Identification of Tomato SET Domain Group Gene Family and Function Analysis Under Temperature Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Chromosomal Location of SlSDG Family Members
2.2. Phylogenetic Tree Construction of SDG Proteins
2.3. Conserved Motif and Gene Structure Analysis of SlSDG Family
2.4. Analysis of Cis-Acting Elements of SlSDG in Tomato
2.5. Plant Material and Growth Condition
2.6. Total RNA Extraction and qRT-PCR Analysis
2.7. Protein Extraction and Western Blot Assay
2.8. Virus-Induced Gene Silencing (VIGS) Experiment
2.9. Temperature Stress Treatment and Phenotype Identification
2.10. Data Statistics and Analysis
3. Results
3.1. Identification and Chromosomal Location of SlSDG Gene Family
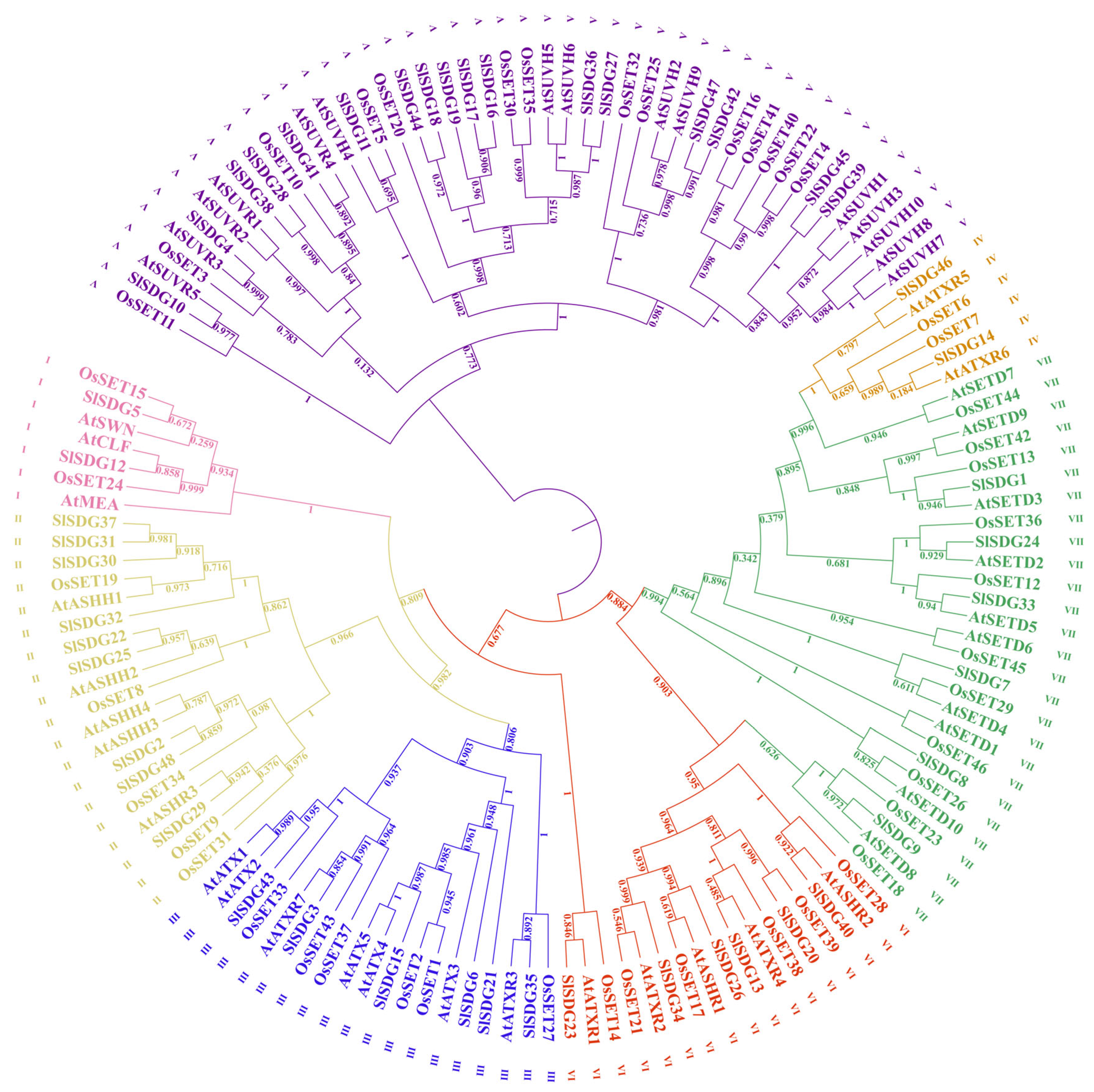
3.2. Phylogenetic Analysis of SlSDG Genes
3.3. Conserved Motif and Gene Structure Analysis of SlSDG Genes
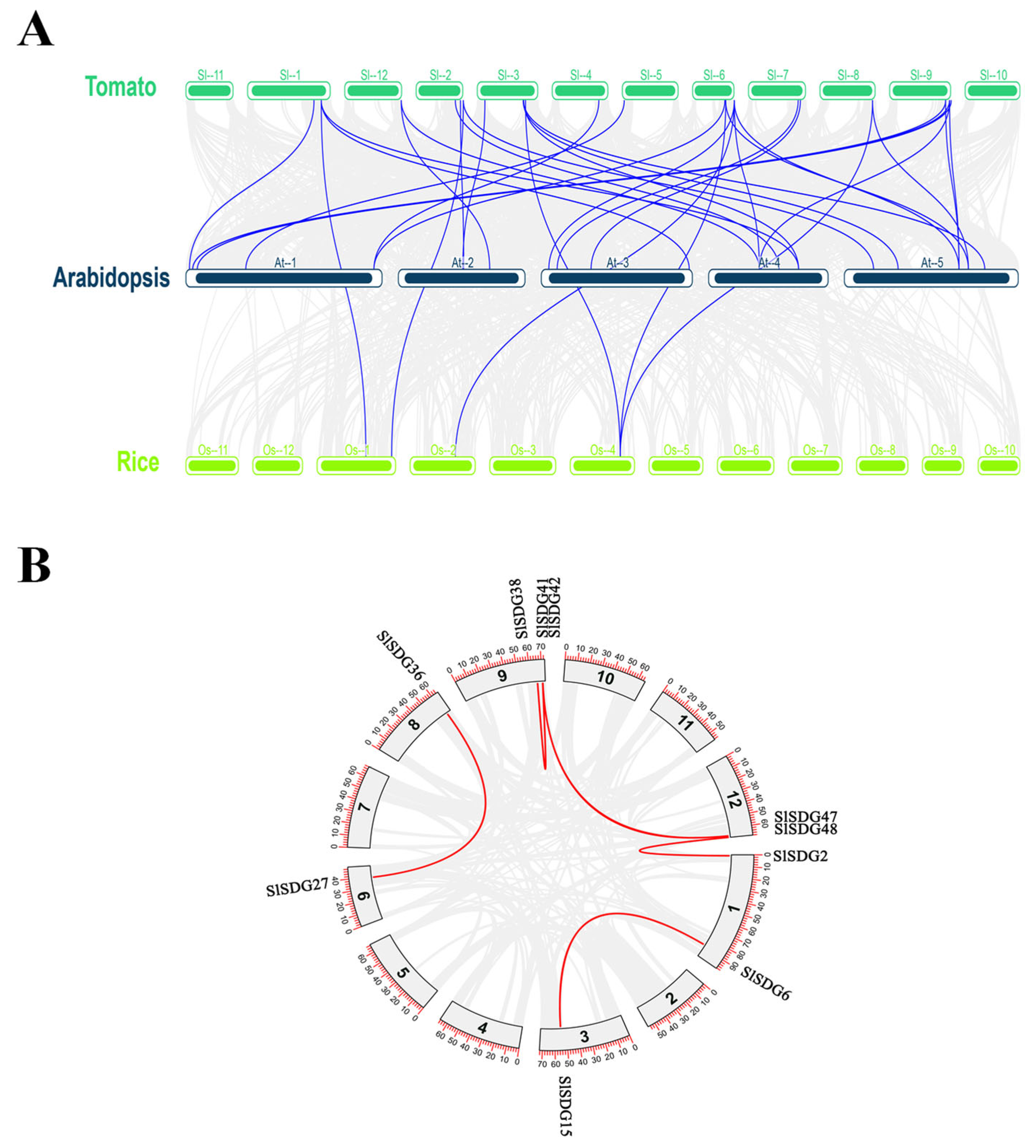
3.4. Collinearity and Duplication Analysis of SlSDG Gene
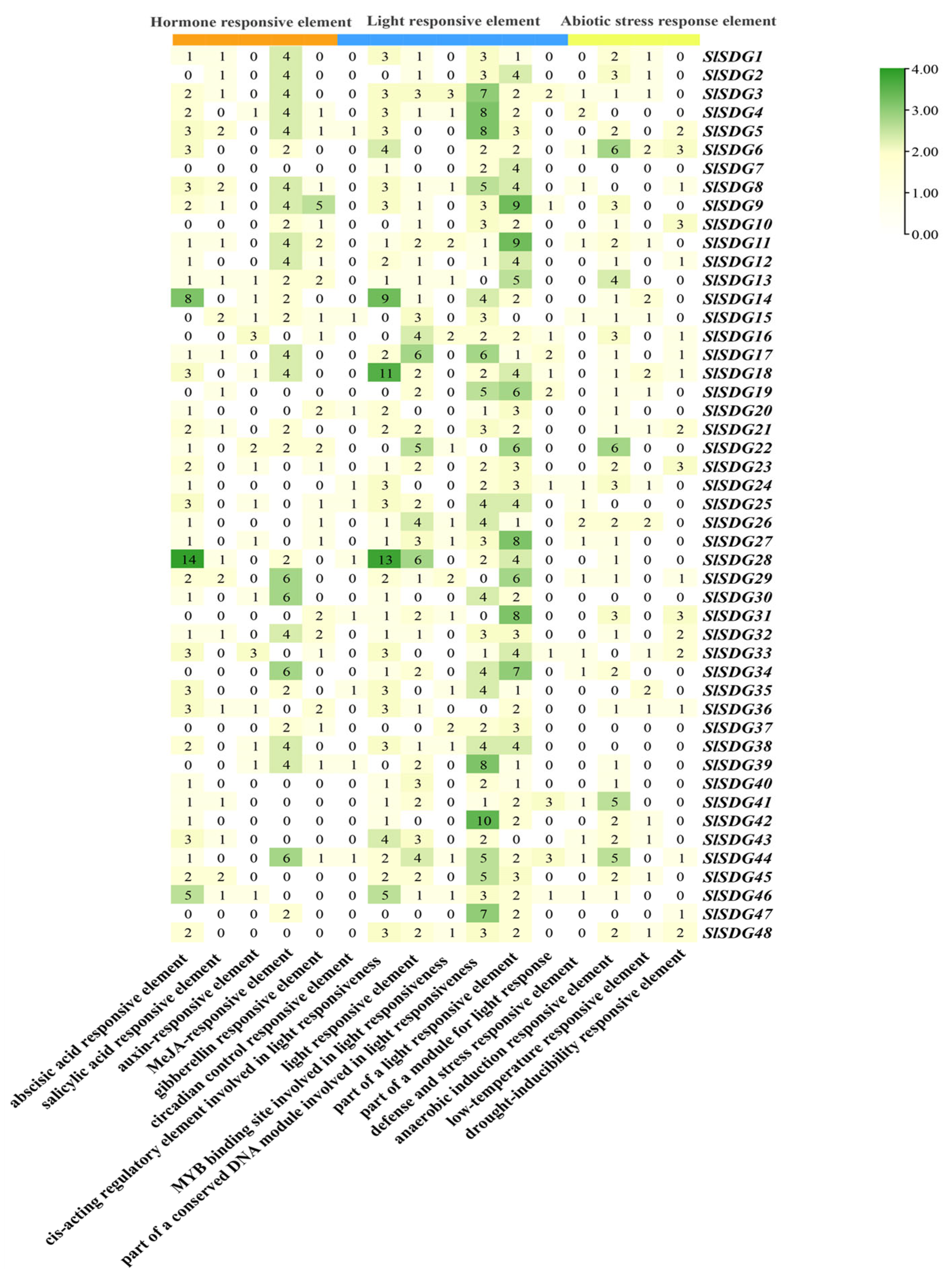
3.5. Cis-Element Analysis of SlSDG Gene Promoters
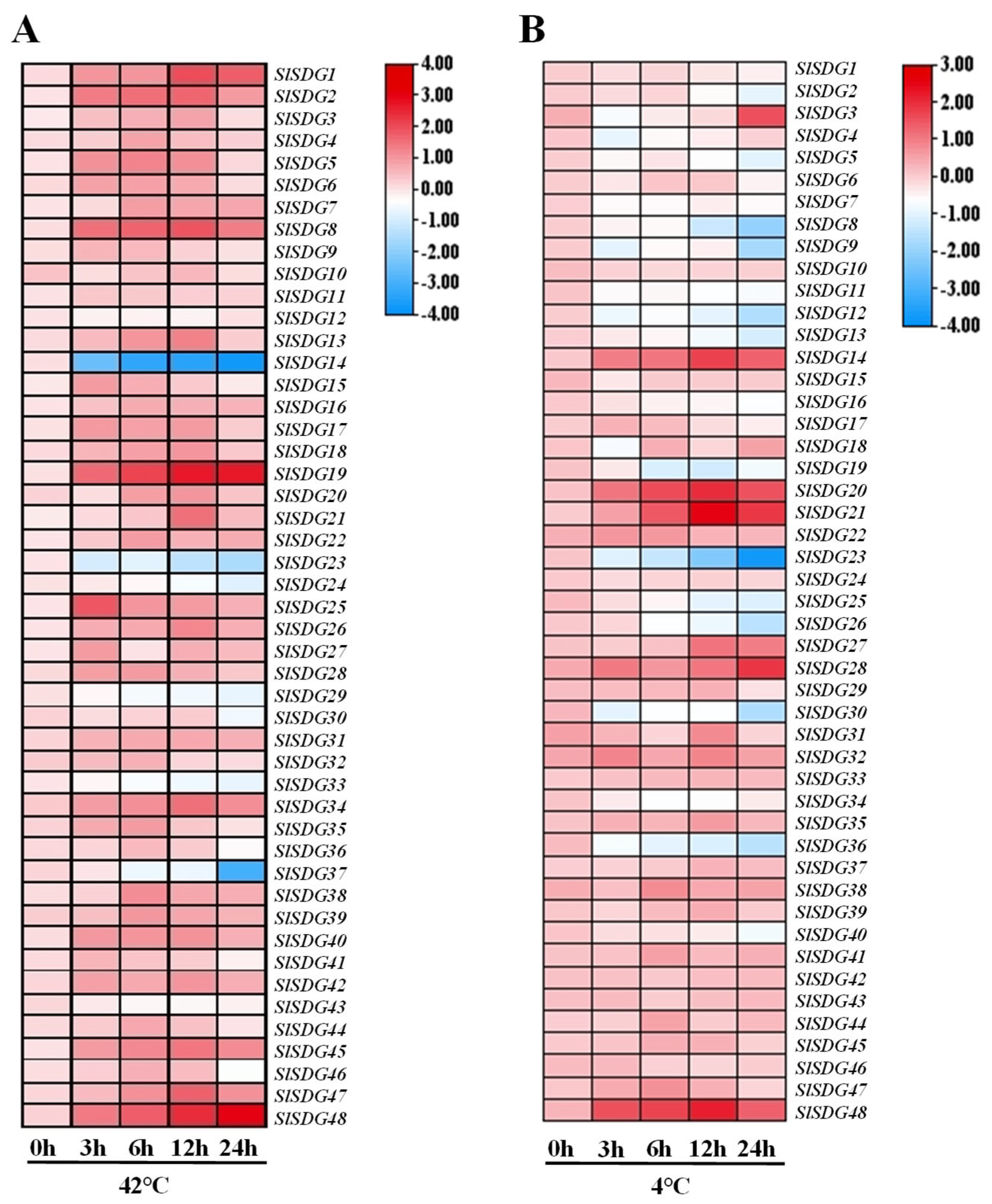
3.6. Expression Patterns of SlSDG Genes Under Temperature Stress
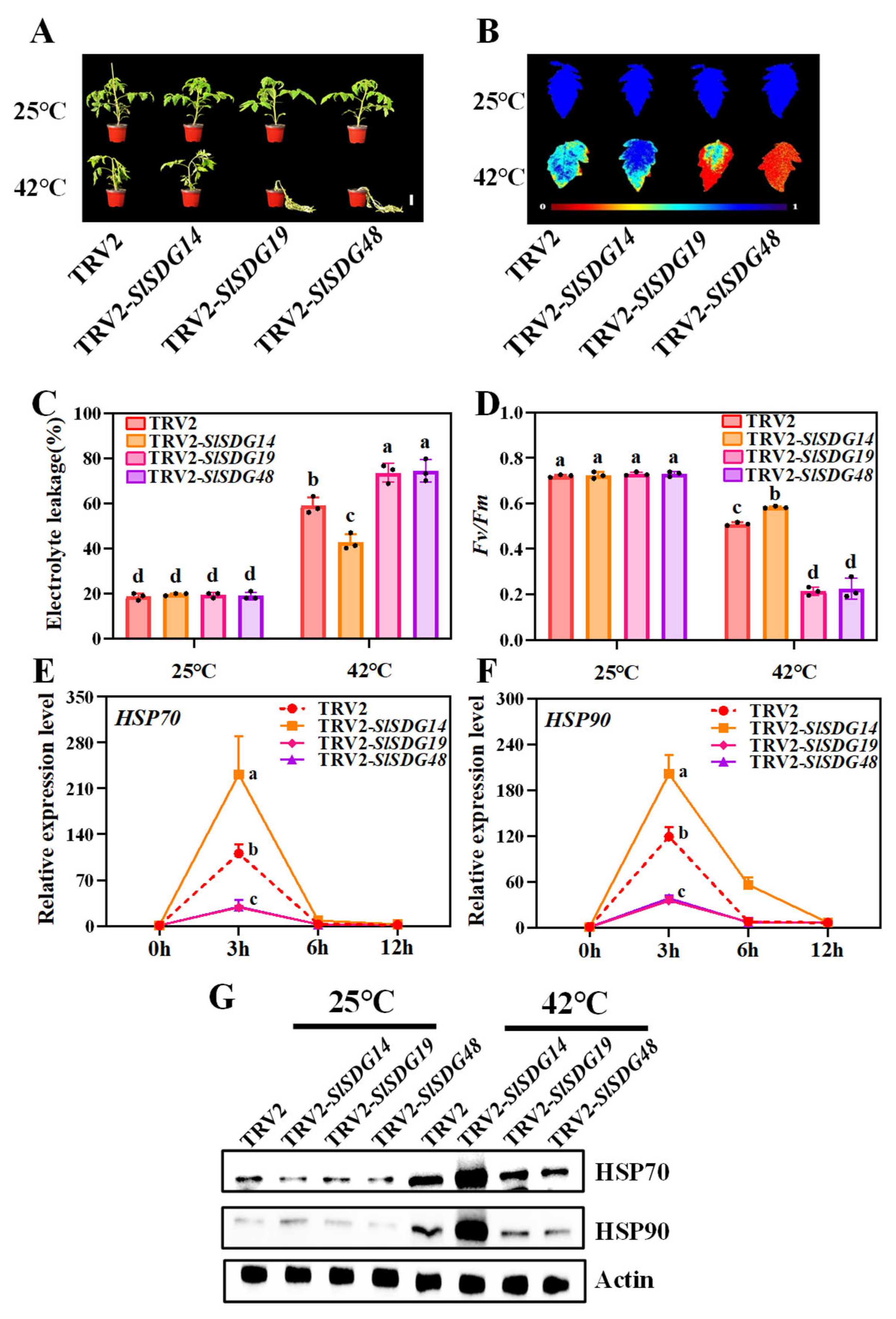
3.7. SlSDG19 and SlSDG48 Positively Regulate and SlSDG14 Negatively Regulates Tomato Heat Tolerance
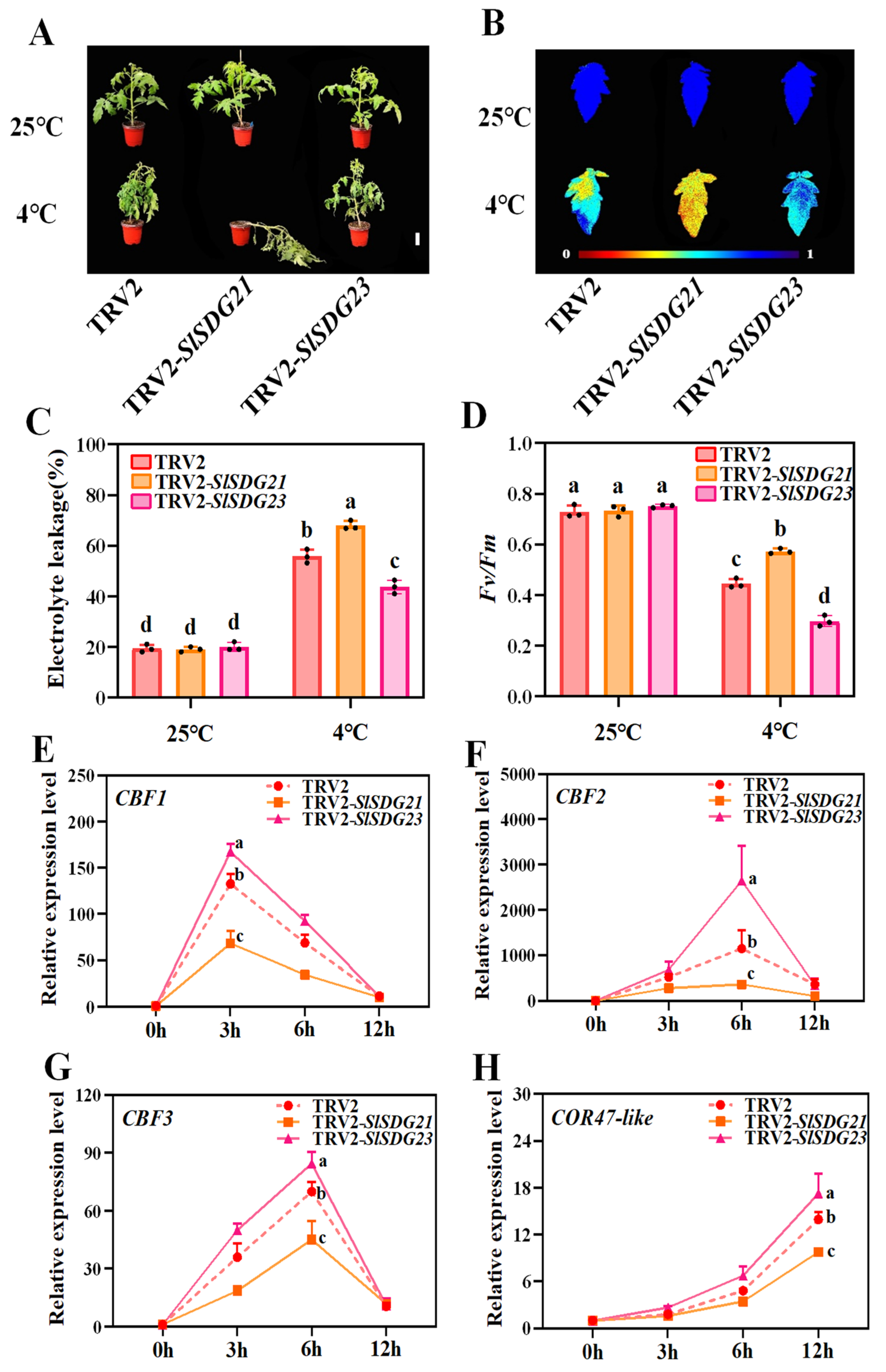
3.8. SlSDG21 Positively Regulates and SlSDG23 Negatively Regulates Tomato Cold Tolerance
4. Discussion
4.1. The Structure, Characteristics and Evolutionary Relationships of Tomato SlSDG Genes
4.2. Transcript and Genetic Analysis Reveal That SlSDGs Regulate Tomato Heat Stress by Influencing the HSP System
4.3. Tomato SlSDG21/23 Modulates Cold Tolerance by Mediating the CBFs-CORs Signaling Pathway
