Abstract
Fritillaria unibracteata is a rare and endangered medicinal plant in the Liliaceae family, whose bulbs have been used in traditional Chinese traditional medicine for over 2000 years. The mevalonate (MVA) pathway is involved in the growth, development, response to environmental stress, and active ingredient production of plants; however, the functional characterization of MVA-pathway genes in the Liliaceae family remains poorly documented. In this study, an Acetyl-CoA C-acetyltransferase gene (FuAACT) was first cloned from F. unibracteata. It exhibited structural features of the thiolase family and showed the highest sequence identity with the Dioscorea cayenensis homolog. The Km, Vmax, and Kcat of the recombinant FuAACT were determined to be 3.035 ± 0.215 μM, 0.128 ± 0.0058 μmol/(min·mg), and 1.275 ± 0.0575 min−1, respectively. The optimal catalytic conditions for FuAACT were ascertained to be 30 °C and pH 8.9. It was stable below 50 °C. His361 was confirmed to be a key amino acid residue to enzymatic catalysis by site-directed mutagenesis. Subsequent subcellular localization experiments demonstrated that FuAACT was localized in chloroplasts and cytoplasm. FuAACT-overexpressing transgenic Arabidopsis thaliana plants showed higher drought tolerance than wild-type plants. This phenotypic difference was corroborated by significant differences in seed germination rate, lateral root number, plant height, and leaf number (p < 0.05). Furthermore, the FuAACT transgenic plants resulted in the formation of a more developed fibrous root system. These results indicated that the FuAACT gene revealed substantial biological activity in vitro and in vivo, hopefully providing the basis for its further research and application in liliaceous ornamental and medicinal plants.
1. Introduction
The Liliaceae family is composed of a variety of economically ornamental and medicinal plants. Among them, Fritillaria unibracteata Hsiao et K.C.Hsia (F. unibracteata) is a perennial herb that belongs to the genus Fritillaria, which is a resource of ornamental plants and pharmaceutical metabolites [1]. It is one of the original plants of the important medicinal herb “Chuan Beimu” [2]. It was originally distributed in alpine areas of western Sichuan, southern Qinghai, and southern Gansu with their altitudes mainly ranging from 3200 to 4500 m [3,4]. Furthermore, temperature and water supply play significant roles in the growth, development, and distribution of F. unibracteata [4]. F. unibracteata is rich in diverse steroidal alkaloids, terpenoids, and steroidal saponins, which are associated with significant pharmacological activities, such as the ability to promote expectoration, suppress coughing, and relieve asthma [5,6]. Subject to long-term wild resource exploitation, specific environmental needs, and a 4-5-year perennial life cycle, F. unibracteata has become endangered and been designated a Level II protected species in China. Although artificial cultivation has been applied to partly alleviate the pressure on wild resources, the limited increase in supply has not met the tremendous clinical demand [7]. Therefore, the functional identification of genes related to the biosynthesis of active ingredients in F. unibracteata will provide candidate genes for the metabolic engineering of active ingredients, with which wild resources of F. unibracteata will be eventually protected.
The biosynthesis of steroidal alkaloids, terpenoids, and steroidal saponins relies on the mevalonate (MVA) pathway in the cytoplasm and the 2-methyl-D-erythritol 4-phosphate (MEP) pathway in the plastid [5,8], which are involved in the formation of skeleton. Acetyl-CoA C-acetyltransferase (AACT), a member of the thiolase family, is responsible for the initial step of the MVA pathway, which catalyzes the condensation of two acetyl-CoA molecules to form acetoacetyl-CoA. The expression of AACT genes from various species has revealed species-specific expression patterns. For instance, Bacopa monniera AACT (BmAACT) is enriched in roots and petals [9], A. thaliana AACT2 (AtAACT2) is strongly expressed in seedlings, roots and inflorescences, while it is weaker in stems and leaves [10], and Houttuynia cordata AACT (HcAACT) has the highest transcriptional abundance in stems and rhizomes [11]; however, Ginkgo biloba AACT (GbAACT) is enriched in fruits and leaves [12]. More importantly, the thiolase family directly affects the biosynthesis of sterols, terpenoids, and phytohormones [13,14,15]. Positive correlation between AACT levels and saponin production was observed in Platycodon grandiflorus and Rehmannia glutinosa [16,17]. In Ganoderma lucidum, AACT transcript levels showed a similar significant positive correlation with triterpenoid production. This finding suggests that AACT plays a central role in regulating terpenoid metabolic pathways [18]. Moreover, Sando et al. [19] found that overexpression of the HbAACT gene promoted yeast growth, and Soto et al. [20] found that overexpression of the MsAACT1 gene enhanced both salt tolerance and squalene production in transgenic plants. Another study confirmed that the overexpression of AACT could improve the stress tolerance of B. monniera [9]. Collectively, these results suggest that AACT displays divergent expression profiles and functional roles in different plant species, making functional investigations of AACT across various plant lineages essential.
In our previous work, a total of five candidate FuAACT unigenes were obtained from F. unibracteata by de novo sequencing [5], and therefore, it is essential to carry out functional characterization of these candidate members. In this study, our main aims were to clone one FuAACT gene from F. unibracteata and to investigate its expression pattern in various tissues first. Second, the heterologous expression of the FuAACT gene in E. coli was measured, and the kinetic parameters of recombinant FuAACT were determined. Third, His361 was substituted with Ala, thereby confirming the essential role of His361 in condensation reaction. In addition, the subcellular localization of FuAACT was determined. Finally, transgenic A. thaliana plants overexpressing the FuAACT gene were constructed, and their drought tolerance was subsequently evaluated. To our knowledge, this paper is among the few studies concerning the MVA pathway genes in liliaceous plants. This work will contribute to providing a basis for further verification of other FuAACTs and other genes encoding MVA pathway enzymes.
2. Materials and Methods
2.1. Materials
2.1.1. Strain and Plasmid
E. coli strains (BL21, DH5α™) harboring pET28a-FuAACT, pGreenII0229-GFP, and pCambia2301 plasmids, along with Agrobacterium tumefaciens GV3101, were stored in the laboratory. All strains were purchased from Sangon Biotech (Shanghai, China), while study primers designed by Primer Premier 5 were synthesized by Tsingke Biotechnology (Beijing, China).
2.1.2. Reagents and Plant Materials
This study used a Plasmid Mini Kit I D6943 (OMEGA Bio-tek, Guangzhou, China), 1-5TM 2× High Fidelity Master Mix (Tsingke, Beijing, China), Mut Express II Fast Mutagenesis Kit V2 (Vazyme, Nanjing, China), Gel Extraction Kit D2500 (OMEGA Bio-tek, Guangzhou, China), BCA Protein Assay Kit (Sangon, Shanghai, China), and Prime STAR HS DNA Polymerase (Takara, Dalian, China).
Fresh F. unibracteata samples from Qinghai Lvkang Biol. Dev. Co., Huzhu, Xining, Qinghai, China, were rinsed with distilled water and stored at −80 °C for analyses. Wild-type A. thaliana (ecotype Columbia-0) and Nicotiana benthamiana (N. benthamiana, ecotype LAB) seeds were kept in our laboratory.
