |
| RF | 25.40 | 28.90 | 0.77 | 0.66 | 330.76 | 345.17 | |
| NN | 28.95 | 29.80 | 0.70 | 0.64 | 346.40 | 360.81 | |
| SVM | 35.65 | 29.85 | 0.54 | 0.64 | 346.84 | 361.25 | |
| Cubist | 24.35 | 29.10 | 0.82 | 0.76 | 286.60 | 301.01 | |
| RMSE | R2 | ||||||
|---|---|---|---|---|---|---|---|
| Attributes | Model | Train | Test | Train | Test | AIC | BIC |
| Chla (mg m−2) | LR | 37.58 | 30.50 | 0.49 | 0.40 | 844.85 | 859.26 |
| PLS | 37.55 | 30.26 | 0.49 | 0.41 | 836.54 | 845.19 | |
| Gboost | 40.25 | 33.14 | 0.41 | 0.29 | 884.98 | 893.63 | |
| RF | 39.39 | 30.75 | 0.44 | 0.39 | 845.18 | 853.83 | |
| NN | 37.80 | 31.36 | 0.48 | 0.37 | 854.63 | 863.28 | |
| SVM | 48.68 | 37.42 | 0.14 | 0.09 | 949.81 | 958.46 | |
| Cubist | 29.07 | 35.13 | 0.55 | 0.25 | 911.61 | 926.03 | |
| Chlb (mg m−2) | LR | 18.55 | 14.49 | 0.42 | 0.39 | 646.54 | 655.19 |
| PLS | 18.56 | 14.39 | 0.42 | 0.39 | 644.71 | 653.36 | |
| Gboost | 22.07 | 18.28 | 0.18 | 0.03 | 773.11 | 787.52 | |
| RF | 19.67 | 14.48 | 0.35 | 0.39 | 650.36 | 664.77 | |
| NN | 18.66 | 14.99 | 0.41 | 0.35 | 667.88 | 682.29 | |
| SVM | 22.58 | 16.76 | 0.14 | 0.18 | 728.01 | 742.43 | |
| Cubist | 14.41 | 15.92 | 0.43 | 0.31 | 666.44 | 680.85 | |
| Car (mg m−2) | LR | 11.64 | 9.36 | 0.52 | 0.48 | 514.10 | 528.51 |
| PLS | 11.65 | 9.29 | 0.52 | 0.48 | 508.12 | 516.77 | |
| Gboost | 13.73 | 10.12 | 0.33 | 0.39 | 551.78 | 560.43 | |
| RF | 12.44 | 8.93 | 0.45 | 0.52 | 487.12 | 495.77 | |
| NN | 11.68 | 9.71 | 0.52 | 0.44 | 529.57 | 538.22 | |
| SVM | 13.74 | 10.70 | 0.33 | 0.31 | 582.76 | 591.41 | |
| Cubist | 8.61 | 11.22 | 0.60 | 0.42 | 576.36 | 590.78 | |
| Anc (nmol m−2) | LR | 11.47 | 11.38 | 0.52 | 0.60 | 531.06 | 545.47 |
| PLS | 12.40 | 12.63 | 0.44 | 0.51 | 581.36 | 590.01 | |
| Gboost | 9.01 | 14.51 | 0.70 | 0.36 | 653.24 | 661.89 | |
| RF | 8.44 | 13.69 | 0.74 | 0.43 | 622.60 | 631.24 | |
| NN | 10.26 | 8.98 | 0.62 | 0.75 | 402.49 | 411.13 | |
| SVM | 11.71 | 13.54 | 0.50 | 0.44 | 617.35 | 626.00 | |
| Cubist | 2.87 | 9.24 | 0.97 | 0.66 | 454.61 | 469.02 | |
| Flv (nmol m−2) | LR | 24.86 | 28.72 | 0.48 | 0.50 | 804.90 | 819.32 |
| PLS | 25.67 | 29.64 | 0.45 | 0.47 | 816.92 | 825.57 | |
| Gboost | 31.21 | 27.20 | 0.18 | 0.55 | 772.64 | 781.29 | |
| RF | 26.69 | 26.58 | 0.40 | 0.57 | 760.55 | 769.20 | |
| NN | 23.97 | 26.95 | 0.52 | 0.56 | 767.24 | 775.89 | |
| SVM | 27.85 | 34.19 | 0.35 | 0.29 | 893.22 | 901.87 | |
| Cubist | 19.57 | 29.55 | 0.64 | 0.41 | 769.06 | 783.48 | |
| Phe (mg GAE m−2) | LR | 28.25 | 25.40 | 0.71 | 0.74 | 261.26 | 275.68 |
| PLS | 28.50 | 25.60 | 0.71 | 0.73 | 264.32 | 272.96 | |
| Gboost | 30.85 | 31.25 | 0.66 | 0.60 | 368.85 | 377.49 | |
| RF | 29.80 | 32.35 | 0.68 | 0.57 | 387.53 | 396.17 | |
| NN | 28.10 | 25.10 | 0.72 | 0.74 | 254.13 | 262.78 | |
| SVM | 31.80 | 26.35 | 0.63 | 0.72 | 276.74 | 285.39 | |
| Cubist | 24.10 | 31.00 | 0.82 | 0.62 | 363.95 | 378.37 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gonçalves, J.V.F.; Falcioni, R.; Rutz, T.; Silva, A.L.B.R.d.; Furlanetto, R.H.; Crusiol, L.G.T.; Oliveira, K.M.d.; Oliveira, C.A.d.; Vedana, N.G.; Demattê, J.A.M.; et al. Mapping of Leaf Pigments in Lettuce via Hyperspectral Imaging and Machine Learning. Horticulturae 2025, 11, 1077. https://doi.org/10.3390/horticulturae11091077
Gonçalves JVF, Falcioni R, Rutz T, Silva ALBRd, Furlanetto RH, Crusiol LGT, Oliveira KMd, Oliveira CAd, Vedana NG, Demattê JAM, et al. Mapping of Leaf Pigments in Lettuce via Hyperspectral Imaging and Machine Learning. Horticulturae. 2025; 11(9):1077. https://doi.org/10.3390/horticulturae11091077
Chicago/Turabian StyleGonçalves, João Vitor Ferreira, Renan Falcioni, Thiago Rutz, Andre Luiz Biscaia Ribeiro da Silva, Renato Herrig Furlanetto, Luís Guilherme Teixeira Crusiol, Karym Mayara de Oliveira, Caio Almeida de Oliveira, Nicole Ghinzelli Vedana, José Alexandre Melo Demattê, and et al. 2025. "Mapping of Leaf Pigments in Lettuce via Hyperspectral Imaging and Machine Learning" Horticulturae 11, no. 9: 1077. https://doi.org/10.3390/horticulturae11091077
APA StyleGonçalves, J. V. F., Falcioni, R., Rutz, T., Silva, A. L. B. R. d., Furlanetto, R. H., Crusiol, L. G. T., Oliveira, K. M. d., Oliveira, C. A. d., Vedana, N. G., Demattê, J. A. M., & Nanni, M. R. (2025). Mapping of Leaf Pigments in Lettuce via Hyperspectral Imaging and Machine Learning. Horticulturae, 11(9), 1077. https://doi.org/10.3390/horticulturae11091077