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Liu, C.Y.; Lu, F.L.; Cui, X.; Cao, X.F. Histone methylation in higher plants. Annu. Rev. Plant Biol. 2010, 61, 395–420. [Google Scholar] [CrossRef] [PubMed]
- Jenuwein, T.; Allis, C.D. Translating the histone code. Science 2001, 293, 1074–1080. [Google Scholar] [CrossRef]
- Deng, X.; Song, X.W.; Wei, L.Y.; Liu, C.Y.; Cao, X.F. Epigenetic regulation and epigenomic landscape in rice. Natl. Sci. Rev. 2016, 3, 309–327. [Google Scholar] [CrossRef]
- Xiao, J.; Lee, U.S.; Wagner, D. Tug of war: Adding and removing histone lysine methylation in Arabidopsis. Curr. Opin. Plant Biol. 2016, 34, 41–53. [Google Scholar] [CrossRef]
- Klose, R.J.; Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nat. Rev. Mol. Cell Biol. 2007, 8, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Ng, D.W.K.; Wang, T.; Chandrasekharan, M.B.; Aramayo, R.; Kertbundit, S.; Hall, T.C. Plant SET domain-containing proteins: Structure, function and regulation. Biochim. Biophys. Acta (BBA)—Gene Struct. Expr. 2007, 1769, 316–329. [Google Scholar] [CrossRef]
- Sehrish, S.; Sumbal, W.; Xie, M.L.; Zhao, C.J.; Zuo, R.; Gao, F.; Liu, S.Y. Genome-wide identification and characterization of SET domain family genes in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 1936. [Google Scholar] [CrossRef]
- Zhou, H.Y.; Liu, Y.H.; Liang, Y.W.; Zhou, D.; Li, S.F.; Lin, S.; Dong, H.; Huang, L. The function of histone lysine methylation related SET domain group proteins in plants. Protein Sci. 2020, 29, 1120–1137. [Google Scholar] [CrossRef]
- Thorstensen, T.; Grini, P.E.; Aalen, R.B. SET domain proteins in plant development. Biochim. Biophys. Acta (BBA)—Gene Regul. Mech. 2011, 1809, 407–420. [Google Scholar] [CrossRef]
- Liu, Y.T.; Wang, J.; Liu, B.; Xu, Z.Y. Dynamic regulation of DNA methylation and histone modifications in response to abiotic stresses in plants. J. Integr. Plant Biol. 2022, 64, 2252–2274. [Google Scholar] [CrossRef] [PubMed]