2.1.3. Experimental Apparatus
Thus study employed a VeritiTM96 Well Fast Thermal Cycler (Thermo Fisher, Waltham, MA, USA) and Laser confocal microscopy A1R+ (Nikon 1774059S, Tokyo, Japan).
2.2. Experimental Methods
2.2.1. Cloning and Bioinformatics Analysis of FuAACT
Transcriptome data of F. unibracteata tissues were previously obtained [5]. Based on the de novo sequence of FuAACT from the transcriptome data, the coding region of FuAACT was amplified using forward primer FuAACT—F1: 5′-GGATCCATGGCAAGCCTGCA-3′ (BamHI site in italics) and reverse primer FuAACT—R1: 5′-GAATTCTTAAATCAGTTCCAGAACCAGTGC-3′ (EcoRI site in italics). The amplified products were separated by agarose gel electrophoresis, gel-purified, digested with BamHI and EcoRI, and cloned into pET28a. The recombinant plasmid pET28a-FuAACT was sequenced to confirm that no substitutions or deletions occurred.
AACTs from various plants were downloaded from the National Center for Biotechnology Information (NCBI) database. The isoelectric point and molecular weight were calculated on the ExPASy website (https://web.expasy.org/protparam/, accessed on 28 January 2025). The homology modeling of FuAACT was followed with the SWISS-MODEL tool (http://swissmodel.expasy.org/, accessed on 10 April 2025). The final model of FuAACT was docked with the substrates using Autodock vina 1.5.6 [21]. Secondary structure prediction was performed using the Prabi online tool (https://npsa-prabi.ibcp.fr/, accessed on 10 June 2025). Multiple sequence alignment was performed on the AACTs from various species using MEGA 12.0 software. Following, a phylogenetic tree was constructed using the maximum likelihood (ML) approach on MEGA 12.0 software. The domains of the predicted encoded proteins were analyzed using InterPro (https://www.ebi.ac.uk/interpro/, accessed on 28 January 2025). Shannon entropy allows for the quantification of protein variation, whose value (H(X)) was calculated using the following equation in Python 3.13 software:
where p(xi) represents the probability of a random variable xi.
2.2.2. Protein Expression and Purification
The recombinant pET28a-FuAACT plasmid was transformed into E. coli BL21 (DE3) using the heat shock protocol. A single colony was subjected to rotation culturation at 37 °C in LB liquid medium containing 50 μg/mL kanamycin until OD600 reached 0.6. After induction with 0.1 mM Isopropyl-β-d-thiogalactoside (IPTG) overnight at 23 °C at 100 rpm, cells were pelleted, resuspended in binding buffer (10 mM Tris-HCl, pH 7.9, 500 mM NaCl, 20 mM imidazole), and sonicated at 30% power for 45 min (3 s on/3 s off). The supernatant was loaded onto Ni2+-NTA resin and eluted with 200 mM imidazole buffer to yield purified recombinant FuAACT.
The eluted recombinant proteins underwent sequential dialysis against 10% glycerol, 250 mM NaCl, and 2 mM Tris-HCl (pH 7.4) at 4 °C for 4 h, followed by 150 mM NaCl (same buffer) overnight. Crude and purified protein samples (10 μg each) were resolved by 12.5% SDS-PAGE and visualized via Coomassie Blue staining.
2.2.3. Enzymatic Assay of FuAACT
FuAACT (1 mL) was incubated with acetyl-CoA in 100 mM Tris-HCl (pH 8.8) containing MgCl2 at 30 °C for 12 min, and acetoacetyl-CoA formation was monitored spectrophotometrically at 303 nm (the Mg2+-enolate complex; Thermo Fisher). Various temperatures, including 27 °C, 30 °C, 37 °C, 43 °C, and 50 °C, were adopted to measure the optimal temperature. Meanwhile, the relative activities of FuAACT at various pH environments, such as 6.4, 7.8, 8.4, 8.9, 9.4, and 9.9, were measured, and the given pH exhibiting the highest activity was determined as the optimal pH.
Thermal stability was assessed by pre-incubating FuAACT at 40, 50, 60, and 70 °C for 1 h, followed by residual activity assays under optimal conditions (pH 8.8, 30 °C).
2.2.4. Site Mutagenesis
To ascertain the role of His361 in the catalytic function of FuAACT, the His361 residue was replaced with an Ala residue using a pair of primers FuAACT-361H-A:5′-TAGTCTGGGTGCACCGCTGGGTTGCA-3′ and FuAACT-361H-A: 5′-CAGCGGTGCACCCAGACTAACTGCA-3′. According to the procedure of the Point Mutation Kit (Tiandz, China), pET28-FuAACT was used as a template to generate the mutated pET28-FuAACTH361A plasmid [22]. Transformation, expression, purification, and activity assays of the mutant were conducted as previously described (Section 2.2.2 and Section 2.2.3).
2.2.5. Subcellular Localization of FuAACT
The prediction of protein subcellular localization was performed using various tools, including SignalP v4.1 (https://services.healthtech.dtu.dk/services/SignalP-4.1/, for the prediction of signal peptides, accessed on 28 July 2025), TMHMM Server v2.0 (https://services.healthtech.dtu.dk/services/TMHMM-2.0/, for the prediction of transmembrane helices, accessed on 28 July 2025) and WoLF PSORT (https://wolfpsort.hgc.jp/, for the prediction of subcellular localization, accessed on 28 July 2025) [23,24].
Using the pET28a-FuAACT plasmid as a template, primers FuAACT -F2: 5′-GAATTCATGGCAGCCGGGAGCGTT-3′ (EcoRI site in italics) and FuAACT -R2: 5′-CGCGGATCCGCGAATCAGTTTATGAATGG-3′ (BamHI site in italics) were applied to amplify the FuAACT fragment. Following digestion with EcoRI and BamHI, the PCR product was ligated into the corresponding sites of the pGreenII0229-GFP vector to generate the pGreenII0229-FuAACT-GFP translational fusion. The pGreenII0229-FuAACT-GFP plasmid was transformed into A. tumefaciens GV3101 using the freeze–thawing method and incubated in LB solid medium (containing 50 μg/mL gentamicin and 50 μg/mL kanamycin) to generate recombinant A. tumefaciens GV3101.
A. tumefaciens GV3101 carrying pGreenII0229-FuAACT-GFP was infiltrated into N. benthamiana leaves (1 mL) and incubated under low light for 2 days prior to confocal imaging. Chloroplast autofluorescence and GFP were excited at 640 nm (emission at 675 nm) and 488 nm (emission at 510 nm), respectively.
2.2.6. Expression Pattern of FuAACT Gene
The quantitative expression of the FuAACT gene in flowers, leaves, stems, and bulbs of F. unibracteata was analyzed by real-time PCR. Gene-specific primer pairs (5′-GCTGAAGAAACTCCGACC-3′ and 5′-CAAGAATGCGAGCACCACT-3′) were designed, adopting the 18S rRNA gene as the reference gene [25]. Each reaction consisted of 1 μL cDNA template in 14 μL reaction buffer, with thermal cycling conditions as follows: 1 cycle at 95 °C for 60 s, followed by 40 cycles at 95 °C for 10 s, 50 °C for 10 s, and finally at 72 °C for 10 s. Each reaction was repeated three times. Following the reaction, fluorescence curves were analyzed, and the relative expression of the FuAACT gene was calculated using the 2−ΔΔCt method.