- Alvarez Venegas, R.; Pien, S.; Sadder, M.; Witmer, X.; Grossniklaus, U.; Avramova, Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 2003, 13, 627–637. [Google Scholar] [CrossRef]
- Zheng, J.; Chen, F.Y.; Wang, Z.; Cao, H.; Li, X.Y.; Deng, X.; Soppe, W.J.J.; Li, Y.; Liu, Y.X. A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 2012, 193, 605–616. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Park, O.S.; Choi, C.Y.; Seo, P.J. ARABIDOPSIS TRITHORAX 4 facilitates shoot identity establishment during the plant regeneration process. Plant Cell Physiol. 2019, 60, 826–834. [Google Scholar] [CrossRef]
- Gu, D.C.; Ji, R.J.; He, C.M.; Peng, T.; Zhang, M.Y.; Duan, J.; Xiong, C.Y.; Liu, X.C. Arabidopsis histone methyltransferase SUVH5 is a positive regulator of light-mediated seed germination. Front. Plant Sci. 2019, 10, 841. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Song, Z.T.; Zhang, L.L.; Han, J.J.; Zhou, M.; Liu, J.X. Histone H3K4 methyltransferases SDG25 and ATX1 maintain heat-stress gene expression during recovery in Arabidopsis. Plant J. 2021, 105, 1326–1338. [Google Scholar] [CrossRef]
- Kwon, C.S.; Lee, D.; Choi, G.; Chung, W.I. Histone occupancy-dependent and -independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. Plant J. 2009, 60, 112–121. [Google Scholar] [CrossRef]
- Zhang, X.; Ménard, R.; Li, Y.; Coruzzi, G.M.; Heitz, T.; Shen, W.H.; Berr, A. Arabidopsis SDG8 potentiates the sustainable transcriptional induction of the pathogenesis-related genes PR1 and PR2 during plant defense response. Front. Plant Sci. 2020, 11, 277. [Google Scholar] [CrossRef] [PubMed]
- Berr, A.; McCallum, E.J.; Alioua, A.; Heintz, D.; Heitz, T.; Shen, W.H. Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 2010, 154, 1403–1414. [Google Scholar] [CrossRef]
- Tieman, D.; Zhu, G.T.; Resende, M.F.R.; Lin, T.; Nguyen, C.; Bies, D.; Rambla, J.L.; Beltran, K.S.O.; Taylor, M.; Zhang, B.; et al. A chemical genetic roadmap to improved tomato flavor. Science 2017, 355, 391–394. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, K.; Chen, D.G.; Zhang, Z.Q.; Li, B.Q.; El-Mogy, M.M.; Tian, S.P.; Chen, T. Solanum lycopersicum, a model plant for the studies in developmental biology, stress biology and food science. Foods 2022, 11, 2402. [Google Scholar] [CrossRef]
- Xia, X.-J.; Gao, C.-J.; Song, L.-X.; Zhou, Y.-H.; Shi, K.; Yu, J.-Q. Role of dynamics in brassinosteroid-induced stomatal closure and opening in Solanum lycopersicum. Plant Cell Environ. 2014, 37, 2036–2050. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef]
- Sui, P.F.; Jin, J.; Ye, S.; Mu, C.; Gao, J.; Feng, H.Y.; Shen, W.H.; Yu, Y.; Dong, A.W. H3K36 methylation is critical for brassinosteroid-regulated plant growth and development in rice. Plant J. 2012, 70, 340–347. [Google Scholar] [CrossRef]
- Strahl, B.D.; Ohba, R.; Cook, R.G.; Allis, C.D. Methylation of histone H3 at lysine 4 is highly conserved and correlates with transcriptionally active nuclei in Tetrahymena. Proc. Natl. Acad. Sci. USA 1999, 96, 14967–14972. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.H.; Huang, X.L.; Ouyang, Y.D.; Yao, J.L. Genome-wide identification, phylogenetic and co-expression analysis of OsSET gene family in rice. PLoS ONE 2013, 8, e65426. [Google Scholar] [CrossRef]
- Lei, L.; Zhou, S.L.; Ma, H.; Zhang, L.S. Expansion and diversification of the SET domain gene family following whole-genome duplications in Populus trichocarpa. BMC Evol. Biol. 2012, 12, 51. [Google Scholar] [CrossRef] [PubMed]