2.2.7. Construction of Transgenic A. thaliana Overexpressing FuAACT
The FuAACT gene fragment was amplified with primers FuAACT-F4: 5′-GCGGGTCGACGGTACCATGGCAAGCCTGCAGGCC-3′ and FuAACT-R4: 5′-TAGACATATGGGTACCTTAAATCAGTTCCAGAACCAGTG-3′ (KpnI site in italics). The amplified product and pCambia2301 plasmid were ligated via the homologous recombination method to generate the recombinant pCambia2301- FuAACT plasmid.
The plasmid pCambia2301-FuAACT was then transformed into A. tumefaciens GV3101 followed by the transformation of Arabidopsis thaliana using the leaf dip method. Transgenic lines were screened on half-strength MS medium supplemented with 50 μg/mL kanamycin. The DNA-based identification of transgenic lines was performed by PCR using primers 5′-GACGCACAATCCCACTATCC-3′ and 5′-GCTTCCGGCTCGTATGTTG-3′. The RNA-based identification of transgenic lines was performed by PCR using primers 5′-TATCTGGCAGAAGCACGCAA-3′ and 5′-TCTGCACACACACCCATACC-3′.
2.2.8. Statistical Analysis
All experimental values represent the mean ± SD of triplicates. An independent t-test or one-way ANOVA test was used to determine whether there was a statistically significant difference between the two groups. Statistical significance was defined as p < 0.05.
3. Results
3.1. Sequence Analysis of FuAACT Gene
The FuAACT gene was successfully cloned from the rhizome of F. unibracteata and submitted to NCBI under accession number PV706214. The FuAACT gene consists of 1218 bp with a GC content of 46%, encoding a 405-amino-acid peptide with a molecular weight of 41.79 kD. To evaluate the evolutionary relationship of FuAACT, 60 AACTs from various species were collected, including 54 sequences from angiosperms, 1 sequence from moss, 1 sequence from algae, and 4 sequences from fungi. The maximum likelihood (ML) method was used to construct a phylogenetic tree, with bootstrap values exceeding 70 for 80.05% of nodes, confirming its high reliability (Figure 1). In the ML tree, fungi AACTs formed a root branch with high bootstrap (99 BP), indicating that they might represent ancestral genes for AACTs in algae, muscus, and spermatophyte. The ingroup corresponding to spermatophyte was strongly supported (100 BP), in which angiosperm was monophyletic (99 BP) and a sister to Ginkgo biloba from gymnosperm, suggesting that AACT from angiosperms might originate from AACT from gymnosperm. Furthermore, dicots, as the largest plant group, were paraphyletic, with the majority falling into a moderately supported clade (95 BP). FuAACT clustered closely with monocot AACTs, supported by relatively high bootstrap values (100 BP). Since FuAACT and Dioscorea cayenensis AACTs formed the closest clade (98 BP), a close genetic relationship between the two species was proposed.
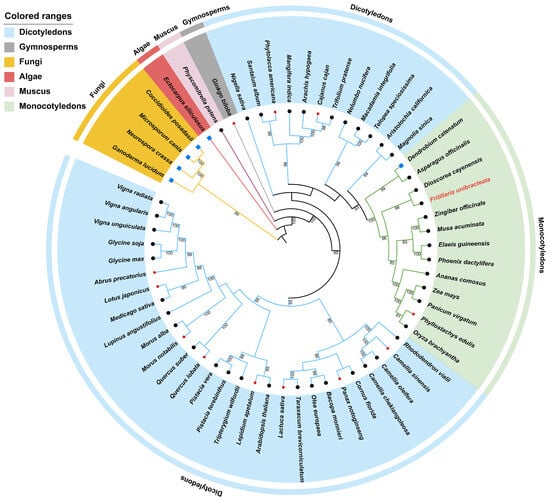
Figure 1.
Phylogenetic relationship of 60 species inferred from maximum likelihood tree based on nucleotide sequence. Numbers above nodes are supporting values with ML bootstrap values. Bootstrap values lower than 70 are not displayed. Species from dicotyledons, gymnosperms, fungi, algae, muscus, and monocotyledons are marked with light blue, gray, yellow, pink, purple, and green, respectively.
3.2. Conserved Motif Analysis of AACT
InterPro analysis identified two conserved domains in the FuAACT protein: IPR020616 (N-terminal; 15–274 amino acids) and IPR020617 (C-terminal; 283–403 amino acids). Two motifs involved in the catalytic function, NVHGGAVSLGHPLGCSG (blue box in Supplementary Material S2) and GVASVCNGGGASA (green box in Supplementary Material S2), were significant and were also detected in the C-terminal regions of FuAACT. The core heptapeptide CNGGGAS catalyzes the Claisen condensation of acetyl-CoA to acetoacetyl-CoA [26,27]. Moreover, several key residues such as Cys99, His361, and Cys391 were absolutely conserved in all AACTs (Supplementary Material S2). Specifically, Cys99 is involved in the formation of acylase intermediates, while Cys391 and His361 are responsible for the deprotonation and stabilization of the transition state, respectively [28]. Conserved motif analysis indicated that a total of 11 motifs (Figure 2) were distributed in AACTs with p-values ranging from 1.69 × 10−99 to 3.48 × 10−194 (Supplementary Material S3), confirming high reliability of this analysis. Among them, motifs 2, 3, 4, 5, 6, 9, and 10 were conserved across all AACTs. Two key functional motifs, NVHGGAVSLGHPLGCSG and GVASVCNGGGASA, were distributed in motif 10 and motif 11, respectively. Thus, whether D. catenatum AACT lacks thiolase activity due to the absence of motif 11 needs to be verified in the future. Overall, these conserved motifs and key residues observed in FuAACT imply that FuAACT might share enzymatic activity similar to that of other AACTs.
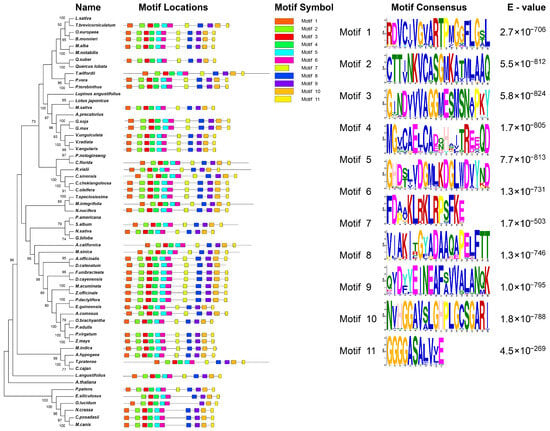
Figure 2.
Conserved motif analysis of 48 AACT proteins.
3.3. Protein Variability Analysis of AACT
The quantification of protein variation was achieved by calculating Shannon entropy between FuAACT and AACT proteins from monocot and dicot plants. The mean Shannon entropy of AACTs in monocots (0.256 ± 0.222) was significantly lower than in dicots (0.469 ± 0.362), reflecting greater sequence divergence between FuAACT and dicot AACTs compared to monocot AACTs (Supplementary Material S4). Whereas monocots showed minimal variability (Shannon entropy ≥ 2.0), dicot AACTs possessed six highly variable N-terminal loci (Shannon entropy ≥ 2.0), demonstrating that the N-terminal region underlies sequence variability. The top five conserved regions within the monocotyledonous and dicotyledonous AACT proteins were identified with an average Shannon entropy ranging from 0.034 to 0.157 (Table 1), where residues from Thr373 to Gly393 contained the core catalytic Cys391-Asn392-Gly393 tripeptide.