- Aiese Cigliano, R.; Sanseverino, W.; Cremona, G.; Ercolano, M.R.; Conicella, C.; Consiglio, F.M. Genome-wide analysis of histone modifiers in tomato: Gaining an insight into their developmental roles. BMC Genom. 2013, 14, 57. [Google Scholar] [CrossRef]
- Lin, T.; Zhu, G.T.; Zhang, J.H.; Xu, X.Y.; Yu, Q.H.; Zheng, Z.; Zhang, Z.H.; Lun, Y.Y.; Li, S.; Wang, X.X.; et al. Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef]
- Su, X.; Wang, B.; Geng, X.L.; Du, Y.F.; Yang, Q.Q.; Liang, B.; Meng, G.; Gao, Q.; Yang, W.C.; Zhu, Y.F.; et al. A high-continuity and annotated tomato reference genome. BMC Genom. 2021, 22, 898. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Z.Y.; Bao, Z.G.; Li, H.B.; Lyu, Y.Q.; Zan, Y.J.; Wu, Y.Y.; Cheng, L.; Fang, Y.H.; Wu, K.; et al. Graph pangenome captures missing heritability and empowers tomato breeding. Nature 2022, 606, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; He, Q.; Wang, J.; Wang, B.; Zhao, J.T.; Huang, S.Y.; Yang, T.; Tang, Y.P.; Yang, S.B.; Aisimutuola, P.; et al. Super-pangenome analyses highlight genomic diversity and structural variation across wild and cultivated tomato species. Nat. Genet. 2023, 55, 852–860. [Google Scholar] [CrossRef]
- Zhang, L.S.; Ma, H. Complex evolutionary history and diverse domain organization of SET proteins suggest divergent regulatory interactions. New Phytol. 2012, 195, 248–263. [Google Scholar] [CrossRef]
- Chen, D.H.; Qiu, H.L.; Huang, Y.; Zhang, L.; Si, J.P. Genome-wide identification and expression profiling of SET DOMAIN GROUP family in Dendrobium catenatum. BMC Plant Biol. 2020, 20, 40. [Google Scholar] [CrossRef]