Table 1.
Analysis of top five conserved regions of AACT proteins.
3.4. Homology Modeling and Important Catalytic Sites of FuAACT
The Prabi online tool predicted that α-helix was the main secondary structure in FuAACT, comprising 178 residues (43.95%), followed by random coils (31.85%), extended strand (15.06%), and β-turn (9.14%). This conformational feature is consistent with those of typical thiolases [29]. To identify key residues critical for enzymatic activity, homology modeling was conducted using SWISS-MODEL with template 5xyj.1.A (53.92% identity to FuAACT). Structural visualization using PyMOL software confirmed that α-helix was the predominant secondary structure in FuAACT, consistent with the secondary structure prediction. FuAACT composed four identical monomers (Figure 3A), and its monomer structure was similar to the 3D conformation of the thiolase reported by Wiesenborn et al. [30]. Each monomer contained canonical N-terminal (15–273, purple) and C-terminal (283–403, green) regions (Figure 3B).
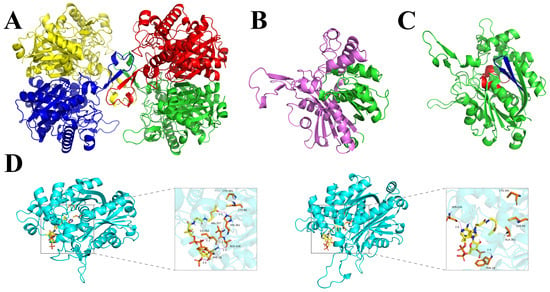
Figure 3.
Homology modeling and molecular docking of FuAACT. (A) Quaternary structure of AACT. The four subunits of FuAACT are in different colors; (B) subunit structure in which the N-terminal domain is colored purple, while the C-terminal domain is colored green; (C) αβαβα sandwich structure. Two key functional motifs, NVHGGAVSLGHPLGCSG and GVASVCNGGGGASA, are colored red and green, respectively; (D) molecular docking between FuAACT and substrate, in which the wild type is on the left, while the mutant is on the right. The hydrogen bonds between FuAACT and Acetyl-CoA were shown as purple dashed lines. These images were generated using the PyMOL Molecular Graphics System, version 1.
Notably, the C-terminal domain protruded from the N-terminal domain and contributed to most of the active sites (Figure 3C). Multiple sequence alignment and structural analysis revealed that Cys99, His361, and Cys391 formed a fully conserved catalytic triad, with His361 situated in the loop region containing the active site. Additionally, His361 was part of the thiolase-conserved motif NVHGGAVSLGHPLGCSG (residues 351 to 367) in the C-terminal region. Molecular dynamics simulation showed that the binding free energy of the FuAACT–substrate complex decreased significantly from −7.0 kcal/mol to −5.1 kcal/mol, and the key hydrogen bond interaction between His361 and the substrate was completely disrupted (Figure 3D). These results highlight the potential role of His361 in the catalytic function of FuAACT.
3.5. Heterologous Expression, Purification, and Activity Assay of FuAACT
The recombinant FuAACT protein was successfully expressed in E. coli BL21 and purified by Ni-agarose resin. Its molecular weight was 45 kDa, higher than the predicted mass (41.79 kDa) due to the N-terminal His-tag fusion peptide (Figure 4). Before determining kinetic parameters, the optimal temperature, pH, and thermal stability of purified FuAACT were determined. FuAACT exhibited optimal activity at 30 °C and pH 8.9. After thermal treatment at 50 °C for 3 h, FuAACT retained 63.08% of the initial activity, whereas activity decreased significantly to 6.04% at 60 °C, indicating that FuAACT exhibits good thermal stability below 50 °C.
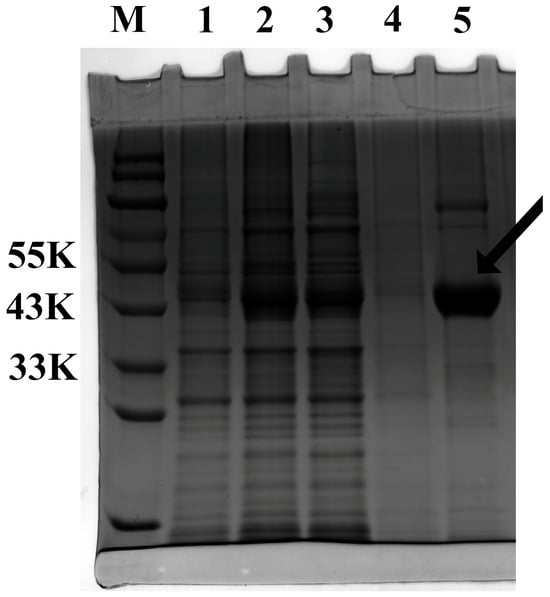
Figure 4.
SDS-PAGE of recombinant FuAACT protein. Lane M represents the protein marker. Lane 1 represents the culture supernatant of E. coli containing empty vector pET28a. Lane 2 represents the culture supernatant of E. coli containing pET28a-FuAACT. Lanes 3 and 4 represent fractions that were eluted with binding buffer two times, respectively. Lane 5 represents the purified FuAACT protein eluted by 200 mM imidazole. The arrow indicates the location of purified FuAACT protein.
Then, under the optimal reaction conditions, the Km, Vmax, and Kcat/Km values of FuAACT were determined as 3.035 μM, 0.128 μmol/(min·mg), and 0.421 μM−1·min−1, respectively. To investigate the role of His361 in FuAACT catalysis, this residue underwent site-directed mutagenesis. As shown in Table 2, this point mutation significantly affected FuAACT catalysis. For FuAACTH361A, the Km, Vmax, and Kcat/Km values changed to 2.41 μM, 0.006 μmol/(min·mg), and 0.029 μM−1·min−1, respectively (Table 2). Compared with wild-type FuAACT, FuAACTH361A exhibited decreases of 95.3% and 93.1% in Vmax and Kcat/Km, respectively. This indicates that the mutant has increased substrate affinity but significantly decreased catalytic efficiency. His361 is a conserved residue in the catalytic core of the thiolase family, and its nitrogen-containing heterocycle structure is essential for maintaining the active site conformation. Upon mutation to alanine, loss of the heterocycle structure may disrupt π-π interactions with neighboring residues (e.g., Cys99 and Cys391), leading to the distortion of the active site structure and subsequent impairment of catalytic efficiency.

Table 2.
Kinetic parameters of FuAACT and FuAACT-H361A.
The results show that FuAACT catalyzes the condensation of two molecules of acetyl-CoA to acetoacetyl-CoA, and its activity depends on the integrity of the conserved residue His361. Through functional validation and site-directed mutagenesis, these findings clarify the key role of His361 in the enzymatic catalytic mechanism, providing a theoretical foundation for further investigating the molecular regulation of steroid alkaloid synthesis in F. unibracteata bulbs.
3.6. Analysis on Sucellular Localization of FuAACT
SignalP v4.1 and TMHMM Server v2.0 predicted that the FuAACT protein lacked signal peptides and transmembrane domains, suggesting its role as an intracellular protein. To confirm its subcellular localization, the amino acid sequence of FuAACT was analyzed using WoLF PSORT. The results indicate that FuAACT was predicted to localize to the chloroplast, cytoplasm, and nucleus.
To verify the prediction, plasmids pGreenII0229-GFP and pGreenII0229-FuAACT-GFP were constructed and transiently introduced into N. benthamiana. Under laser confocal scanning (Figure 5), the control GFP showed an indiscriminate green fluorescence, while the chloroplast revealed red fluorescence. The overlapping of FuAACT-GFP fluorescence with chloroplast autofluorescence produced yellow fluorescence, indicating co-localization. This result indicates that the FuAACT protein is predominantly localized to chloroplasts and the cytoplasm, where the MVA pathway exists.
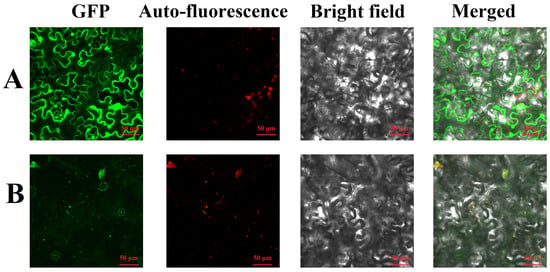
Figure 5.
Subcellular localization of FuAACT in tobacco: (A) represents 35S: GFP; (B) represents 35S: FuAACT-GFP.
3.7. Expressional Pattern of FuAACT Gene
To further investigate the expression pattern of the FuAACT gene, real-time PCR was used to detect its expression in the stem, leaf, flower, and bulb of F. unibracteata. The results (Figure 6) show that FuAACT expression was tissue-specific, with the highest levels in the leaf (10.56), followed by the stem (4.7), bulb (1.0), and flower (0.99) (p < 0.001). These results suggest that high FuAACT expression in the leaf may supply sufficient intermediates for the synthesis of downstream metabolites.
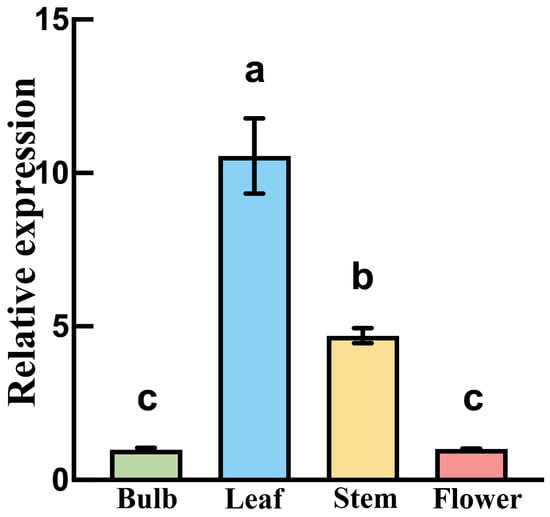
Figure 6.
FuAACT transcripts in different tissues of F. unibracteata. Different lowercase letters indicate significant differences between pairs (p < 0.05).
3.8. Phenotypic Analysis of Transgenic FuAACT A. thaliana Under Drought Stress
The PCR verification of transgenic plants was performed using DNA as a template (Figure 7A). Meanwhile, real-time PCR was carried out to evaluate the expression data of the FuAACT gene in transgenic Arabidopsis, by which substantial expression of the FuAACT gene was observed in the transgenic Arabidopsis, while a negative result was observed in the wild-type Arabidopsis (Figure 7B). The results confirm that the transgenic plants overexpressing the FuAACT gene were constructed successfully. In the seed germination experiment, seeds of both transgenic and wild-type A. thaliana began to germinate on day 1, followed by a rapid germination phase between days 1 and 3, reaching maximal germination on day 5. Statistical results showed that the final germination rate of transgenic A. thaliana was 67.20%, which was significantly higher than that of the wild type (57.21%) (Figure 8A), suggesting that overexpression of the FuAACT gene may promote germination. It is hypothesized that FuAACT may promote acetoacetyl-CoA by activating the MVA pathway, thereby upregulating the accumulation of downstream plant hormones, such as cytokinins and gibberellin (GA), to improve seed germination efficiency.
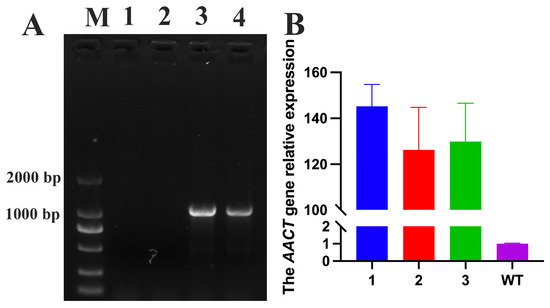
Figure 7.
The verification of transgenic plants. (A) The PCR verification of transgenic plants using DNA as a template. Lane M represents DNA marker. Lanes 1 and 2 represent wild-type plants, while Lanes 3 and 4 represent transgenic plants. (B) The real-time PCR of the FuAACT gene in wild-type and transgenic plants. Lanes 1-3 represent the transgenic plants.
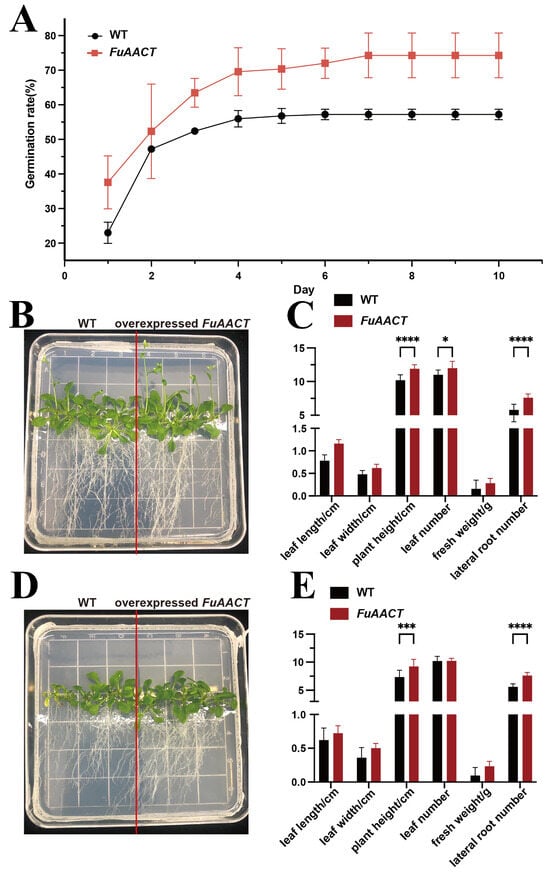
Figure 8.