- Zhu, X.Y.; Ma, H.; Chen, Z.D. Phylogenetics and evolution of Su(var)3-9 SET genes in land plants: Rapid diversification in structure and function. BMC Evol. Biol. 2011, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.H.; Hu, S.L.; Guo, F.; Zhao, H.; Wang, M.L.; Ni, D.J.; Wang, Y.; Wang, P. Characterization of the SET DOMAIN GROUP gene family members in Camellia sinensis and functional analysis of the SDG43 gene in abiotic stresses. Environ. Exp. Bot. 2021, 182, 104306. [Google Scholar] [CrossRef]
- Li, Z.X.; Yao, Z.P.; Ruan, M.Y.; Wang, R.Q.; Ye, Q.J.; Wan, H.J.; Zhou, G.Z.; Cheng, Y.; Guo, S.J.; Liu, C.C.; et al. The PLA gene family in tomato: Identification, phylogeny, and functional characterization. Genes 2025, 16, 130. [Google Scholar] [CrossRef] [PubMed]
- Pang, X.; Chen, J.; Xu, Y.; Liu, J.; Zhong, Y.; Wang, L.; Zheng, J.; Wan, H. Genome-wide characterization of ascorbate peroxidase gene family in pepper (Capsicum annuum L.) in response to multiple abiotic stresses. Front. Plant Sci. 2023, 14, 1189020. [Google Scholar] [CrossRef]
- Li, W.J.; Yan, J.J.; Wang, S.C.; Wang, Q.Y.; Wang, C.X.; Li, Z.X.; Zhang, D.H.; Ma, F.W.; Guan, Q.M.; Xu, J.D. Genome-wide analysis of SET-domain group histone methyltransferases in apple reveals their role in development and stress responses. BMC Genom. 2021, 22, 283. [Google Scholar] [CrossRef]
- Huang, Y.; Mo, Y.J.; Chen, P.Y.; Yuan, X.L.; Meng, F.N.; Zhu, S.W.; Liu, Z. Identification of SET Domain-containing proteins in Gossypium raimondii and their response to high temperature stress. Sci. Rep. 2016, 6, 32729. [Google Scholar] [CrossRef] [PubMed]
- Yancoskie, M.N.; Maritz, C.; van Eijk, P.; Reed, S.H.; Naegeli, H. To incise or not and where: SET-domain methyltransferases know. Trends Biochem. Sci. 2023, 48, 321–330. [Google Scholar] [CrossRef]
- Liu, Y.T.; Zhang, A.; Yin, H.; Meng, Q.X.; Yu, X.M.; Huang, S.Z.; Wang, J.; Ahmad, R.; Liu, B.; Xu, Z.Y. Trithorax-group proteins ARABIDOPSIS TRITHORAX4 (ATX4) and ATX5 function in abscisic acid and dehydration stress responses. New Phytol. 2018, 217, 1582–1597. [Google Scholar] [CrossRef]