Phenotypic analysis of wild-type and FuAACT transgenic plants. (A) Germination rate of FuAACT transgenic and wild A. thaliana under normal conditions; (B) phenotypes of FuAACT transgenic and wild-type plants cultivated on culture medium under normal conditions; (C) the various characteristics of transgenic FuAACT and wild A. thaliana under normal conditions; (D) phenotypes of FuAACT transgenic and wild-type plants cultivated on culture medium under drought stress; (E) the various characteristics of transgenic FuAACT and wild A. thaliana under drought stress. One-way ANOVA was used to perform statistical analysis. All experiments had at least three biological replicates. *, ***, and **** represent statistical significance of p < 0.05, p < 0.001, and p < 0.0001, respectively.
Under normal conditions, the transgenic A. thaliana plants grew slightly better than the wild type, while the transgenic plants showed stronger adaptability to drought stress (Figure 8). Under drought stress, the leaves of transgenic plants remained green, while those of wild-type plants exhibited slight etiolation. Additionally, transgenic plants exhibited a significantly higher number of lateral roots, greater plant height, and increased leaf number compared to wild-type plants (p < 0.05). The improved roots were conducive to nutrient and water absorption to absorb nutrients and water from soil, while the improved leaves might enhance photosynthesis to provide sufficient carbohydrates. Overall, these results identify FuAACT as a candidate gene for drought tolerance engineering, despite the underlying molecular mechanisms remaining unclear.
4. Discussion
Vollack and Bach (1996) first cloned the AACT gene in the plant cytoplasm and confirmed its enzymatic activity [31], which promoted the subsequent in-depth research on plant metabolic engineering and stress tolerance. Current studies have highlighted the role of AACTs in isoprenoid synthesis (e.g., squalene, natural rubber, triterpene saponins) by regulating precursors in dicotyledonous plants such as Medicago sativa [20], Helianthus annuus L. [14], H. brasiliensis [19], and B. monniera [9]. However, the functional identification of AACT in the Liliaceae family has not been reported to date.
In this study, the FuAACT gene was cloned for the first time. Its amino acid length was similar to those of BmAACT [9], HbAACT [19], SaAACT [32], TaAACT [33], and other dicots (365–437 AA). Sequence alignment showed the conserved key catalytic residues (Cys99, His361, and Cys391) along with the motif NVHGGAVSLGHPLGCSG (motif 10) and an active site GVASVCNGGGGASA in the C-terminal region. These conserved sites and motifs implied that the construction of the active site of AACT remained canonical during plant evolution, consistent with the report of Xiong et al. [33]. Compared with AtAACT2 [10] and BmAACT [9], FuAACT exhibited higher affinity (lower Km) but lower catalytic efficiency (Kcat/Km), indicating that the enzymatic properties of AACTs varied in different plants. Such differences in kinetic properties might result from the variation in amino acids in the binding sites, including His353, Ala388, Ser389, and Val390. Furthermore, the results also suggest that AACTs in different plants might control the metabolic flux through the mutation of key residues at precise positions of key residues. In comparison with F. unibracteata mevalonate kinase (FuMK) [5], FuAACT exhibited a lower catalytic rate due to its lower Vmax, suggesting that FuAACT may play a more critical role than FuMK in steroid alkaloid biosynthesis in F. unibracteata.
The conserved His361 residue in the active site of FuAACT was proposed to be the catalytic residue [28]. Notably, in existing thiolase studies, the His348 residue forms a hydrogen bond with nucleophilic cysteine (Cys89) to enhance its nucleophilicity. Mutants H348A or H348N lost 99–99.9% of their original activities [34]. A similar hydrogen bond was also observed in the FuAACT-acetyl CoA complex using homology modeling, and the subsequent substitution of H361 residue by Ala residue caused substantial loss in catalytic efficiency but improved affinity. In combination with site-directed mutagenesis and model observation, it was speculated that the mutation enhanced the hydrophobicity in the substrate pocket, thereby improving affinity. However, the disruption of the hydrogen bond between His361 and Cys89 residues destroyed the electron transfer and reduced the nucleophilicity of Cys89 residue. In addition, based on the results of amino acid sequence alignment (Supplementary Material S2) and molecular docking (Figure 3D), the conserved Phe28, Ileu262, and Ser358 residues also participate in the catalytic process, whose roles in catalysis will be verified in future work.
Diverse subcellular localization of AACT has been observed in different plants, although the MVA pathway is canonically localized to the cytoplasm. Indeed, the cytoplasm, endoplasmic reticulum, nucleus, and peroxisomes have been reported as AACT localization sites [1,10,35,36]. In this study, FuAACT was found to be localized in both plastids and cytoplasm, suggesting that various subcellular compartmentalization was proposed to be involved in the biosynthetic pathway of terpenoids. This finding echoes the reported involvement of AACT in chloroplast metabolism in A. thaliana [10], strengthening the role of chloroplast in the isoprenoid synthesis. Another possible reason is that AACT might carry non-enzymatic functions, similar to glycolytic enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH). GAPDH is abundant in the cytosol but is also found in the nucleus where it plays roles via non-enzymatic binding [37]. However, this simultaneous localization of FuAACT must be further verified, and hence, a plastid-localized marker, such as ribulose bisphosphate carboxylase, will be used in future work [38]. Tissue-specific expression analysis further revealed that FuAACT expression was significantly higher in leaves than in roots, bulbs, and flowers. A similar leaf-enriched distribution of AACTs was observed in G. biloba [12] and Isodon rubescens [39]. Combined with its subcellular localization, we speculate that FuAACT may play a significant role in leaf tissue. Previous studies have confirmed that the enhancing expression of MVA pathway genes in the leaf tissue drove the accumulation of terpenoids such as carotenoids [40], an important pigment in photosynthesis. Therefore, the role of FuAACT in the biosynthesis of steroidal alkaloids in bulbs of F. unibracteata might be indirect, or its role is being replaced with other FuAACT homologs. However, the above speculation needs further experimental verification.
The overexpressed FuAACT gene promoted seed germination and drought tolerance of A. thaliana, which might be due to the increased synthesis of phytohormones. However, the accumulation of various phytohormones needs to be measured in future studies to validate this hypothesis. Additionally, to elucidate the role of FuAACT in stress tolerance, future studies should determine (1) whether subcellular localization of FuAACT changes under drought and salinity stress; (2) the expression pattern of FuAACT under these stresses; (3) the change in metabolites, such as isoprenoid precursor and downstream terpenoids, between transgenic and wild-type plants; and (4) the crystal structure of FuAACT to clarify its catalytic mechanism.
5. Conclusions
In this study, the FuAACT gene was first cloned and characterized in vitro and in vivo. The His361 residue was verified as a key catalytic residue, which lays the foundation for understanding the catalytic mechanism and modification of FuAACT. Overexpression of FuAACT improved tolerance to drought and salinity stress, although the detailed mechanism remained unclear. In conclusion, this work extended the functional AACT members of the Liliaceae family, rich with economically ornamental and medicinal plants, but also enlarged the pool of candidate genes for plant stress resistance.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11080913/s1, Supplementary Material S1: codon-optimized FuAACT; Supplementary Material S2: Amino acid sequence alignment of FuAACT with relative AACTs; Supplementary Material S3: The p-values of AACTs from various species; Supplementary Material S4: Shannon Protein Variability of A) with in monocotyledonous plants B) with in Dicotyledonous plants; Supplementary Material S5: The original data of Figure 6; Supplementary Material S6: The map of pCambia2301 vector; Supplementary Material S7: The original data of Figure 7B; Supplementary Material S8: The original map of Figure 7A.
Author Contributions
Z.M., Q.A., X.H., H.L. (Hongting Liu) and F.G. performed the experiments. Z.M., Q.A., X.H., H.L. (Hongting Liu), F.G. and H.Y. analyzed the data. Z.M., Q.A., X.H., H.L. (Hongting Liu), F.G., H.Y. and J.Z. contributed to the reagent/material/analysis tools. H.L. (Hai Liao) and J.Z. wrote and are responsible for the paper. All authors have read and agreed to the published version of the manuscript.
Funding
This work was co-funded by the National Natural Science Foundation of China (No. 32270410), Sichuan Science and Technology Program (No. 2018SZ0061), and Fundamental Research Funds for the Central Universities (GX202516011).
Data Availability Statement
The full-length coding sequence (CDS) of FuAACT was submitted to GenBank databases with accession number PV706214. In this study, the codon-optimized FuAACT, Ct raw data of real-time PCR, supplementary figures, and other original data are included in the Supplementary Materials.
Acknowledgments
We would like to thank Analysis and Testing Center of Southwest Jiaotong University for their assistance with data analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Wang, M.; Wang, D.; Zhang, Q.; Chai, J.; Peng, Y.; Cai, X. Identification and cytochemical immunolocalization of acetyl-CoA acetyltransferase involved in the terpenoid mevalonate pathway in Euphorbia helioscopia laticifers. Bot. Stud. 2017, 58, 62. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Chen, J.; Yan, H.; Huang, X.; Chen, J.; Ma, Z.; Zhou, J.; Liao, H. Functional Identification of the Isopentenyl Diphosphate Isomerase Gene from Fritillaria unibracteata. Horticulturae 2024, 10, 887. [Google Scholar] [CrossRef]
- Deng, C.; Li, J.; Tao, S.; Jin, Y.; Peng, F. Identifying Suitable Regions for Fritillaria unibracteata Cultivation Without Damage from the Pest Eospalax baileyi. Plants 2025, 14, 674. [Google Scholar] [CrossRef]
- Jiang, R.; Zou, M.; Qin, Y.; Tan, G.; Huang, S.; Quan, H.; Zhou, J.; Liao, H. Modeling of the Potential Geographical Distribution of Three Fritillaria Species Under Climate Change. Front. Plant Sci. 2021, 12, 749838. [Google Scholar] [CrossRef]
- Liao, H.; Quan, H.; Huang, B.; Ji, H.; Zhang, T.; Chen, J.; Zhou, J. Integrated transcriptomic and metabolomic analysis reveals the molecular basis of tissue-specific accumulation of bioactive steroidal alkaloids in Fritillaria unibracteata. Phytochemistry 2023, 214, 113831. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, M.; Yang, T.; Wai Ming, T.; Wai Gaun, T.K.; Ye, B. LC-MS/MS coupled with chemometric analysis as an approach for the differentiation of bulbus Fritillaria unibracteata and Fritillaria ussuriensis. Phytochem. Anal. 2021, 32, 957–969. [Google Scholar] [CrossRef]
- Zhao, D.; Wang, J.; Dai, W.; Ye, K.; Chen, J.; Lai, Q.; Li, H.; Zhong, B.; Yu, X. Effects of climate warming and human activities on the distribution patterns of Fritillaria unibracteata in eastern Qinghai-Tibetan Plateau. Sci. Rep. 2023, 13, 15770. [Google Scholar] [CrossRef]
- Wang, G.; Wan, X.; Li, X.; Ou, J.; Li, G.; Deng, H. Transcriptome-based analysis of key functional genes in the triterpenoid saponin synthesis pathway of Platycodon grandiflorum. BMC Genomic Data 2024, 25, 83. [Google Scholar] [CrossRef]
- Vishwakarma, R.K.; Ruby; Somesh, S.; Sonawane, P.D.; Srivastava, S.; Kumari, U.; Santosh Kumar, R.J.; Khan, B.M. Molecular cloning, biochemical characterization, and differential expression of an Acetyl-CoA C-Acetyltransferase Gene (AACT) of Brahmi (Bacopa monniera). Plant Mol. Biol. Rep. 2013, 31, 547–557. [Google Scholar] [CrossRef]
- Jin, H.; Song, Z.; Nikolau, B.J. Reverse genetic characterization of two paralogous acetoacetyl CoA thiolase genes in Arabidopsis reveals their importance in plant growth and development. Plant J. Cell Mol. Biol. 2012, 70, 1015–1032. [Google Scholar] [CrossRef]
- Yao, Y.Z.; Li, X.Y.; Wei, L.; Wu, X.J.; Tang, Y.L. Cloning, expression, and bioinformatics analysis of acetyl-CoA C-acetyltransferase gene in Houttuynia cordata. Chin. Tradit. Herbal Drugs 2015, 46, 107–111. [Google Scholar]
- Chen, Q.; Yan, J.; Meng, X.; Xu, F.; Zhang, W.; Liao, Y.; Qu, J. Molecular cloning, characterization, and functional analysis of acetyl-CoA C-acetyltransferase and mevalonate kinase genes involved in terpene trilactone biosynthesis from Ginkgo biloba. Molecules 2017, 22, 74. [Google Scholar] [CrossRef]
- Rohmer, M.; Knani, M.; Simonin, P.; Sutter, B.; Sahm, H. Isoprenoid biosynthesis in bacteria: A novel pathway for the early steps leading to isopentenyl diphosphate. Biochem. J. 1993, 295, 517–524. [Google Scholar] [CrossRef]
- Dyer, J.H.; Maina, A.; Gomez, I.D.; Cadet, M.; Oeljeklaus, S.; Schiedel, A.C. Cloning, expression and purification of an acetoacetyl CoA thiolase from sunflower cotyledon. Int. J. Biol. Sci. 2009, 5, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Van Moerkercke, A.; Schauvinhold, I.; Pichersky, E.; Haring, M.A.; Schuurink, R.C. A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 2009, 60, 292–302. [Google Scholar] [CrossRef]
- Liu, M.; Yu, H.; Li, J.; Dong, N.; Chen, B.; Xu, R.; Wu, J.; Chang, X.; Wang, J.; Peng, H.; et al. Cloning, Expression, and Functional Analysis of the Full-Length cDNA of Acetyl-CoA C-acetyltransferase (AACT) Genes Related to Terpenoid Synthesis in Platycodon grandiflorus. Protein Pept. Lett. 2022, 29, 1061–1071. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.N.; Han, J.W.; Yang, S.H.; Lee, S.M. Co-Expression Analysis Reveals Differential Expression of Homologous Genes Associated with Specific Terpenoid Biosynthesis in Rehmannia glutinosa. Genes 2022, 13, 1092. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Shi, L.; Xu, Y.J.; Zhao, M.W. Cloning of a sterol 14α-demethylase gene and the effects of over-expression of the gene on biological synthesis of triterpenes in Ganoderma lucidum. Mycosystema 2011, 30, 7. [Google Scholar]