- Liu, Y.T.; Chen, X.; Xue, S.Y.; Quan, T.Y.; Cui, D.; Han, L.Z.; Cong, W.X.; Li, M.T.; Yun, D.J.; Liu, B.; et al. SET DOMAIN GROUP 721 protein functions in saline–alkaline stress tolerance in the model rice variety Kitaake. Plant Biotechnol. J. 2021, 19, 2576–2588. [Google Scholar] [CrossRef]
- Askari Khorasgani, O.; Pessarakli, M. Protective roles of plant proteins in conferring tolerance to heat stress. J. Plant Nutr. 2019, 42, 1114–1123. [Google Scholar] [CrossRef]
- Yamaguchi, N.; Matsubara, S.; Yoshimizu, K.; Seki, M.; Hamada, K.; Kamitani, M.; Kurita, Y.; Nomura, Y.; Nagashima, K.; Inagaki, S.; et al. H3K27me3 demethylases alter HSP22 and HSP17.6C expression in response to recurring heat in Arabidopsis. Nat. Commun. 2021, 12, 3480. [Google Scholar] [CrossRef] [PubMed]
- Potok, M.E.; Zhong, Z.H.; Picard, C.L.; Liu, Q.K.; Do, T.; Jacobsen, C.E.; Sakr, O.; Naranbaatar, B.; Thilakaratne, R.; Khnkoyan, Z.; et al. The role of ATXR6 expression in modulating genome stability and transposable element repression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2022, 119, e2115570119. [Google Scholar] [CrossRef] [PubMed]
- Bvindi, C.; Lee, S.; Tang, L.; Mickelbart, M.V.; Li, Y.; Mengiste, T. Improved pathogen and stress tolerance in tomato mutants of SET domain histone 3 lysine methyltransferases. New Phytol. 2022, 235, 1957–1976. [Google Scholar] [CrossRef]
- Ding, Y.L.; Shi, Y.T.; Yang, S.H. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704. [Google Scholar] [CrossRef] [PubMed]
- Song, C.B.; Yang, Y.Y.; Yang, T.W.; Ba, L.J.; Zhang, H.; Han, Y.C.; Xiao, Y.Y.; Shan, W.; Kuang, J.F.; Chen, J.Y.; et al. MaMYB4 recruits histone deacetylase MaHDA2 and modulates the expression of ω-3 fatty acid desaturase genes during cold stress response in banana fruit. Plant Cell Physiol. 2019, 60, 2410–2422. [Google Scholar] [CrossRef]