- Sando, T.; Takaoka, C.; Mukai, Y.; Yamashita, A.; Hattori, M.; Ogasawara, N.; Fukusaki, E.; Kobayashi, A. Cloning and characterization of mevalonate pathway genes in a natural rubber producing plant, Hevea brasiliensis. Biosci. Biotechnol. Biochem. 2008, 72, 2049–2060. [Google Scholar] [CrossRef]
- Soto, G.; Stritzler, M.; Lisi, C.; Alleva, K.; Pagano, M.E.; Ardila, F.; Mozzicafreddo, M.; Cuccioloni, M.; Angeletti, M.; Ayub, N.D. Acetoacetyl-CoA thiolase regulates the mevalonate pathway during abiotic stress adaptation. J. Exp. Bot. 2011, 62, 5699–5711. [Google Scholar] [CrossRef]
- Zhang, C.; Ji, Z.; Xu, N.; Yuan, J.; Zeng, W.; Wang, Y.; He, Q.; Dong, J.; Zhang, X.; Yang, D.; et al. Integrating network pharmacology and experimental validation to decipher the pharmacological mechanism of DXXK in treating diabetic kidney injury. Sci. Rep. 2024, 14, 22319. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Chen, A.; Zhu, J.; Yan, Z.; An, Q.; Zhou, J.; Liao, H.; Yu, Y. Structure basis of the caffeic acid O-methyltransferase from Ligusiticum chuanxiong to understand its selective mechanism. Int. J. Biol. Macromol. 2022, 194, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Zeng, R.; Gao, S.; Xu, L.; Liu, X.; Dai, F. Prediction of pathogenesis-related secreted proteins from Stemphylium lycopersici. BMC Microbiol. 2018, 18, 191. [Google Scholar] [CrossRef]
- Qin, Y.; Li, Q.; An, Q.; Li, D.; Huang, S.; Zhao, Y.; Chen, W.; Zhou, J.; Liao, H. A phenylalanine ammonia lyase from Fritillaria unibracteata promotes drought tolerance by regulating lignin biosynthesis and SA signaling pathway. Int. J. Biol. Macromol. 2022, 213, 574–588. [Google Scholar] [CrossRef]
- Arakawa, H.; Takiguchi, M.; Amaya, Y.; Nagata, S.; Hayashi, H.; Mori, M. cDNA-derived amino acid sequence of rat mitochondrial 3-oxoacyl-CoA thiolase with no transient presequence: Structural relationship with peroxisomal isozyme. EMBO J. 1987, 6, 1361–1366. [Google Scholar] [CrossRef]
- Sun, S.; Kang, X.P.; Tian, Y.S.; Zheng, S.W.; Xing, G.M. Cloning and bioinformatic analysis of a novel Thiolase II Gene (BPLTHI2) from Betula platyphylla. Biotechnol. Biotechnol. Equip. 2013, 27, 4167–4171. [Google Scholar] [CrossRef]
- Zeng, J.; Li, D. Expression and purification of His-tagged rat mitochondrial 3-ketoacyl-CoA thiolase wild-type and His352 mutant proteins. Protein Expr. Purif. 2004, 35, 320–326. [Google Scholar] [CrossRef]
- Bhaskar, S.; Steer, D.L.; Anand, R.; Panjikar, S. Structural basis for differentiation between two classes of thiolase: Degradative vs biosynthetic thiolase. J. Struct. Biol. X 2020, 4, 100018. [Google Scholar] [CrossRef]
- Wiesenborn, D.P.; Rudolph, F.B.; Papoutsakis, E.T. Thiolase from Clostridium acetobutylicum ATCC 824 and its role in the synthesis of acids and solvents. Appl. Environ. Microbiol. 1988, 54, 2717–2722. [Google Scholar] [CrossRef]
- Vollack, K.U.; Bach, T.J. Cloning of a cDNA encoding cytosolic acetoacetyl-coenzyme A thiolase from radish by functional expression in Saccharomyces cerevisiae. Plant Physiol. 1996, 111, 1097–1107. [Google Scholar] [CrossRef]
- Niu, M.; Yan, H.; Xiong, Y.; Zhang, Y.; Zhang, X.; Li, Y.; Da Silva, J.a.T.; Ma, G. Cloning, characterization, and functional analysis of acetyl-CoA C-acetyltransferase and 3-hydroxy-3-methylglutaryl-CoA synthase genes in Santalum album. Sci. Rep. 2021, 11, 1082. [Google Scholar] [CrossRef] [PubMed]
- Xiong, F.; Zhu, X.; Luo, C.; Liu, Z.; Zhang, Z. The cytosolic acetoacetyl-CoA thiolase TaAACT1 is required for defense against Fusarium pseudograminearum in wheat. Int. J. Mol. Sci. 2023, 24, 6165. [Google Scholar] [CrossRef] [PubMed]
- Harijan, R.K.; Dalwani, S.; Kiema, T.R.; Venkatesan, R.; Wierenga, R.K. Thiolase: A versatile biocatalyst employing coenzyme A–thioester chemistry for making and breaking C–C bonds. Annu. Rev. Biochem. 2023, 92, 351–384. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zheng, Z.; Tian, Z.; Zhang, H.; Zhu, C.; Yao, X.; Yang, Y.; Cai, X. Molecular Cloning and Analysis of an Acetyl-CoA C-acetyltransferase Gene (EkAACT) from Euphorbia kansui Liou. Plants 2022, 11, 1539. [Google Scholar] [CrossRef]
- Wu, J.; Liu, W.; Lu, J.; Xu, R.; Xie, J.; Zha, L. Cloning, Prokaryotic Expression, and Purification of Acetyl-CoA C-Acetyltransferase from Atractylodes lancea. Protein Pept. Lett. 2022, 29, 156–165. [Google Scholar] [CrossRef]
- Guo, J.S.; Wang, J.Y.; Chen, S.H.; Deng, Y.P.; Gao, Q.Y.; Liu, Z.X.; Liu, J.; Lv, K.; Liu, N.; Bai, G.Y.; et al. The natural product micheliolide promotes the nuclear translocation of GAPDH via binding to Cys247 and induces glioblastoma cell death in combination with temozolomide. Biochem. Pharmacol. 2025, 233, 116759. [Google Scholar] [CrossRef]
- Scofield, S.R.; Jones, D.A.; Harrison, K.; Jones, J.D. Chloroplast targeting of spectinomycin adenyltransferase provides a cell-autonomous marker for monitoring transposon excision in tomato and tobacco. Mol. Gen. Genet. 1994, 244, 189–196. [Google Scholar] [CrossRef]
- Zhu, Y.H.; Su, X.H.; Dong, C.M.; Chen, S.Q.; Shao, Y.Y.; Zhang, F.B. Cloning and expression analysis of acetyl-COA C-acetyltransferase gene in Isodon rubescens. J. Chin. Med. Mater. 2016, 39, 37–41. [Google Scholar]
- Andersen, T.B.; Llorente, B.; Morelli, L.; Torres-Montilla, S.; Bordanaba-Florit, G.; Espinosa, F.A.; Rodriguez-Goberna, M.R.; Campos, N.; Olmedilla-Alonso, B.; Llansola-Portoles, M.J.; et al. An engineered extraplastidial pathway for carotenoid biofortification of leaves. Plant Biotechnol. J. 2021, 19, 1008–1021. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).