- Liu, L.J.; Song, Y.; Xu, J.; Li, D.M.; Li, G.P.; An, L.Z. Differential expression by chromatin modifications of alcohol dehydrogenase 1 of Chorispora bungeana in cold stress. Gene 2017, 636, 1–16. [Google Scholar] [CrossRef]
- To, T.K.; Nakaminami, K.; Kim, J.-M.; Morosawa, T.; Ishida, J.; Tanaka, M.; Yokoyama, S.; Shinozaki, K.; Seki, M. Arabidopsis HDA6 is required for freezing tolerance. Biochem. Biophys. Res. Commun. 2011, 406, 414–419. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lu, C.; Cheng, Y.; Wan, H.; Yao, Z.; Ruan, M.; Wang, R.; Ye, Q.; Zhou, G.; Wang, H.; Liu, C. Identification of Tomato SET Domain Group Gene Family and Function Analysis Under Temperature Stress. Horticulturae 2025, 11, 958. https://doi.org/10.3390/horticulturae11080958
Lu C, Cheng Y, Wan H, Yao Z, Ruan M, Wang R, Ye Q, Zhou G, Wang H, Liu C. Identification of Tomato SET Domain Group Gene Family and Function Analysis Under Temperature Stress. Horticulturae. 2025; 11(8):958. https://doi.org/10.3390/horticulturae11080958
Chicago/Turabian StyleLu, Chuanlong, Yuan Cheng, Hongjian Wan, Zhuping Yao, Meiying Ruan, Rongqing Wang, Qingjing Ye, Guozhi Zhou, Huasen Wang, and Chenxu Liu. 2025. "Identification of Tomato SET Domain Group Gene Family and Function Analysis Under Temperature Stress" Horticulturae 11, no. 8: 958. https://doi.org/10.3390/horticulturae11080958
APA StyleLu, C., Cheng, Y., Wan, H., Yao, Z., Ruan, M., Wang, R., Ye, Q., Zhou, G., Wang, H., & Liu, C. (2025). Identification of Tomato SET Domain Group Gene Family and Function Analysis Under Temperature Stress. Horticulturae, 11(8), 958. https://doi.org/10.3390/horticulturae11080958






